Abstract
Almost 15 years of careful study have established the related bHLH transcription factors Hand1 and Hand2 as critical for heart development across evolution. Hand factors make broad contributions, revealed through animal models, to the development of multiple cellular lineages that ultimately contribute to the heart. They perform critical roles in ventricular cardiomyocyte growth, differentiation, morphogenesis, and conduction. They are also important for the proper development of the cardiac outflow tract, epicardium, and endocardium. Molecularly, they function both through DNA-binding and through protein-protein interactions, which are regulated transcriptionally, post-transcriptionally by microRNAs, and post-translationally through phospho-regulation, Although direct Hand factor transcriptional targets are progressively being identified, confirmed direct targets of Hand factor transcriptional activity in the heart are limited. Identification of these targets will be critical to model the mechanisms by which Hand factor bHLH interactions affect developmental pathways. Improved understanding of Hand factor-mediated transcriptional cascades will be necessary to determine how Hand factor disregulation translates to human disease phenotypes. The following review summarizes the insight animal models have provided into the regulation and function of these factors during heart development, and the recent findings that suggest roles for HAND1 and HAND2 in human congenital heart disease.
Keywords: Basic helix-loop-helix, HAND1, HAND2, dimerization, Transcription, DNA-binding, Cardiogenesis, Heart development, Congenital heart defect
Introduction
Cardiac development involves the coordination of multiple cell types and differentiation programs
The heart is composed of disparate cell types, which progressively assume divergent cell fates as cardiogenesis advances. For example, cells derived from the epicardium ultimately differentiate into both cardiac fibroblasts and the smooth muscle of the coronary vasculature. However, in certain instances, these distinct cell types must assume similar characteristics to function as a cohesive unit. For example, the left and right ventricles are derived from distinct cell populations, termed the primary and secondary heart fields (PHF and SHF), respectively, and exhibit unique morphological characteristics, such as chamber shape and wall thickness. However, these two chambers also express common metabolic, cytoskeletal and cell-cell junction proteins that enable myocytes to synchronize contractile function. These conserved muscle gene programs may reflect the influence of pan-cardiac transcription factors, such as Nkx2.5 and Gata4, which are expressed in both forming ventricles. Interestingly, closely related factors such as Hand1 and Hand2, which perform both unique and redundant transcriptional functions, may both draw distinctions between the developing ventricular chambers and regulate a common coterie of cardiac myogenic and morphogenic promoters within the defined primary and secondary cardiac fields. Further, these factors may operate in similar unique and redundant manners within other developing heart tissues, such as the cardiac outflow tract (OFT) and the proepicardium (PE).
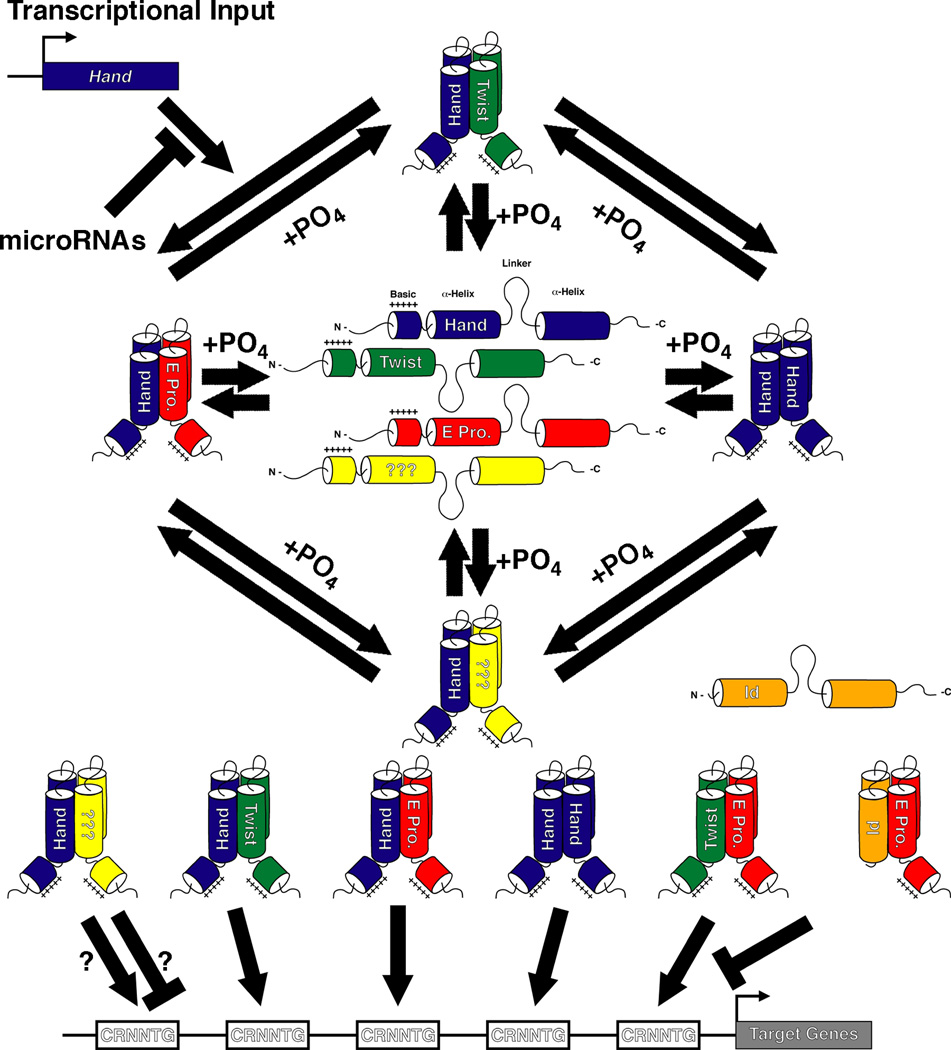
Regulation of Hand factor function
Hand1 and Hand2, members of the Twist-family of basic Helix-Loop-Helix (bHLH) proteins, function as either homo- or heterodimers, binding consensus elements termed E-boxes (CANNTG), or related, degenerate sequences termed D-boxes (CGNNTG), and regulating downstream effector genes (Barnes and Firulli, 2009; Conway et al. 2010; Firulli, 2003). They possess two characterized mechanisms of regulation. Twist-family member function can be regulated through adjustment of relative gene expression levels (Fig. 1). For example, Hand2 and Twist1, a related Twist-family bHLH transcription factor, perform antagonistic functions in the developing limb, and proper anterior-posterior limb patterning depends upon maintenance of relative expression levels between these two factors (Firulli et al. 2005). It is likely that Twist-family members function in evolutionarily established equilibrium not only in the limb, but also in other tissues in which they display overlapping expression domains, such as those of the developing heart.
Figure 1.
Hand1 and Hand2 function is also dictated by dimer partner choice, and this choice can be influenced via phosphorylation of evolutionarily conserved threonine and serine residues within the bHLH domain (Fig. 1; (Firulli et al. 2003). Twist-family members are characterized by conserved serine and threonine residues located in helix I. Phosphorylation of these residues alters both the dimerization affinity of these factors and, consequently, their DNA binding specificity (Firulli et al. 2007). Additionally, this phosphoregulation may direct Hand factor subcellular localization, consequently affecting dimerization (Martindill et al. 2007).
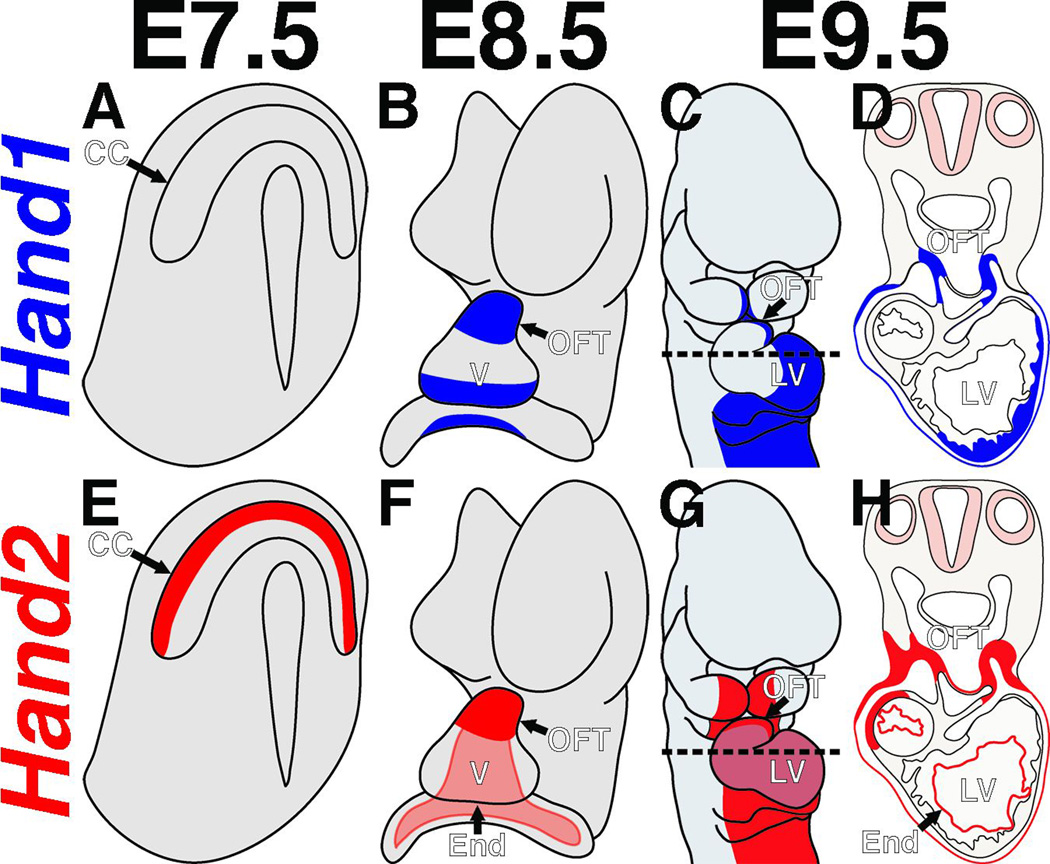
Both Hand1 and Hand2 play critical roles during cardiogenesis, as has been demonstrated through systemic and tissue-specific gene ablation studies. Hand factors contribute to the maturation of several substructures within the developing heart, namely the cardiomyocytes of the ventricles, myocardial and cNCC contributions to the OFT, and the mesothelial cell population forming the epicardium (Barnes et al. 2010). Hand2, potentially in conjunction with Twist1, also likely functions within the endothelial populations of the endocardium and cardiac vales. In the primitive heart tube, Hand genes are upregulated as the early heart tube undergoes looping, with Hand2 expression marking myocardium in the outflow tract (OFT), and right ventricle (RV), and Hand1 expression making in the outer curvature of the left ventricle (LV) and, to a more restricted degree than Hand2, the OFT myocardial cuff (Fig. 2). Hand1 and Hand2 are also dynamically expressed in the cNCCs in partially overlapping patterns. They are both strongly expressed within the NCCs that populate the pharyngeal arches, with Hand2 more broadly expressed and Hand1 more restricted to the medio-ventral cNCCs proximal to the OFT, which will ultimately contribute to the OFT cushions (Fig. 2). They are further expressed in some, but not all, of the cNCCs invading the OFT cushions. Recent studies also indicate that Hand1 and Hand2 are expressed complementarily in the progenitors of the epicardium (Barnes et al. 2010). Hand1 is expressed in mesothelial cell populations in the E8.5 mouse embryo, including the septum transversum (Barnes et al. 2010). Genetic lineage trace analyses indicate that as Hand1 is downregulated in these cells, they contribute to the proepicardial organ, where cells express Hand2. The cells of the proepicardial organ progressively migrate to envelop the heart, ultimately undergoing EMT and contributing to the cardiac fibroblasts and the smooth muscle cells of the coronary vasculature. Thus Hand1 and Hand2 are expressed in spatiotemporally distinct manners as these cells mature and differentiate and likely play a role in defining these extra-cardiac cell populations.
Figure 2.
As relative Twist family member expression levels dictate their cellular function, unraveling the mechanisms by which Hand1 and Hand2 expression is regulated in these tissues is pivotal to understanding their roles in cardiogenesis. Thus far, studies identifying the direct transcriptional regulators of Hand1 and Hand2 in the heart are limited. A Hand2 cis-regulatory element that is bound and activated by Gata factors and that drives gene expression in the RV and OFT has been isolated (McFadden et al. 2000). The cis-regulatory elements governing Hand1 expression in the heart remain elusive.
Cellular protein levels can additionally be controlled posttranscriptionally by microRNAs (miRNAs) (Ambros, 2004; Kloosterman and Plasterk, 2006; Zhao and Srivastava, 2007). As miRNAs can serve to “fine-tune” protein synthesis (Baek et al. 2008; Selbach et al. 2008), they are strong candidate regulators of the precise cellular Twist family member levels required to properly implement developmental programs. The closely related miRNAs, mirR-1-1 and mirR-1-2, are both expressed in the developing heart where, as shown through in vivo overexpression studies, they negatively regulate ventricular cardiomyocyte proliferation (Zhao et al. 2005). In silico analyses identified Hand2 as a putative target of these miRNAs, and indeed, Hand2 protein levels are reduced in miR-1 overexpressing hearts, while RNA levels remain unchanged (Zhao et al. 2005). Conversely, targeted deletion of mirR-1-2 leads to a 4-fold upregulation of Hand2 protein expression (Zhao et al. 2007). Currently, no microRNAs have been associated in regulating Hand1 protein levels, although in silco analysis indentifies several seed-sequences within its 3’ UTR. Thus, cardiac Hand factor expression levels are partially controlled by microRNAs.
Hand factors regulate specific aspects of cardiomyocyte growth, differentiation, morphogenesis, and conduction
Disruption of Hand1 function in the mouse heart causes myocardial defects
Insight into the functional roles Hand1 and Hand2 play during cardiogenesis stems largely from animal models. Cardiovascular defects are observed when the Drosophila Hand factor orthologue, Hand, is disrupted, indicating an evolutionarily conserved requirement for these factors during cardiogenesis (Lo et al. 2007). Systemic ablation of Hand1 in mice results in embryonic lethality at E9.5, most likely owing to defects in trophonoblast differentiation (Firulli et al. 1998; Riley et al. 1998) and extra-embryonic vascularization (Morikawa and Cserjesi, 2004). Hand1−/− KO embryos experience growth arrest at E7.5, and fail to undergo turning. Heart development in these mutants arrest and the primitive heart tube fails to undergo looping (Firulli et al. 1998; Riley et al. 1998). Rescue of extra-embryonic phenotypes through tetraploid aggregation extends embryonic viability to E10.5, and while these rescued embryos undergo turning normally, the heart tube nonetheless fails to loop (Riley et al. 1998). These mutant hearts display diminished cardiomyocyte cell numbers and an absence of ventricular trabeculation (Riley et al. 1998). Hypomorphic alleles of Hand1 enable analyses of mutant heart development at later stages (Firulli et al. 2010). Embryos homozygous for each of these two alleles express Hand1 mRNA at either 40% or 30%, and survive to E12.5 and E10.5, respectively (Firulli et al. 2010). Although the hearts in these mutants loop normally, they display a thin LV myocardium, hypotrabeculation, and diminished expression of the LV-specific markers, Nppa, Chisel, and Cited1 (Firulli et al. 2010). Hand1 has also been ablated tissue-specifically to analyze associated cardiac phenotypes in further detail. Deletion of Hand1 specifically in the myocardium, with the exception of a few escapers (<2%) in the case of αMHC-Cre, results in perinatal lethality (McFadden et al. 2005). These mice display a spectrum of congenital heart defects, including membranous ventricular septal defects (VSDs), overriding aorta, hyperplastic atrioventricular (AV) valves, and double outlet right ventricle (DORV). Although LV chamber size is reduced in these mutants at mid gestation (E11.5–E13.5), ventricular size recovers by birth. However, the muscular ventricular septum is noticeably thickened in these mutants at this stage, which may indicate that the “rescue” of ventricular size in these mutants occurs through a hypertrophic mechanism.
Misexpression of Hand1 in the mouse heart causes myocardial defects
Overexpression studies have, in conjunction with loss-of-function analyses, provided insight into the role of Hand1 during cardiogenesis. Overexpression of Hand1 within its endogenous expression domain results in a range of ventricular phenotypes, including a variable reduction in ventricular expansion and aberrant cardiomyocyte differentiation (Risebro et al. 2006). Cardiomyocytes in these mutants display elevated myocyte density and hypertrophy. These gain-of-function phenotypes are strikingly similar to those associated with loss of Hand1 function. As perturbation of the cellular bHLH factor balance results in inappropriate dimer formation, potentially derailing normal development, comparable structural abnormalities could result from either too much or too little Hand1.
Hand1 misexpression studies have also uncovered a role for Hand1 during interventricular septum formation. Hand1 misexpression throughout both ventricles, achieved by targeting a Hand1 expression cassette to the Mlc2v locus, leads to an increased expansion of the outer curvatures of the right and left ventricles, although the myocardium itself is thin (Togi et al. 2004). Interestingly, no interventricular groove or septum forms in these knock-in embryos, although Tbx5 expression remains chamber-specific. This loss of septum phenotype is not recapitulated when Hand1 misexpression is confined to the RV. Hand1-lineage analysis reveals that Hand1-marked cells are largely excluded from the septum, suggesting that forced ectopic Hand1 expression causes this phenotype (Barnes et al. 2010). Thus, in addition to its role in ventricular cardiomyocyte expansion, Hand1 may function to establish the boundary of the nascent ventricular septum.
In addition to ventricular proliferation and differentiation, Hand1 influences cardiac conduction. Additional genetic studies have used a doxycycline inducible tet-on system to overexpress Hand1 roughly six-fold in adult mouse myocardium (Breckenridge et al. 2009). One month after transgene induction, these Hand1-overexpressers display a mild ventricular hypertrophy (~10% increase in cell diameter). Despite displaying no overt change in their behavior or cardiac pressure/volume relationships, Hand1-overexpressing mice displayed an increased 12-month mortality compared to control animals. Surface ECGs revealed that, compared to controls, Hand1-overexpressers displayed significantly prolonged PR, QRS and RR intervals (but not QTc) both 28 days and 250 days after induction. Pacing experiments designed to assess the ventricular tachycardia (VT) threshold of Hand1-overexpressing hearts revealed a significantly prolonged sinus node recovery time (SNRT600). These hearts showed an increased susceptibility to VT, and VT lasted longer than in control mice. Thus, these mice likely die from ventricular arrhythmia. Immunohistohemistry reveals that Connexin43 (Gja1, Cx43), a major component of the ventricular cardiomyocyte gap junction, is downregulated in the ventricular cardiac intercalated discs of Hand1 overexpressers. However, qRT-PCR reveals that Gja1 mRNA is not similarly downregulated, indicating a post-transcriptional mechanism of regulation. Conversely, β-catenin shows both increased protein levels within the intercalated discs and increased mRNA levels in Hand1 overexpressing ventricles. Gja1 downregulation has been linked with ventricular arrhythmias and sudden death in both transgenic models (Gutstein et al. 2001; Litchenberg et al. 2000) and human patients (Dupont et al. 2001; Kostin et al. 2004; Sepp et al. 1996), whereas β-catenin protein upregulation at the intercalated disc has been associated with human hypertrophic cardiomyopathy (Masuelli et al. 2003). These data provide molecular evidence that cardiomyocyte electrical coupling is compromised in Hand1 overexpressers. Importantly, these molecular and physiological defects could be ameliorated through the withdrawal of doxycycline.
Confirmed direct transcriptional targets of Hand1 are rare. The actin monomer-binding protein Thymosin β4 was identified in a differential screen of wild-type and Hand1−/− embryoid bodies as a downstream target of Hand1 (Smart et al. 2002). Further studies have confirmed that Thymosin β4 is a direct transcriptional target of Hand1 (Smart et al. 2010). Although the contribution of this regulatory relationship to cardiogenesis has not been defined, evidence suggests Thymosin β4 may stimulate vascular and myocardial growth while inhibiting myocardial cell death (Shrivastava et al. 2010).
Disruption of Hand2 function in the mouse generates cardiac phenotypes
Ablation of Hand2 in mice similarly generates cardiac defects and results in early embryonic lethality. Hand2−/− mutants die by E10.5, displaying hypoplasia of both the ventricles and the pharyngeal arches (Srivastava et al. 1997; Thomas et al. 1998), as well as vascular defects (Yamagishi et al. 2000). Like Hand1, it has been proposed that Hand2 also regulates cardiomyocyte differentiation, specifically in the right ventricle (Srivastava et al. 1997). Alternatively, it is thought that Hand2 inhibits apoptosis in the developing right ventricle (Srivastava, 1999; Yamagishi et al. 1999). Although expression of the ventricle-specific marker myosin light chain 2v (Mlc2v) is maintained in Hand2−/− mutants (Srivastava et al. 1997), the domain of a transgene driving lacZ under the control of the Hand2 RV-specific enhancer is reduced (McFadden et al. 2000), suggesting either that the RV is reduced in these mutants, or that Hand2 regulates its own expression. The Hand2 RV-specific enhancer does contain highly conserved E-box elements; however, as mentioned, it has been shown to be dependent on the presence of conserved Gata cis-elements (McFadden et al. 2000). Conditional alleles of Hand2 are now available (Hendershot et al. 2007; Morikawa et al. 2007). Myocardial deletion of Hand2 using the cardiac troponin Cre (cTNTCre; (Morikawa and Cserjesi, 2008) shows a near phenocopy of the Hand2 systemic knockout. Further studies incorporating the various heart-specific Cre drivers available will be important to better understand the etiology of the Hand2−/− myocardial phenotype.
Functional interactions and redundancy between Hand1 and Hand2
As Hand1 and Hand2 exhibit similar functional properties, both sharing dimer partners and binding to E- and D-boxes, it is thought that they may perform redundant functions. Treatment of chick hearts, which express Hand1 and Hand2 in uniform and completely overlapping patterns, with antisense oligonucleotides against both factors causes heart development to arrest during looping. When either Hand1 or Hand2 antisense oligonucleotides are used individually, heart development proceeds normally (Srivastava et al. 1995). These results indicate that, in the chick, Hand1 and Hand2 can functionally compensate for each other’s absence.
Studies in the mouse have provided additional evidence that Hand1 and Hand2 share biological functions. As mentioned, conditional ablation of Hand1 in the myocardium causes a reduction of the LV during mid-gestation (McFadden et al. 2005). Breeding these embryos onto a Hand2+/− haploinsufficient background exacerbates this phenotype. Unlike Hand1 conditional knockouts, Hand1fx/−;Hand2+/−;Nkx2.5-Cre(+) embryos are not recovered after E10.5 (McFadden et al. 2005) and present a comparatively thin, poorly trabeculated myocardium. Thus, Hand1 and Hand2 can perform redundant functions, or at least participate within the same molecular networks, and a cumulative critical gene dosage of both factors is necessary for proper heart development. Hand1 and Hand2 can form heterodimers with each other (Firulli et al. 2000; Firulli et al. 2003), and their respective contribution to such a heterodimer would by default be defined as functionally redundant. Rigorous assessment of functional redundancy between Hand1 and Hand2 necessitates gene replacement strategies, which have yet to be reported. Conversely, breeding a Hand1 conditional allele onto a Hand2 null background (Hand1fx/+;Hand2−/−;Nkx2.5-Cre[+]) led to an increased deposition of cardiac jelly, the dense extracellular matrix (ECM) that lies between the myocardium and endocardium (McFadden et al. 2005). As these embryos die at E9.5, however, the ultimate developmental impact of this increased matrix deposition cannot be assessed.
Hand factors function independently of DNA binding in developing cardiomyocytes
Given that Hand1 and Hand2 encode transcription factors, it would be convenient to suppose that the loss-of-function phenotypes observed for each mutant reflect disruption of Hand factor-mediated transcriptional programs. Studies using a targeted allele of Hand2 in which basic domain function has been abolished have shown that Hand2 is able to partially function in vivo in the absence of DNA binding (Liu et al. 2009). Indeed, although the RV and OFT of systemic Hand2 mutants are severely hypoplastic, causing lethality by E10.5, the heart is structurally normal, if somewhat smaller, in these DNA-binding mutants. Somewhat similarly to the phenotypes observed in Hand1 misexpressing mutants, Hand2 DNA-binding mutants display abnormal myocardial thickness and ventricular septum formation. At E11.5 these mutants have a thin left ventricular myocardium, and though the interventricular groove begins to form, it is developmentally delayed and its myocytes fail to thicken. Although histologically similar to wild-type counterparts at E9.5, expression of the ventricle-specific homeobox transcription factor Irx4 is slightly decreased in Hand2 DNA-binding mutants, suggesting that Hand2-mediated regulation of Irx4 expression requires DNA binding. These findings indicate that, during cardiogenesis, Hand2 has both DNA-binding dependent and independent functions, both of which mechanistically rely on dimerization. Furthermore, it is important to consider that when Hand factors are expressed in ectopic locations that their presence will have profound effects on the bHLH dimer pool within those cells. To fully understand relevant gain-of-function future efforts will require faithful upregulation only within cardiomyocytes that express either Hand1 or Hand2.
Given that, together, Hand factors regulate multiple aspects of ventricular development, the possibility arises that different gene sets, and thereby different developmental processes, are regulated mechanistically through distinct modes of Hand factor function, It has been shown that, in the developing sympathetic nervous system, Hand2 synergistically, with the homeoprotein Phox2a, upregulates transcription from the dopamine β-hydroxylase promoter independent of DNA binding (Rychlik et al. 2003; Xu et al. 2003). In the heart, Hand2 may similarly interact with other DNA-bound trans-activators to regulate transcription as a component of a larger protein complex. Alternatively, Hand2 may heterodimerize with negative-regulatory bHLH factors, for example, the HES-related transcription factors (HRT; also referred to as Hey, Hesr, HERP, or CHF; (Firulli et al. 2000), providing relief of repression for certain genes. Indeed Hand2 has been shown to interact with Nkx2.5 synergistically to activate transcription of the Nppa promoter through a DNA-binding independent mechanism (Thattaliyath et al. 2002). These possibilities add an additional layer of complexity to Hand factor function. In the context of human birth defects, it is important to bear in mind that, while partial or full loss of Hand factor function may be detrimental, mutations which cause these two factors to interact with protein binding partners in aberrant ways may also lead to congenital disease.
Insights into hand function during cardiomyocyte growth and morphogenesis from zebrafish
In addition to cardiomyocyte maturation, studies in zebrafish have demonstrated that Hand factors regulate cardiac morphogeneis. In contrast to the mammalian heart, which develops from the cardiac crescent, he zebrafish heart originates as two bilateral cardiogenic fields that migrate medially, fusing to form the primitive heart tube.
Unlike medaka, tetradon and stickleback, which have both hand1 and hand2 genes (ENSEMBL), the zebrafish genome only contains hand2. Similar to what is seen in mice, Twist proteins are expressed in the endocardial cushions, but not the myocardium (Yeo et al. 2009). Thus, functional studies in zebrafish are not clouded by potential functional redundancy between factors. The sole zebrafish hand factor is disrupted in Hands off (han) mutants.
Although their precardiac mesoderm is specified normally, han mutants have reduced myocardium (Yelon et al. 2000). The myocardium that does form, rather than fusing to form the medial heart tube, remains laterally displaced. Differentiation of ventricular cardiomyocytes, as assessed through ventricular myosin heavy chain (vmhc) expression, is strongly affected, with han mutants showing either no vmhc-expressing cells, or few, irregularly distributed vmhc-expressing cells. Although the majority of cardiac transcription factors examined, including mef2c, gata4, gata5, gata6, and the Tbx20 ortholog, hrt, were all expressed normally in han mutant cardiomyocytes, expression of the T-box transcription factor tbx5 is not maintained (Yelon et al. 2000).
han mutants display deficient cardiogenic lateral plate mesoderm (LPM) expansion mediolaterally within the embryo, as visualized through gata4 expression, potentially by regulating cell movement and proliferation. These observations indicate that in zebrafish, hand2 regulates not only cardiac chamber-specific gene expression, but also a more general role in the early morphogenesis and differentiation of the LPM (Schoenebeck et al. 2007).
In zebrafish, hand2 is thought to play a permissive, rather than instructive, role in potentiating cardiomyocyte differentiation. Ectopic hand2 expression does not generate ectopic cardiomyocytes (Yelon et al. 2000). Dimensions of the heart-forming region, as gauged by the relative domains of nkx2.5, which marks cardiogenic progenitors, and scl, which designates vascular and hematopoetic progenitors, are unchanged by a loss of hand2. However, fewer of these progenitors (~60%) ultimately contribute to the heart in han mutants. It is thought that a portion of cardiac progenitors initiates myocardial differentiation, but fail to complete the program.
han mutant cells behave indistinguishably from wild-type cells when transplanted into wild-type zebrafish embryos, migrating medially and integrating into the heart, while wild-type cells transplanted into han mutant embryos fail to move independently towards the midline (Garavito-Aguilar et al. 2010). Hand2 function during zebrafish cardiac field fusion is, therefore, non-cell autonomous. This interesting observation could suggest defects in extracellular matrix (ECM) that alter cell migration. Interestingly, only about 60% of these han mutant-derived donor cells differentiate into cardiomyocytes, consistent with the ~40% reduction of cardiomyocyte production typical of the han mutant heart field (Schoenebeck et al. 2007), while wild-type donor-derived cells differentiate into cardiomyocytes at comparable frequencies when implanted in either wild-type or han mutant hosts. Therefore, unlike its influence on cardiac cell migration, hand2-mediated potentiation of cardiomyocyte cell fate is likely cell-autonomous. Zebrafish hand2 thus performs independent functions to both potentiate myocardial differentiation cell-autonomously, and to promote cardiac fusion through an extracellular mediator.
Strong evidence that hand2 modulates ECM has recently been reported. Fibronectin1 (fn1) is a large, adhesive glycoprotein that occupies the extracellular matrix and functions during certain forms of cell migration as an ECM substrate, chemotactic promoter, and an integrin signaling activator (Yamada, 2000). Fn1 gene expression is upregulated and fn1 protein deposition is disorganized in han mutants (Garavito-Aguilar et al. 2010; Trinh et al. 2005). Conversely, embryos overexpressing hand2 display reduced fn1 expression and deposition. These hand2 overexpressers phenocopy certain aspects of fn1 mutant zebrafish (Trinh and Stainier, 2004), such as scattered cardiomyocytes, a disorganized myocardial monolayer, and delayed cardiac field fusion.
Further supporting antagonistic roles for hand2 and fn1 during early cardiogenesis, heterozygosity of fn1 rescues cardiac fusion (but not cardiomyocyte production) in han mutants. As both excess and absence of fn1 leads to cardia bifida, these data strongly suggest that hand2 enables cardiac fusion through fn1 modulation.
han mutant cardiomyocytes also exhibit polarity defects. Normal cardiomyocytes display distinct cellular polarity and defined subcellular localization of β-catenin (basolaterally), aPKC (apically), and ZO-1 (laterally). han mutants lack all of these polarized features, and their cardiomyocytes aggregate into clusters, rather than forming a monolayer. However, fn1 heterozygous rescue partially restores cardiomyocytes to a monolayer conformation, although it fails to restore other features of cellular polarity.
Thus, hand2 plays an integral role in zebrafish cardiogenesis regulating gene expression during ventricular cardiomyocyte specification, ECM deposition during cardiac field migration and fusion, and apicobasal cell polarity necessary for heart tube extension.
Hand2 and the TALE (Three Amino acid Loop Extension)-class homeodomain proteins pbx2 and pbx4, as revealed by morpholino knockdown experiments, regulate a common set of downstream targets and share similar myocardial differentiation and morphogenesis reduction-of-function phenotypes (Maves et al. 2009). Hand2 expression is upregulated in the cardiogenic fields of pbx2;pbx4 morphants. Morphants in which hand2, pbx2 and pbx4 have all been knocked down phenocopy the cardia bifida associated with hand2 null embryos, potentially revealing that the primary role of pbx proteins in the heart is to facilitate hand2 function.
Zebrafish are a powerful model organism, in that their hearts have the capacity to regenerate following mechanical damage, and the growth of adult fish can be modulated experimentally depending upon age and aquarium density. Unlike mammalian cardiomyocytes, which typically generate postnatal cardiac growth through the hypertrophy of existing cells, zebrafish cardiomyocytes are proliferative. During zebrafish heart regeneration, hand2-expressing cells appear along the apical ridge of the wound where they remain until regeneration is complete (Wills et al. 2008). Interestingly, myocardial hand2 expression is greater in adult zebrafish undergoing slow growth than those maintaining a static size. This expression increases to levels comparable to those seen during cardiomyocyte regeneration in fish undergoing rapid growth. These results suggest that adult zebrafish cardiomyocytes reactivate factors that are active during early development, such as hand2, to generate new cardiomyocytes during periods of rapid growth and regeneration.
Animal models of Hand1 and Hand2 function in the OFT (cNCC and SHF)
In addition to their contributions to ventricular cardiomyocyte expansion and heart chamber morphogenesis, Hand factors play integral roles in the development of extracardiac tissues important for formation of the functional heart, among them the cardiac neural crest cells (cNCCs). The cNCCs originate from the dorsal lip of the neural tube caudal to the otic placode, at the axial level of rhombomeres 5–7. These cells delaminate from the neural tube, migrating ventrally and invading the caudal pharyngeal arches (arches III, IV and VI), where they surround the pharyngeal arch arteries (Scholl and Kirby, 2009). They subsequently migrate into the cardiac jelly-filled lumen intervening the OFT myocardium and endocardium. Through extensive remodeling, the outflow tract is septated into aortic and pulmonary components, and the pharyngeal arch arteries become the great vessels of the aortic arch. The cNCCs themselves contribute to the aorticopulmonary septum and differentiate into the smooth muscle cells of the great arteries. Neural crest cell dysfunction disrupts various aspects of this remodeling process.
Hand1 and Hand2 are both expressed dynamically in cNCCs that have migrated into the caudal pharyngeal arches (Cserjesi et al. 1995; Srivastava et al. 1997; Vincentz et al. 2008). Hand1 continues to be expressed, although not uniformly, in cNCCs occupying the OFT cushions (Vincentz et al. 2008). Additionally, both Hand1 and Hand2 are expressed strongly in the developing myocardial cuff (Cserjesi et al. 1995; Srivastava et al. 1997; Vincentz et al. 2008). Thus, the OFT is one of the only regions of the developing heart in which Hand1 and Hand2 are strongly expressed in overlapping domains.
A broad spectrum of CHDs, including VSDs, aortic arch artery patterning defects, and aorticopulmonary valve defects, which represent a substantial proportion of all birth defects, can be attributed to cNCC dysfunction (reviewed in (Lie-Venema et al. 2007; Mitchell et al. 2007; Obler et al. 2008; Snarr et al. 2008; Srivastava and Olson, 2000; Stoller and Epstein, 2005; Waldo et al. 1998). Although Hand2−/− systemic mutants die too early to assess formation of the OFT, the general hypoplasia of the OFT myocardium (Srivastava et al. 1997) and pharyngeal arches (Thomas et al. 1998; Yamagishi et al. 2000) characteristic of these mutants strongly indicates that disruption of Hand2 would recapitulate a subset of these disease phenotypes. Loss of the catecholamine norepinepherine in the neural crest cell-derived sympathetic nervous system causes embryonic lethality at mid-gestation (Lim et al. 2000). Hand2 is required in the developing sympathetic nervous system to transcriptionally regulate biosynthetic enzymes, such as tyrosine hydroxylase (TH) and dopamine β-hydroxylase (DBH), necessary to generate norepinephrine (Morikawa et al. 2007). Conditional ablation of Hand2 in NCCs, consequently, causes embryonic lethality at E12.5 (Morikawa et al. 2007). However, these Hand2 cNCC-CKOs embryos can be rescued to birth by supplementing the water of pregnant dams with the adrenoceptor agonist, isoproterenol (Morikawa and Cserjesi, 2008). Hand2 cNCC-conditional knockouts (CKOs) then succumb, perinatally, to various OFT patterning defects, including pulmonary stenosis, B-type interrupted aortic arch (IAA-B), aberrant origin of the right subclavian artery (AORSA), often including retroesophageal right subclavian artery, and membranous VSDs coupled with double outlet right ventricle (DORV; (Holler et al. 2010; Morikawa and Cserjesi, 2008). Conflicting lineage trace analyses make it difficult to assess whether cNCCs contribute to the Hand2 cNCC-CKO OFT qualitatively at levels below that of (Holler et al. 2010) or comparable to (Morikawa and Cserjesi, 2008) control embryos. Although vascular smooth muscle cell differentiation is dysfunctional in Hand2−/− systemic mutants (Yamagishi et al. 2000), differentiation of Hand2 conditionally null cNCCs into smooth muscle cells is unimpeded (Morikawa and Cserjesi, 2008). Hand2 conditionally null cNCCs do display proliferative defects (Holler et al. 2010). Expression of the semaphorin3C (Sema3C) receptor, plexinA2 (PlxnA2), is downregulated in Hand2 cNCC-CKO OFTs, raising the possibility that Hand2 null cNCCs are incapable of responding to Sema3C signaling, resulting in a partial phenocopy of the Sema3C−/− mutant phenotype (Morikawa and Cserjesi, 2008). Extensive microarray analyses of Hand2 conditionally null cNCCs have identified a number of factors regulated by Hand2 and potentially important for cNCC migration (Holler et al. 2010), including decreased Pdgfα (Morrison-Graham et al. 1992; Orr-Urtreger and Lonai, 1992) and Gja5 (Cx40), and increased integrin α9 (Itgα9; (Huang et al. 2005; Young et al. 2001) and integrin α4 (Itgα4; (Pinco et al. 2001; Sheppard et al. 1994).
Hand2 cNCC-CKOs, surprisingly, display an enlarged RV (Morikawa and Cserjesi, 2008). This phenotype is likely due to increased proliferation in the trabecular zone of the RV, reflective of a failure of trabecular cardiomyocytes to exit the cell cycle. The trabecular zone marker, atrial natriuretic peptide (Nppa), is downregulated in the RV, but not LV of Hand2 cNCC-CKOs. Together, these data suggest that Hand2 functions non-cell autonomously in the cNCCs to, prior to the onset of Nppa expression at E10.5, to regulate SHF-derived cardiomyocyte proliferation vs. differentiation. This finding is intriguing, as it provides a model for the congenital disorder hyper-trabeculation/ventricular noncompaction (VHT), whose prevalence has only recently begun to be appreciated through advancements in imaging technology (Weiford et al. 2004). Although the genetic causes of this disorder are unknown, it is often associated with facial dysmorphism (Chin et al. 1990; Ichida et al. 1999) and other cardiac defects attributable to cNCC dysfunction (Kenny et al. 2007). Thus, NCC defects may ultimately prove to comprise the etiology of VHT.
No OFT phenotypes have yet been reported for Hand1 conditional ablation in cNCCs. Genetic interactions have been reported between Hand1 and Hand2 in cranial NCCs (Barbosa et al. 2007). As mentioned previously, Hand1 and Hand2 expression overlaps in the cNCCs. Genetic interaction studies would be warranted and necessary to determine whether Hand1 and Hand2 perform redundant functions during OFT morphogenesis.
Observations of NCCs in Twist1−/− OFTs have suggested that Hand1 and Hand2 may perform unique functions within specific subpopulations of these cells. Among the normally compacted cNCC-derived ectomesenchyme of the Twist1−/− OFT cushions are amorphic cellular aggregates (Vincentz et al. 2008). These aggregates are of NCC origin, but differ from surrounding, phenotypically normal NCC ectomesenchyme in that they express high levels of Hand1 and Hand2 (Vincentz et al. 2008). Thus, the subpopulation of Hand1/Hand2 co-expressing NCCs in the OFT cushions are uniquely affected by a loss of Twist1 function. Whether Hand1 and Hand2 actively function to contribute to the phenotype of these mutant cells remains to be seen.
Hand2−/− mutants have diminished SHF-derived myocardium, most likely due to cardiomyocyte apoptosis (Thomas et al. 1998). Conditional ablation of Hand2 in differentiated cardiomyocytes, via the cTnt-Cre mouse line, causes embryonic lethality between E9.5 and E13.5. These mice display RV and OFT hypoplasia with variable expressivity, with severely affected mutants phenocopying Hand2−/− systemic mutants and later-surviving mutants, despite also suffering from hypoplasia, displaying two distinct ventricles. These data confirm that Hand2 regulates the development of differentiated, SHF-derived cardiomyocytes.
Conversely, overexpression of Hand1 in the myocardial cuff generates an OFT 1.5 – 2 fold longer than that of a wild-type embryo (Risebro et al. 2006). Markers of OFT cardiomyocyte differentiation, Nppa and Wnt11, are downregulated in the OFTs of these mutants. Immunohistochemistry for phospho-histone H3 reveals that the OFT cardiomyocytes in these mutants are over proliferative. As a whole, these data suggest that Hand factor function governs cell number in the SHF-derived OFT myocardium.
Hand factor function in the epicardium and endocardium
Derivatives of the epicardium and endocardium fulfill important roles in the developing heart. Although Hand factors have been shown to contribute to the development of several heart structures, their respective contributions to the development of the epicardial and endocardial lineages has not been thoroughly studied. Cells of the endocardium undergo EMT, invading the AV cushions and ultimately contributing to the AV valves. In E11.5 Hand1 cardiac-specific conditional knockouts, the AV cushions are hyperplastic in the absence of any detectable cell proliferation or cell death abnormalities (McFadden et al. 2005). The neonatal AV valves of these mutants are comparatively thicker than those of wild-type littermates. This phenotype is intriguing, as Hand1 expression is never observed in the endocardium or AV cushions (Fig. 2). As such, Hand1 function may be required in the myocardium to regulate endocardial cell contribution to the AV cushions in a non-cell autonomous manner. It should be noted, however, that the direct Hand1 transcriptional target Thymosin β4 is expressed within these tissues at E10.0, a time point consistent with an E11.5 phenotype (Smart et al. 2002). Thus, Hand1 may be transiently or weakly expressed in the endocardium and/or AV cushions and has yet to be detected and reported, although lineage trace analyses using the Hand1EGFPCre knock-in allele fail to identify a Hand1-expressing lineage in the endocardium (Barnes et al. 2010) and provide evidence against this hypothesis. Similarly, E11.5 Hand2 DNA binding-deficient mutants have enlarged and disorganized endocardial cushions (Liu et al. 2009). Unlike Hand1, Hand2 is strongly expressed in the endocardium, and may be directly required, through a DNA binding-independent mechanism, to regulate AV cushion development. More so than the limited amount that is known about Hand factor function in the endocardium, the function of Hand1 and Hand2 in the epicardium is completely unexplored, although recent studies confirm that Hand1-expressing cells ultimately form the epicardium and its derivatives (Barnes et al. 2010). Additionally, development of the proepicardium is disrupted in zebrafish han mutants (Liu and Stainier). Extensive tissue-specific gene ablation studies will be required to unravel the cell autonomous and non-cell autonomous functions of Hand1 and Hand2 in the developing endocardium and, potentially, epicardium.
HAND factors in human CHD
The respective reciprocal expression domains in the left and right ventricle are well-characterized features of Hand1 and Hand2 in the heart, marking them both as candidate contributing factors to human CHDs that affect the developing heart asymmetrically. For example, as the name suggests, Hypoplastic Left Heart Syndrome (HLHS), a severe form of CHD affecting 0.16–0.36 of every 1000 live births (Talner, 1998), specifically affects components of the left side of the heart, including the aorta, LV and both the aortic and mitral valves. The genetic causes of HLHS are an almost complete black box; however, one of the two human genes implicated in HLHS is HAND1.
Reamon-Buettner, et. al, sequenced the HAND1 gene in 31 unrelated hypoplastic heart syndrome patients. 24 of these 31 patients exhibited a common frameshift mutation (A126fs) in the loop region of the bHLH domain, resulting in a truncated protein featuring has only a single complete α-helix. This truncated HAND1 is incapable of modulating transcription from D- or E-boxes either alone or in conjunction with bHLH binding partners. Thus, disruption of HAND1 function is associated with human cardiac hypoplasia.
Gja1 is also implicated in HLHS. Gja1 protein expression is downregulated by overexpressing Hand1 in mice (Breckenridge et al. 2009); however, no change in mRNA levels is reported, indicating that, while a regulatory relationship exists between Gja1 and Hand1 in cardiomyocytes, current evidence suggests that it is not directly transcriptional. Hand1 and Hand2 may both transcriptionally regulate the related protein Gja5 (Cx40; (McFadden et al. 2005), although cardiomyocyte expression of this factor is limited to the LV embryonically. Expression of Gja5 is slightly decreased in Hand1 cardiac-CKO embryos and is not detected in the ventricular myocardium of Hand1fx/-;Hand2+/−;Nkx2.5-Cre(+) embryos. Further expression studies in Hand2 cardiac-CKO embryos, coupled with in vitro DNA binding and trans-activation assays indicate that Gja5 is a direct transcriptional target of Hand2 in these cells (Holler et al. 2010). Thus, regulation of Gja proteins may be an evolutionarily conserved mechanism by which Hand factors contribute to cardiogenesis and congenital disease.
The association of HAND2 with human CHDs is not as well characterized; however, recent studies have provided tantalizing evidence that HAND2 may contribute to human CHDs. HAND2 maps to chromosome 4q33. A high incidence of CHDs, including VSDs, septal defects, pulmonary atresia, coarctation of the aorta, and tetralogy of Fallot, is associated with genomic duplications or deletions of 4q33 (Borochowitz et al. 1997; Byatt et al. 1997). Studies of 131 ethnic Han Chinese children with CHDs, such as tetralogy of Fallot, pulmonary stenosis, atrioventricular septal defects, and VSDs with DORV, identified three HAND2 missense mutations, one isonymous mutation, and three mutations within the HAND2 5’ and 3’ untranslated regions in 12 of these patients (Shen et al. 2010). It should be noted that one of the missense mutations, S36N, is not in an evolutionarily conserved residue, is not predicted in silico to affect HAND2 protein function, and was also detected in one of the 250 healthy controls. Although this specific mutation is not a strong candidate causative factor in human CHDs, the relatively high frequency of HAND2 mutations revealed by this study suggests that HAND2 dysfunction may contribute to CHDs affecting the RV and OFT. It will be interesting to determine whether these mutations, whether through altered protein-protein interactions or DNA-binding, or through altered mRNA stability or translation, result in HAND2 hypo- or hyper-activity. Indeed, increased HAND2 mRNA expression has been associated with tetralogy of Fallot, while increased HAND1 mRNA expression has been associated with hypertrophic obstructive cardiomyopathy (Ritter et al. 1999), echoing the idea that either too much or too little HAND factor function may be detrimental to cardiac growth and morphogenesis. Functional validation of these mutant HAND2 variants will be critical to elucidating their contribution to congenital heart disease.
Acknowledgements
We would like thank all the laboratories who contributed publications that fill this review. Infrastructural support at the Herman B Wells Center for Pediatric Research is in part supported by the generosity of the Riley Children’s Foundation and Division of Pediatric Cardiology. This work is supported by the NIH RO1HL061677-11 1P01HL085098-04 (ABF), and AHA 0815426G (RMB).
Literature Cited
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AC, Funato N, Chapman S, McKee MD, Richardson JA, Olson EN, Yanagisawa H. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev Biol. 2007;310(1):154–168. doi: 10.1016/j.ydbio.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. Int J Dev Biol. 2009;53(7):909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli BA, Conway SJ, Vincentz JW, Firulli AB. Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Dev Dyn. 2010;239(11):3086–3097. doi: 10.1002/dvdy.22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borochowitz Z, Shalev SA, Yehudai I, Bar-el H, Dar H, Tirosh E. Deletion (4)(q33 --> qter): a case report and review of the literature. J Child Neurol. 1997;12(5):335–337. doi: 10.1177/088307389701200510. [DOI] [PubMed] [Google Scholar]
- Breckenridge RA, Zuberi Z, Gomes J, Orford R, Dupays L, Felkin LE, Clark JE, Magee AI, Ehler E, Birks EJ, Barton PJ, Tinker A, Mohun TJ. Overexpression of the transcription factor Hand1 causes predisposition towards arrhythmia in mice. J Mol Cell Cardiol. 2009;47(1):133–141. doi: 10.1016/j.yjmcc.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Byatt SA, Baker E, Richards RI, Roberts C, Smith A. Unbalanced t(4;11)(q32;q23) in a 34-year-old man with manifestations of distal monosomy 11q and trisomy 4q syndromes. Am J Med Genet. 1997;70(4):357–360. [PubMed] [Google Scholar]
- Chin TK, Perloff JK, Williams RG, Jue K, Mohrmann R. Isolated noncompaction of left ventricular myocardium. A study of eight cases. Circulation. 1990;82(2):507–513. doi: 10.1161/01.cir.82.2.507. [DOI] [PubMed] [Google Scholar]
- Conway SJ, Firulli B, Firulli AB. A bHLH code for cardiac morphogenesis. Pediatr Cardiol. 2010;31(3):318–324. doi: 10.1007/s00246-009-9608-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserjesi P, Brown D, Lyons GE, Olson EN. Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev Biol. 1995;170(2):664–678. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- Dupont E, Matsushita T, Kaba RA, Vozzi C, Coppen SR, Khan N, Kaprielian R, Yacoub MH, Severs NJ. Altered connexin expression in human congestive heart failure. J Mol Cell Cardiol. 2001;33(2):359–371. doi: 10.1006/jmcc.2000.1308. [DOI] [PubMed] [Google Scholar]
- Firulli AB. A HANDful of questions: the molecular biology of the heart and neural crest derivatives (HAND)-subclass of basic helix-loop-helix transcription factors. Gene. 2003;312:27–40. doi: 10.1016/s0378-1119(03)00669-3. [DOI] [PubMed] [Google Scholar]
- Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18(3):266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Hadzic DB, McDaid JR, Firulli AB. The basic helix-loop-helix transcription factors dHAND and eHAND exhibit dimerization characteristics that suggest complex regulation of function. J Biol Chem. 2000;275(43):33567–33573. doi: 10.1074/jbc.M005888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Howard MJ, McDaid JR, McIlreavey L, Dionne KM, Centonze VE, Cserjesi P, Virshup DM, Firulli AB. PKA, PKC, and the protein phosphatase 2A influence HAND factor function: a mechanism for tissue-specific transcriptional regulation. Mol Cell. 2003;12(5):1225–1237. doi: 10.1016/s1097-2765(03)00425-8. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37(4):373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, McConville DP, Byers JS, 3rd, Vincentz JW, Barnes RM, Firulli AB. Analysis of a Hand1 hypomorphic allele reveals a critical threshold for embryonic viability. Dev Dyn. 2010;239(10):2748–2760. doi: 10.1002/dvdy.22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Redick BA, Conway SJ, Firulli AB. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J Biol Chem. 2007;282(37):27536–27546. doi: 10.1074/jbc.M702613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavito-Aguilar ZV, Riley HE, Yelon D. Hand2 ensures an appropriate environment for cardiac fusion by limiting Fibronectin function. Development. 2010;137(19):3215–3220. doi: 10.1242/dev.052225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutstein DE, Morley GE, Vaidya D, Liu F, Chen FL, Stuhlmann H, Fishman GI. Heterogeneous expression of Gap junction channels in the heart leads to conduction defects and ventricular dysfunction. Circulation. 2001;104(10):1194–1199. doi: 10.1161/hc3601.093990. [DOI] [PubMed] [Google Scholar]
- Hendershot TJ, Liu H, Sarkar AA, Giovannucci DR, Clouthier DE, Abe M, Howard MJ. Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev Dyn. 2007;236(1):93–105. doi: 10.1002/dvdy.20989. [DOI] [PubMed] [Google Scholar]
- Holler KL, Hendershot TJ, Troy SE, Vincentz JW, Firulli AB, Howard MJ. Targeted deletion of Hand2 in cardiac neural crest-derived cells influences cardiac gene expression and outflow tract development. Dev Biol. 2010;341(1):291–304. doi: 10.1016/j.ydbio.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Bridges LC, White JM. Selective modulation of integrin-mediated cell migration by distinct ADAM family members. Mol Biol Cell. 2005;16(10):4982–4991. doi: 10.1091/mbc.E05-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida F, Hamamichi Y, Miyawaki T, Ono Y, Kamiya T, Akagi T, Hamada H, Hirose O, Isobe T, Yamada K, Kurotobi S, Mito H, Miyake T, Murakami Y, Nishi T, Shinohara M, Seguchi M, Tashiro S, Tomimatsu H. Clinical features of isolated noncompaction of the ventricular myocardium: long-term clinical course, hemodynamic properties, and genetic background. J Am Coll Cardiol. 1999;34(1):233–240. doi: 10.1016/s0735-1097(99)00170-9. [DOI] [PubMed] [Google Scholar]
- Kenny D, Hares D, Uzun O. Ventricular non-compaction in the setting of double-outlet right ventricle (tetralogy of Fallot type) with doubly committed subarterial ventricular septal defect. Heart. 2007;93(5):647. doi: 10.1136/hrt.2006.090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kostin S, Dammer S, Hein S, Klovekorn WP, Bauer EP, Schaper J. Connexin 43 expression and distribution in compensated and decompensated cardiac hypertrophy in patients with aortic stenosis. Cardiovasc Res. 2004;62(2):426–436. doi: 10.1016/j.cardiores.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Lie-Venema H, van den Akker NM, Bax NA, Winter EM, Maas S, Kekarainen T, Hoeben RC, deRuiter MC, Poelmann RE, Gittenberger-de Groot AC. Origin, fate, and function of epicardium-derived cells (EPDCs) in normal and abnormal cardiac development. Scientific World Journal. 2007;7:1777–1798. doi: 10.1100/tsw.2007.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KC, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat Genet. 2000;25(2):209–212. doi: 10.1038/76080. [DOI] [PubMed] [Google Scholar]
- Litchenberg WH, Norman LW, Holwell AK, Martin KL, Hewett KW, Gourdie RG. The rate and anisotropy of impulse propagation in the postnatal terminal crest are correlated with remodeling of Cx43 gap junction pattern. Cardiovasc Res. 2000;45(2):379–387. doi: 10.1016/s0008-6363(99)00363-6. [DOI] [PubMed] [Google Scholar]
- Liu J, Stainier DYR. Tbx5 and Bmp signaling are essential for proepicardium specification in zebrafish. Circulation Research. 106(12):1818–1828. doi: 10.1161/CIRCRESAHA.110.217950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Barbosa AC, Chapman SL, Bezprozvannaya S, Qi X, Richardson JA, Yanagisawa H, Olson EN. DNA binding-dependent and -independent functions of the Hand2 transcription factor during mouse embryogenesis. Development. 2009;136(6):933–942. doi: 10.1242/dev.034025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo PC, Zaffran S, Senatore S, Frasch M. The Drosophila Hand gene is required for remodeling of the developing adult heart and midgut during metamorphosis. Dev Biol. 2007;311(2):287–296. doi: 10.1016/j.ydbio.2007.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindill DM, Risebro CA, Smart N, Franco-Viseras Mdel M, Rosario CO, Swallow CJ, Dennis JW, Riley PR. Nucleolar release of Hand1 acts as a molecular switch to determine cell fate. Nat Cell Biol. 2007;9(10):1131–1141. doi: 10.1038/ncb1633. [DOI] [PubMed] [Google Scholar]
- Masuelli L, Bei R, Sacchetti P, Scappaticci I, Francalanci P, Albonici L, Coletti A, Palumbo C, Minieri M, Fiaccavento R, Carotenuto F, Fantini C, Carosella L, Modesti A, Di Nardo P. Beta-catenin accumulates in intercalated disks of hypertrophic cardiomyopathic hearts. Cardiovasc Res. 2003;60(2):376–387. doi: 10.1016/j.cardiores.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Maves L, Tyler A, Moens CB, Tapscott SJ. Pbx acts with Hand2 in early myocardial differentiation. Dev Biol. 2009;333(2):409–418. doi: 10.1016/j.ydbio.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132(1):189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Charite J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127(24):5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- Mitchell ME, Sander TL, Klinkner DB, Tomita-Mitchell A. The molecular basis of congenital heart disease. Semin Thorac Cardiovasc Surg. 2007;19(3):228–237. doi: 10.1053/j.semtcvs.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P. Extra-embryonic vasculature development is regulated by the transcription factor HAND1. Development. 2004;131(9):2195–2204. doi: 10.1242/dev.01091. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P. Cardiac neural crest expression of Hand2 regulates outflow and second heart field development. Circ Res. 2008;103(12):1422–1429. doi: 10.1161/CIRCRESAHA.108.180083. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, D'Autreaux F, Gershon MD, Cserjesi P. Hand2 determines the noradrenergic phenotype in the mouse sympathetic nervous system. Dev Biol. 2007;307(1):114–126. doi: 10.1016/j.ydbio.2007.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison-Graham K, Schatteman GC, Bork T, Bowen-Pope DF, Weston JA. A PDGF receptor mutation in the mouse (Patch) perturbs the development of a non-neuronal subset of neural crest-derived cells. Development. 1992;115(1):133–142. doi: 10.1242/dev.115.1.133. [DOI] [PubMed] [Google Scholar]
- Obler D, Juraszek AL, Smoot LB, Natowicz MR. Double outlet right ventricle: aetiologies and associations. J Med Genet. 2008;45(8):481–497. doi: 10.1136/jmg.2008.057984. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Lonai P. Platelet-derived growth factor-A and its receptor are expressed in separate, but adjacent cell layers of the mouse embryo. Development. 1992;115(4):1045–1058. doi: 10.1242/dev.115.4.1045. [DOI] [PubMed] [Google Scholar]
- Pinco KA, Liu S, Yang JT. alpha4 integrin is expressed in a subset of cranial neural crest cells and in epicardial progenitor cells during early mouse development. Mech Dev. 2001;100(1):99–103. doi: 10.1016/s0925-4773(00)00503-7. [DOI] [PubMed] [Google Scholar]
- Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18(3):271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- Risebro CA, Smart N, Dupays L, Breckenridge R, Mohun TJ, Riley PR. Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development. 2006;133(22):4595–4606. doi: 10.1242/dev.02625. [DOI] [PubMed] [Google Scholar]
- Ritter O, Haase H, Schulte HD, Lange PE, Morano I. Remodeling of the hypertrophied human myocardium by cardiac bHLH transcription factors. J Cell Biochem. 1999;74(4):551–561. doi: 10.1002/(sici)1097-4644(19990915)74:4<551::aid-jcb5>3.3.co;2-0. [DOI] [PubMed] [Google Scholar]
- Rychlik JL, Gerbasi V, Lewis EJ. The interaction between dHAND and Arix at the dopamine beta-hydroxylase promoter region is independent of direct dHAND binding to DNA. J Biol Chem. 2003;278(49):49652–49660. doi: 10.1074/jbc.M308577200. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13(2):254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl AM, Kirby ML. Signals controlling neural crest contributions to the heart. Wiley Interdiscip Rev Syst Biol Med. 2009;1(2):220–227. doi: 10.1002/wsbm.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sepp R, Severs NJ, Gourdie RG. Altered patterns of cardiac intercellular junction distribution in hypertrophic cardiomyopathy. Heart. 1996;76(5):412–417. doi: 10.1136/hrt.76.5.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Li XF, Shen AD, Wang Q, Liu CX, Guo YJ, Song ZJ, Li ZZ. Transcription factor HAND2 mutations in sporadic Chinese patients with congenital heart disease. Chin Med J (Engl) 2010;123(13):1623–1627. [PubMed] [Google Scholar]
- Sheppard AM, Onken MD, Rosen GD, Noakes PG, Dean DC. Expanding roles for alpha 4 integrin and its ligands in development. Cell Adhes Commun. 1994;2(1):27–43. doi: 10.3109/15419069409014200. [DOI] [PubMed] [Google Scholar]
- Shrivastava S, Srivastava D, Olson EN, DiMaio JM, Bock-Marquette I. Thymosin beta4 and cardiac repair. Ann N Y Acad Sci. 2010;1194:87–96. doi: 10.1111/j.1749-6632.2010.05468.x. [DOI] [PubMed] [Google Scholar]
- Smart N, Dube KN, Riley PR. Identification of Thymosin beta4 as an effector of Hand1-mediated vascular development. Nat Commun. 2010;1(4):1–10. doi: 10.1038/ncomms1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Hill AA, Cross JC, Riley PR. A differential screen for putative targets of the bHLH transcription factor Hand1 in cardiac morphogenesis. Gene Expr Patterns. 2002;2(1–2):61–67. doi: 10.1016/s0925-4773(02)00380-5. [DOI] [PubMed] [Google Scholar]
- Snarr BS, Kern CB, Wessels A. Origin and fate of cardiac mesenchyme. Dev Dyn. 2008;237(10):2804–2819. doi: 10.1002/dvdy.21725. [DOI] [PubMed] [Google Scholar]
- Srivastava D. HAND proteins: molecular mediators of cardiac development and congenital heart disease. Trends Cardiovasc Med. 1999;9(1–2):11–18. doi: 10.1016/s1050-1738(98)00033-4. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270(5244):1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407(6801):221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor. dHAND. Nat Genet. 1997;16(2):154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Stoller JZ, Epstein JA. Cardiac neural crest. Semin Cell Dev Biol. 2005;16(6):704–715. doi: 10.1016/j.semcdb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Talner CN. Report of the New England Regional Infant Cardiac Program. Pediatrics. 1998;102(1 Pt 2):258–259. by Donald C. Fyler, MD, Pediatrics, 1980;65(suppl):375–461. [PubMed] [Google Scholar]
- Thattaliyath BD, Firulli BA, Firulli AB. The basic-helix-loop-helix transcription factor HAND2 directly regulates transcription of the atrial naturetic peptide gene. J Mol Cell Cardiol. 2002;34(10):1335–1344. doi: 10.1006/jmcc.2002.2085. [DOI] [PubMed] [Google Scholar]
- Thomas T, Kurihara H, Yamagishi H, Kurihara Y, Yazaki Y, Olson EN, Srivastava D. A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998;125(16):3005–3014. doi: 10.1242/dev.125.16.3005. [DOI] [PubMed] [Google Scholar]
- Togi K, Kawamoto T, Yamauchi R, Yoshida Y, Kita T, Tanaka M. Role of Hand1/eHAND in the dorso-ventral patterning and interventricular septum formation in the embryonic heart. Mol Cell Biol. 2004;24(11):4627–4635. doi: 10.1128/MCB.24.11.4627-4635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6(3):371–382. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- Trinh LA, Yelon D, Stainier DY. Hand2 regulates epithelial formation during myocardial diferentiation. Curr Biol. 2005;15(5):441–446. doi: 10.1016/j.cub.2004.12.083. [DOI] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, Rodgers R, Firulli BA, Conway SJ, Firulli AB. An absence of Twist1 results in aberrant cardiac neural crest morphogenesis. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo K, Miyagawa-Tomita S, Kumiski D, Kirby ML. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Dev Biol. 1998;196(2):129–144. doi: 10.1006/dbio.1998.8860. [DOI] [PubMed] [Google Scholar]
- Weiford BC, Subbarao VD, Mulhern KM. Noncompaction of the ventricular myocardium. Circulation. 2004;109(24):2965–2971. doi: 10.1161/01.CIR.0000132478.60674.D0. [DOI] [PubMed] [Google Scholar]
- Wills AA, Holdway JE, Major RJ, Poss KD. Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development. 2008;135(1):183–192. doi: 10.1242/dev.010363. [DOI] [PubMed] [Google Scholar]
- Xu H, Firulli AB, Zhang X, Howard MJ. HAND2 synergistically enhances transcription of dopamine-beta-hydroxylase in the presence of Phox2a. Dev Biol. 2003;262(1):183–193. doi: 10.1016/s0012-1606(03)00361-0. [DOI] [PubMed] [Google Scholar]
- Yamada KM. Fibronectin peptides in cell migration and wound repair. J Clin Invest. 2000;105(11):1507–1509. doi: 10.1172/JCI10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H, Garg V, Matsuoka R, Thomas T, Srivastava D. A molecular pathway revealing a genetic basis for human cardiac and craniofacial defects. Science. 1999;283(5405):1158–1161. doi: 10.1126/science.283.5405.1158. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Olson EN, Srivastava D. The basic helix-loop-helix transcription factor, dHAND, is required for vascular development. J Clin Invest. 2000;105(3):261–270. doi: 10.1172/JCI8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DY. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127(12):2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- Yeo GH, Cheah FS, Winkler C, Jabs EW, Venkatesh B, Chong SS. Phylogenetic and evolutionary relationships and developmental expression patterns of the zebrafish twist gene family. Dev Genes Evol. 2009;219(6):289–300. doi: 10.1007/s00427-009-0290-z. [DOI] [PubMed] [Google Scholar]
- Young BA, Taooka Y, Liu S, Askins KJ, Yokosaki Y, Thomas SM, Sheppard D. The cytoplasmic domain of the integrin alpha9 subunit requires the adaptor protein paxillin to inhibit cell spreading but promotes cell migration in a paxillin-independent manner. Mol Biol Cell. 2001;12(10):3214–3225. doi: 10.1091/mbc.12.10.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32(4):189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]