Abstract
The focus of this protocol is mouse hepatitis virus (MHV), with occasional references to other coronaviruses. Many of these protocols can be easily adapted to other coronaviruses. Protocols for propagating MHV in DBT and 17CL‐1 cells; the storage and titration of viral stocks; purification of MHV on sucrose gradients; and the generation of recombinant viruses by a cDNA assembly method and by targeted recombination will be presented. Protocols are also included for the propagation of DBT, 17CL‐1, and L2 cells used for growing and titrating MHV, and for the growth of BHK‐R cells and FCWF cells. The latter two cell lines are used for regenerating infectious MHV by an in vitro cDNA assembly protocol and by a targeted recombination protocol, respectively, allowing reverse genetic manipulation of these viruses. An additional protocol for the maintenance of the large plasmids used for generating recombinant MHVs will also be presented. Curr. Protoc. Microbiol. 21:15E.1.1‐15E.1.46. © 2011 by John Wiley & Sons, Inc.
Keywords: coronavirus, reverse genetics, plaque assay, virus purification, targeted recombination, mouse hepatitis virus
Introduction
The coronaviruses encompass a group of enveloped RNA viruses that are widespread in nature and infect a wide variety of animals as well as humans, most commonly causing gastrointestinal or respiratory illnesses in the infected host, although more systemic infections can also occur.
Before the outbreak of severe acute respiratory syndrome (SARS) in 2002 to 2003, the majority of coronaviruses that had been isolated were recovered from humans and domesticated animal species, including laboratory rodents, and were regarded primarily as veterinary pathogens. The rodent coronaviruses, such as the various strains of MHV, provided rodent models for various human diseases. The demonstration that the agent that caused SARS was a novel coronavirus, and the subsequent investigations of the outbreak that indicated that the SARS‐coronavirus (SARS‐CoV) almost certainly entered the human population through zoonotic spread in wild animal markets in China, and subsequently underwent adaptation to humans, greatly increased the interest in this group and spurred efforts to identify and isolate coronaviruses from a large number of species in an attempt to identify the source of the SARS‐CoV. These investigations, and the advent of sensitive RT‐PCR‐based methods (Stephensen et al., 1999) to detect novel as well as known coronaviruses, identified a large number of new coronaviruses in a relatively short period of time. Particularly interesting are the recent findings that bats, a large and diverse group of mammals that account for approximately 20% of extant mammalian species, represent a large and previously unrecognized source of novel coronaviruses, including viruses closely related to the SARS‐CoV (Lau et al., 2005; Li et al., 2005a; Dominguez et al., 2007; Vijaykrishna et al., 2007). Table 1 lists many of the coronaviruses that have been identified, either through viral isolation or by RT‐PCR sequencing studies.
Table 1.
Coronaviruses and Their Natural Host Species
|
Virus |
Hosts |
Cells commonly used |
Disease |
|---|---|---|---|
|
Group Ia |
|||
|
Transmissible Gastroenteritis Virus (TGEV) |
Pigs |
ST, PK‐15 |
Gastroenteritis, respiratory |
|
Porcine Respiratory Coronavirus (PRCoV) |
ST, PK‐15 |
Respiratory |
|
|
Canine Coronavirus (CCoV) |
Dogs |
A‐72 |
Gastroenteritis |
|
Feline Infectious Peritonitis Virus (FIPV) |
Cats |
FCWF |
Enteritis, hepatitis, encephalitis, peritonitis |
|
Feline Coronavirus FeCoV |
Cats |
FCWF |
Enteritis |
|
Group Ib |
|||
|
Human Coronavirus 229E (HCoV‐229E) |
Humans |
MRC‐5, L132 |
Upper respiratory disease, possibly CNS |
|
Human Coronavirus NL63 (HCoV‐NL63) |
Humans |
CaCo‐2, LLC‐MK2 |
Upper and lower respiratory disease |
|
Porcine Epidemic Diarrhea Virus (PEDV) |
Pigs |
Vero (cell adapted) |
Enteritis |
|
Bat Coronavirus (BtCoV)a |
Bats |
NC |
Unknown |
|
Rabbit Coronavirus (RbCoV) |
Rabbits |
NC |
Enteritis, cardiomyopathy |
|
Group IIa |
|||
|
Mouse Hepatitis Virus (MHV) |
Mice |
DBT, L2, 17CL‐1 |
Enteritis, hepatitis, encephalomyelitis, pneumonitis |
|
Bovine Coronavirus (BCoV)b |
Cows |
HRT‐18 |
Enteritis, pneumonitis |
|
Sialodacryoadenitis Virus (SDAV) |
Rats |
L2P‐41.a |
Sialodacryoadenitis, pneumonitis |
|
Rat Coronavirus (RCoV) |
Rats |
L2, L2P‐41.a, LBC |
Pneumonitis |
|
Porcine Hemagglutinating Encephalitis Virus (PHEV) |
Pigs |
SK‐K |
Enteritis, encephalomyelitis, pneumonitis |
|
Human Coronavirus OC43 (HCoV‐OC43) |
Humans |
HRT‐18, RD |
Upper respiratory disease, possibly CNS, possibly enteritis |
|
Human Coronavirus HKU1 (HCoV‐HKU1) |
Humans |
Human airway epithelial (HAE) cells |
Upper and lower respiratory disease |
|
Group IIb |
|||
|
SARS Coronavirus (SARS‐CoV) |
Humans, Civets |
Vero |
Severe respiratory disease, enteritis, hepatitis |
|
Bat SARS‐Like Coronavirus (BtSARS‐CoV) |
Bats |
NC |
Unknown |
|
Group IIc |
|||
|
Bat Coronavirus (BtCoV)a |
Bats |
NC |
Unknown |
|
Group IId |
|||
|
Bat Coronavirus (BtCoV)a |
Bats |
NC |
Unknown |
|
Group III |
|||
|
Avian Infectious Bronchitis Virus (IBV)c |
Chickens |
Eggs, CK, Vero, BHK |
Respiratory disease, enteritis, renal disease |
|
Turkey Coronavirus (TCoV) |
Turkeys |
Turkey eggs |
Enteritis |
|
Bulbul Coronavirus (BuCoV) |
Bullbuls |
ND |
Unknown |
|
Thrush Coronavirus (ThCoV) |
Thrushes |
ND |
Unknown |
|
Munia Coronavirus (MuCoV) |
Munias |
ND |
Unknown |
Bat coronaviruses have been isolated from three different continents and fall into different subgroups based on phylogenetic analyses of their sequences.
Bovine coronavirus‐like viruses (GiCoV) have been isolated from a number of ruminant species such as giraffe.
Only IBV strain Beaudette which has been adapted to these cells can be propagated in Vero and BHK cells.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Many of the more recently identified coronaviruses, particularly the bat coronaviruses, have not been successfully grown in culture as yet, and are known only from RT‐PCR and sequencing studies. For those viruses that have been adapted to grow in cell culture, some of the cell lines commonly used to propagate and study these viruses are listed in Table 1.
The focus here will be on MHV, with occasional references to other coronaviruses. A number of these protocols can be easily adapted to many other coronaviruses. Protocols for propagating MHV in DBT and 17CL‐1 cells; the storage, and titration of viral stocks; purification of MHV on sucrose gradients; and the generation of recombinant viruses by a cDNA assembly method and by targeted recombination will be presented. Protocols are also included for the propagation of DBT, 17CL‐1, and L2 cells used for growing and titrating MHV, and for the growth of BHK‐R cells [a line of BHK cells transformed with a cDNA‐encoding murine CEACAM1a, the MHV receptor (Dveksler et al., 1991)]. 17Cl‐1 and DBT cells are both commonly used to propagate MHV. 17CL‐1 cells typically grow MHV to two‐fold greater titers than DBT cells, often reaching titers of 1 × 109 for MHV strain A59 (MHV‐A59). However, the progression of the infection throughout the culture is more variable, and occasionally low‐multiplicity infections fail to spread. Most strains of MHV produce a characteristic cytopathogenic effect (CPE) that is characterized by cell fusion. Both the higher titer and the occasional failure of the infection to spread throughout the culture in 17CL‐1 cells are probably related to the fact that cell fusion in response to MHV infection is slower in 17CL‐1 cells than in DBT cells and proceeds less rapidly as the cultures become more acidic over time. Plaque titrations to determine infectious virus titer can be performed with L2, DBT, or 17CL‐1 cells. We routinely use L2 cells for this purpose, since MHV produces slightly larger and clearer plaques in these cells than in the other two cell lines. We also provide protocols for the growth of FCWF cells used in targeted recombination studies and BHK‐R cells used in the generation of recombinant viruses by a cDNA assembly method. An additional protocol for the maintenance of the large plasmids used for generating recombinant MHVs will also be presented.
NOTE: All solutions and equipment coming into contact with living cells must be sterile, and aseptic technique should be used accordingly.
NOTE: Unless otherwise stated, prepare all solutions in sterile double‐distilled water.
NOTE: When working with RNA, wear gloves and use RNase‐free water (appendix mca02a) and materials.
Basic Protocol 1. Plaque Assay to Determine Viral Infectivity
The infectivity of MHV stocks can be determined using L2, DBT, or 17CL‐1 cells, by either endpoint dilution or plaque assays. The titer is an important component of many experiments: to optimize the amount of the virus needed for different applications, for characterizing mutants for the ability to grow relative to wild‐type virus, or for experiments in animals where we want to determine if there is a connection between the virus replication and the disease progression. We routinely use L2 cells for plaque assays, since MHV produces slightly larger and clearer plaques in these cells than in the other two cell lines. The size of the plaques produced varies among MHV strains, from MHV‐A59 which produces the largest plaques, ∼2 mm in diameter after 48 hr, to MHV‐Yale and other less vigorous strains which produce tiny plaques. Both the plaque assay and the end‐point dilution assay that are described below are easily adapted to other coronaviruses by using the corresponding susceptible cell lines (see Table 1 for a listing of cell lines used for different coronaviruses).
NOTE: L2 cells should be incubated at 3% CO2.
Materials
L2 cells (not commercially available; cells can be obtained from most investigators working with MHV, including the authors)
DME10 (see recipe)
MHV stock to be titered (Basic Protocol 2)
DME2 (see recipe)
DME0 (see recipe)
1.6% agarose (see recipe)
2× DME2 (see recipe)
0.1% (w/v) crystal violet in 70% ethanol
6‐well tissue culture plates
37°C, 3% CO2 incubator
Sterile tubes with caps, 1.5 to 5 ml in size, for serial dilutions of virus
Platform rocker
-
1
Set up L2 cells in 6‐well plates at 0.8 × 106 cells in 2.5 ml of DME10 medium per well, and incubate at 37°C in 3% CO2 for 2 days, at which time they should be confluent.
-
2
On the day of the plaque assay, rapidly thaw the samples to be titered in a 37°C water bath and place on ice immediately. Make serial 10‐fold dilutions of the virus in DME2, keeping the virus dilutions in an ice water bath to keep them cold.
-
3
Aspirate medium from 6‐well plates, no more than four plates at a time, and wash once with DME0 (DME2 or PBS can alternatively be used).
Be careful during aspiration to leave a small amount of medium (∼0.2 ml) behind to keep the plates from drying out when handling a large number of plates.
-
4
Aspirate DME0 from plates as described above and add 0.2 ml/well of each virus dilution. Start with the most dilute sample.
It is not necessary to change pipets between dilutions of a single sample if this procedure is followed. Each dilution is usually titered in duplicate or triplicate wells.
-
5
Distribute virus by rocking by hand. Incubate the plates at room temperature for 60 min while gently rocking from side to side on a platform rocker. Redistribute virus by hand about once every 10 min.
-
6
Melt 1.6% agarose in a microwave and cool to between 45° and 50°C. Warm up 2× DME2 to 45° to 50°C. Mix equal volumes of 1.6% agarose and 2× DME2 (to prepare the agarose overlay) and place in 50°C water bath in a biosafety cabinet. Add 2.5 ml of overlay to each well, and gently swirl the plate immediately after adding the agarose overlay solution to all of the wells in a plate, to absorb the viral inoculum into the agarose solution. Let plates sit without disturbing until the agarose has solidified.
-
7
Incubate at 37°C for 2 days.
Plaques are normally visible by 2 days.
-
8
If needed (in the case of mutants grown at 34°C, or for very small‐plaque viruses), feed cells with either 2 ml of DME2 or with a second agarose overlay and incubate an extra day.
Plaques can be seen without staining but are scored after crystal violet staining as described in the next step.
-
9
Remove the agarose (flip out into disinfectant solution) and stain with 0.1% crystal violet in 70% ethanol. Allow to remain in stain for ∼30 sec, then gently rinse with water, invert on paper towels to drain, then air dry and count.
Alternatively, plaques can also be visualized by neutral red staining. Make up a 1.0% stock of neutral red in PBS and filter sterilize. Store in a foil‐covered bottle. For each plate, make up a second overlay containing 7.5 ml of 2× DME2, 7.5 ml of 1.6% agarose, and 0.6 ml of neutral red stock solution. Add 2.5 ml/well and incubate overnight. Plaques are best visualized with a green filter but are usually visible against a white background.
-
10
Calculate viral titer by multiplying the mean number of plaques per well by the serial dilution value to determine the concentration in 1 ml of the virus preparation being assayed.
Thus, titer (pfu/ml) = average plaque count from replicate wells × 5 (1/0.2 ml) × dilution factor. Results are expressed as plaque‐forming units (pfu)/ml.
Alternate Protocol 1. Endpoint Dilution Assay to Determine Viral Infectivity
Endpoint dilution assays were used to measure viral infectivity prior to the development of the plaque assay, and it is still used for viruses that do not form plaques. It can be miniaturized and run in Terasaki plates that contain as little as 10 µl of medium per well, but is more commonly performed in 96‐well plates (Robb and Bond, 1979). Serial dilutions of a virus stock are prepared and inoculated onto replicate cell cultures, often in multiwell format (e.g., 96‐well plastic plates). After an appropriate incubation period, wells are scored as either infected or noninfected after microscopic observation for cytopathogenic effect, and a tissue culture infectious dose 50 (TCID50) is calculated. In this assay, the distribution of virus into the wells follows a Poisson distribution, since some wells have a probability of receiving more than one infectious virus particle. The relationship between pfu and TCID50 is as follows: TCID50 × 0.7 gives you the equivalent titer in pfu/ml. An advantage of endpoint dilution assay is that it can be easily adapted to viruses that do not produce clear cytopathogenic effects by utilizing immunofluorescent staining for viral antigens, or by using other indicators of viral infection such as hemadsorption. The procedure presented below utilizes L2 cells. If desired, DBT or 17Cl‐1 cells can be substituted for L2 cells.
Additional Materials (also see Basic Protocol 1)
96‐well tissue culture plates
Inverted phase‐contrast microscope
-
1
In the afternoon of the day prior to performing the assay, seed L2 cells into 96‐well plates at 50,000 cells in a volume of 0.1 ml DME10 per well. Incubate overnight at 37°C in 3% CO2.
-
2
On the morning of the assay, rapidly thaw the samples to be titered in a 37°C water bath and place on ice immediately. Make serial 10‐fold dilutions of the virus in 1.5 ml DME2, keeping the virus dilutions in an ice water bath to keep them cold.
-
3
Using a sterile Pasteur pipet attached to a vacuum aspirator, carefully aspirate the medium from no more than four rows of a 96‐well plate, taking care not to damage the monolayer with the pipet tip. Leave a small volume of medium behind in each well to prevent drying. Replace the medium with 0.2 ml DME0 per well (DME2 or PBS can alternatively be used).
-
4
One row of wells at a time, carefully aspirate the medium, again leaving a small volume of medium behind. Replace the medium with 0.1 ml diluted virus from step 2, devoting one row of 12 wells to one dilution of a virus sample, working from the highest dilution to the lowest dilution for each sample. For each plate, inoculate one row of the plate with DME2 alone to serve as uninfected controls.
-
5
Incubate for 2 days at 37°C in 3% CO2, and score for cytopathogenic effect (CPE) with an inverted phase‐contrast microscope.
For most strains of MHV, CPE means the presence of syncytial giant cells. Strains that do not form syncytia such as MHV‐2 or some mutants of other strains still produce cytopathology, rounding of the cells with detachment from the monolayer.
-
6
Calculate the titer using the method of Reed and Muench (Reed and Muench, 1938).
We give an example of such a calculation below that we have adapted from their work (Table 2). In this example of an endpoint dilution assay, ten wells were infected with each virus dilution. At high dilutions, none of the cell cultures are infected because no infectious particles are present. At low dilutions, every cell culture is infected. We have 7 wells with CPE at 10−4 dilution and 4 wells at 10−5 dilution. The endpoint for 50% of cell infection evidently lies between 10−4 and 10−5 dilution, but nearer the latter. It is assumed that, since the CPE at 10−4 is 30% [(7−4)/10] above that at 10−5 dilution, while the 50% point is 10% [(5−4)/10] above, the endpoint is 10%/30% or one‐third of the distance from 10−5 to 10−4. The formula for the proportionate distance of the endpoint above the dilution giving next below 50% CPE is:
Proportional distance = (50% CPE counts – CPE counts at next lower dilution)/(CPE counts at next higher dilution – CPE counts at next dilution below) = (5 – 4)/(7 – 4)=1/3.
Since dilutions are increasing on a logarithmic scale, it is necessary to obtain the final reading as follows:
Logarithm of 4 (lower dilution) = 0.6020
(1 – Proportional distance) × log 2 (dilution factor) = 0.2107
Sum (log of endpoint) = 0.8127
Therefore, the calculated 50% endpoint dilution is 6.50 × 10−4.
Table 2.
A Hypothetical Data Table for an Endpoint Dilution Assay
|
Virus dilution |
Cytopathic effect |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
10−2 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
10−3 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
10−4 |
+ |
+ |
− |
+ |
+ |
− |
+ |
+ |
+ |
− |
|
10−5 |
− |
+ |
− |
− |
+ |
+ |
− |
− |
+ |
− |
|
10−6 |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
− |
|
10−7 |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Support Protocol 1. Preparation of Monolayer Cultures of DBT, 17CL‐1, L2, BHK‐R, and FCWF Cells
Mouse hepatitis virus (MHV) can grow in many murine cell lines such as DBT, L2, and 17CL‐1 cells. However, MHV can infect different cell types that originated from other mammalian species that have been transformed to express the receptor for MHV (Dveksler et al., 1991). One of the most widely used cells is the BHK‐R cell line that is employed in many reverse genetic systems because of its high efficiency of electroporation. In addition, targeted recombination studies with MHV utilize feline cells (see Basic Protocol 5). Felis catus whole fetus (FCWF) cells are used for this purpose. 17Cl‐1 or DBT cells are generally used for growing MHV. 17CL‐1 generally grow the virus to higher titers, although there are occasional failures, particularly with viruses that are not robust growers. DBT cells allow for greater fusion and more rapid spread of the viral infection, and it may be better for viruses that do not grow well. L2 cells are the preferred cell line for plaque assays.
Materials
Felis catus whole fetus (FCWF) cells growing in culture (ATCC, cat. no. CRL‐2787)
L2 cells (not commercially available; cells can be obtained from most investigators working with MHV, including the authors)
DBT cells (not commercially available; cells can be obtained from most investigators working with MHV, including the authors)
17CL‐1 cells (not commercially available; cells can be obtained from most investigators working with MHV, including the authors)
BHK‐R cells (not commercially available; cells can be obtained from Dr. Kathryn Holmes, University of Colorado Medical School; kathryn.holmes@ucdenver.edu)
DME0 (see recipe)
DME10 (see recipe)
DME10 (see recipe) supplemented with 800 µg/ml G418 (for BHK‐R cells)
DME10/FBS (for FCWF cells; see recipe for DME10 but use FBS where calf serum is called for)
Trypsin/EDTA solution (see recipe)
Inverted tissue culture microscope
25‐ and 75‐cm2 tissue culture flasks with filter caps
15‐ and 50‐ml conical polypropylene tubes with screw cap
Additional reagents and equipment for counting cells using a hemacytometer (appendix mca04a)
NOTE: All culture incubations should be performed in a humidified 37°C, 5% CO2 incubator unless otherwise specified. L2 cells should be incubated in 3% CO2.
NOTE: All cell cultures are cultured in DME10 except for FCWF cells, which are cultured in DME10 prepared with 10% fetal bovine serum in place of the usual calf serum.
Prepare monolayer cultures
The following steps are based on the treatment of a single 75‐cm2 monolayer tissue culture flask. Cells regain subconfluency (75%), typically after 3 to 5 days of incubation, depending upon the cell type (DBT cells grow faster than L2 cells). The cells are routinely seeded at 5 × 105 to 1 × 106 cells per 75‐cm2 flask.
-
1
Remove the spent medium from the confluent tissue culture flask.
-
2
Wash the cells with 5 ml of DME0.
-
3
Add 5 ml trypsin/EDTA solution to the flask, then rock the flask back and forth two to three times to completely cover the monolayer with trypsin/EDTA solution. Aspirate 4 ml of the trypsin/EDTA.
-
4
Incubate the cells at room temperature for 2 to 10 min, periodically checking for loosening of cells from the plastic substrate by microscopic observation. When the cells are beginning to come off of the plastic, vigorously rap the bottom of the flask to accelerate the detachment of the cells from the plastic surface.
-
5
Add 9 ml of DME10 to the trypsinized flask to neutralize the trypsin, and rock the plate to wash cells off the bottom of the flask. Pipet the cell suspension up and down five times to break up cell clumps.
-
6
Remove 0.5 ml of the cell suspension to a tube. Count the cells using a hemacytometer to determine their concentration (cells/ml). Multiply the concentration by 10 to give the total amount of cells in the original flask.
-
7
Calculate the required volume of cell suspension needed to keep the cell inoculum at 5 × 105 to 1 × 106 cells per 75‐cm2 flask.
-
8
Add the calculated amount of cell suspension to a new 50‐ml conical polypropylene tube and add DME10 to 15 ml. Mix the cell suspension by pipetting up and down five times with a 10‐ml pipet and transfer the cell suspension to a new 75‐cm2 flask.
-
9
Carefully place the flask in the incubator and incubate until reaching the desired degree of confluence.
Support Protocol 2. Freezing of DBT, 17CL‐1, L2, BHK‐R, and FCWF Cells
Cell cultures can be stored frozen under liquid nitrogen when not in use. Although, cell viability is never 100% when the cells are defrosted, cell cultures can be recovered more successfully by gradual thawing, grown, and refrozen again to ensure a continuous source of cells for virus growth.
Materials
75‐cm2 flask with 80% to 90% confluent cells (Support Protocol 1)
DME0 (see recipe)
10% DMSO/20% serum
Isopropanol
Liquid N2
Tabletop centrifuge (e.g., IEC Clinical)
Cryogenic tubes
Cryo 1°C “Mr. Frosty” Freezing Container (Nalgene)
Liquid N2 freezer
Additional reagents and equipment for culturing and trypsinizing cells (Support Protocol 1)
-
1
Aspirate the medium from a 75‐cm2 flask with 80% to 90% confluent cells.
-
2
Wash the cells with 5 ml DME0.
-
3
Trypsinize the cells with 5 ml of trypsin/EDTA solution as described in Support Protocol 1.
-
4
When the cells are detached from the flask surface, stop the trypsinization reaction by adding 9 ml of DME10.
-
5
Centrifuge the trypsinized cells for 10 min at 2000 rpm at room temperature in a tabletop centrifuge and carefully aspirate the supernatant medium, being careful not to disturb the cell pellet.
-
6
Resuspend cells at a concentration of 3–5 × 106 cells per 1 ml of prechilled (on ice) 10% DMSO/20% serum. Keep on ice.
We freeze cells at this high concentration to take into account that recovery upon thawing is less than 100% and to decrease the amount of time after thawing for the cells to grow to confluence.
-
7
Aliquot cells into cryogenic tubes and place into a Cryo 1°C “Mr. Frosty” Freezing Container filled with isopropanol according to the manufacturer's directions. Place the freezer container on the top shelf of a −80°C freezer.
-
8
After 24 hr, transfer the vials to a liquid nitrogen freezer for long‐term storage.
Support Protocol 3. Thawing Frozen DBT, 17CL‐1, L2, BHK‐R, and FCWF Cells
When thawing, it is critical to add the medium dropwise, gradually and slowly to ensure sufficient time for the cells to recover from the DMSO/serum medium.
NOTE: Cells that are not recovered will eventually lyse and might be seen as debris in the first passage culture.
Materials
Vial of frozen cells (Support Protocol 2)
DME10 (see recipe)
15‐ml conical centrifuge tube (e.g., BD Falcon)
Tabletop centrifuge (e.g., IEC Clinical)
2‐ml disposable pipets
75‐cm2 tissue culture flasks
-
1
Remove the appropriate vial with the frozen cells carefully from the liquid nitrogen container and thaw cells by placing tube in a 37°C water bath until cells just reach a liquid state.
-
2
Remove the frozen cells to a prechilled (on ice) 15‐ml conical centrifuge tube. Add two drops of fresh ice‐cold DME10 medium to the vial, swirl gently, and return to the ice bucket.
-
3
After 20 sec add 4 drops of fresh ice cold DME10, swirl, and return to the ice bucket.
-
4
After additional 20 sec add 0.5 ml fresh ice‐cold DME10, swirl and return to the ice bucket.
-
5
Repeat step 4, doubling the volume of DME10 added, until you reach a total volume of at least 10 ml. Do not forget to swirl and incubate on ice for 20 sec between each addition.
-
6
Centrifuge the thawed cell suspension 5 min at 1000 rpm at room temperature in a tabletop centrifuge, and carefully aspirate the supernatant without disturbing the cell pellet.
-
7
Immerse the cells pellet in 1 ml of fresh DME10; mix the cell suspension by pipetting up and down with a 2‐ml pipet.
-
8
Prepare a 75‐cm2 flask with 9 ml of DME10 and add the 1 ml of cell suspension into the flask. Distribute the cells evenly by rocking the flask back and forth several times.
-
9
Incubate at 37°C until the cells become confluent, replacing the medium if necessary.
Basic Protocol 2. Growth and Characterization of MHV Stock Virus
The preparation of stock virus is an essential procedure for any virology lab. For most experiments, it is advantageous to start with genetically homogeneous stocks. Thus, we perform at least one and often two cycles of plaque cloning of any virus that we receive. We also try to maintain low‐passage‐number stocks in order to avoid selecting mutants that are better adapted to grow in cell culture than the original virus. Stocks are always grown at low multiplicity of infection, both to conserve seed stocks and to avoid the generation of defective interfering particles that can result in lower titers of the stock. For a robust virus such as MHV‐A59, after plaque purification, we generally grow a small amount of P1 (first passage) seed stock and a P2 (second passage) working stock in larger amounts. The P1 stock is subsequently used as to inoculate cultures for P2 stock preparation. P2 stock is used as the working virus stock.
NOTE: All culture incubations should be performed in a humidified 37°C, 5% CO2 incubator unless otherwise specified. L2 cells should be incubated at 3% CO2.
NOTE: All cell cultures are propagated in DME10.
NOTE: Each strain of MHV grows to a different titer. For example MHV‐JHM grows to a titer of ∼106 to 107 pfu/ml; MHV‐3 reaches a titer of ∼4 × 107 pfu/ml; MHV‐1 will grow to ∼2 × 107 pfu/ml; and MHV‐A59 will grow to ∼108 to 109 pfu/ml.
NOTE: MHV titers are generally stable through at least three freeze‐thaw cycles before there is a significant loss in titer. We generally aliquot viral stocks and try to minimize freeze‐thaw cycles.
Materials
Virus‐infected L2 cells (Basic Protocol 1)
DME2 (see recipe)
DME10 (see recipe)
DME0 (see recipe)
DBT cells (not commercially available; cells can be obtained from most investigators working with MHV, including the authors)
Sterile 5‐ml snap‐cap tubes
25‐, 75‐, and 175‐cm2 tissue culture flasks with filter caps
15‐ml and 50‐ml conical polypropylene centrifuge tubes (e.g., BD Falcon)
Cup sonicator
Tabletop centrifuge, 4°C
Additional reagents and equipment for plaque assay (Basic Protocol 1)
Plaque purify virus
-
1
Perform a plaque assay is on L2 cells as described in Basic Protocol 1, following steps 1 to 7.
-
2
On the second day of incubation, pull the plates from the incubator and hold up to the light to visualize the plaques. If plaques are ill‐defined or too small to easily visualize, stain with neutral red as described in the annotation to step 10 of Basic Protocol 1. Once plaques are clearly visible, pick wells that are at the terminal dilutions, and mark the position of well separated plaques on the bottom of the wells with a Sharpie marker.
-
3
Add 1 ml of DME2 to each of five sterile 5‐ml snap‐cap tubes. Place on ice.
-
4
Use a separate sterile plugged Pasteur pipet to pick each plaque into a 5‐ml tube.
Plaques are picked by gently pushing the tip of the Pasteur pipet through the agarose overlay toward the mark at the bottom of the plate that indicates the position of the plaque. The pipet tip is rotated slightly while applying gentle suction with an automatic pipet gun to pull up the overlying agarose and any cells and debris within the plaque. The contents aspirated into the Pasteur pipet are then expelled into a 5‐ml snap‐cap tube. The medium in the tube is pipetted up and down a few times to remove any material that might have adhered to the pipet. We generally pick at least four plaques. Plaques picked in this manner can be stored at −80°C. A second cycle of plaque purification can then be performed on this plaque, if desired.
Grow P1 seed stock
It may be advantageous to grow several different P1 seed stocks from different plaques, especially if these plaques appeared to be heterogeneous in size during plaque purification. These P1 stocks can then be characterized as to plaque size, titer achieved, and other biologic properties prior to growing a larger P2 working stock. Note that the procedure given below inoculates individual plaques into a 25‐cm2 flask. It is possible to grow P1 stocks of many of the strains of MHV that grow to high titers in 75‐cm2 flasks and produce a somewhat larger and higher‐titer P1 stock. However, this is usually not necessary.
-
5
Seed 2 × 106 DBT cells into 25‐cm2 tissue culture flask and incubate in 5 ml of DME10 medium for 24 hr.
-
6
Aspirate the medium from the flask and wash with 1 ml of DME0.
-
7
Inoculate 0.5 ml of virus plaque suspension (from step 4) into the flask. Rock the flask from side to side three times to distribute the virus over the cells. Continue to rock the flasks on a rocker for 60 min at room temperature.
-
8
Add 4.5 ml of DME2 and incubate at 37°C. Observe the cell culture daily for the development of cytopathogenic effect (CPE).
Typically, after 36 to 72 hr of incubation and depending upon the amount of virus in the original plaque and the strain of MHV, the CPE may be expected to reach 95% or more of the cells, and ∼25% of the cells start to detach from the monolayer.
-
9
Freeze the flask at −80°C for at least 1 hr.
-
10
Thaw the cells at 37°C in a water bath, taking care to remove them from the water bath before they are completely thawed. Complete the thawing at room temperature.
-
11
Transfer the cell suspension into 15‐ml polypropylene tube and sonicate on ice in a cup sonicator at 100 W peak envelope power, three bursts of 20 sec, allowing the samples to rest on ice for 20 sec between each burst.
-
12
Clarify the lysate by centrifuging 10 min at 3000 rpm, 4°C. Pour off the supernatant into a fresh tube.
-
13
Aliquot the virus stock into eight 0.5‐ml portions, and several smaller aliquots, and freeze at −80°C.
-
14
Take one of the smaller aliquots and determine the titer of the P1 stock by plaque assay (Basic Protocol 1).
Grow P2 working stock
-
15
Decide how large a stock you would like to grow, seed sufficient 175‐cm2 flasks with 1.5 × 107 DBT cells each in 25 ml of DME10, and incubate for 24 hr at 37°C.
We generally grow stocks that are 100 ml in volume or greater.
-
16
Calculate the amount of virus that you intend to inoculate.
To prepare working stocks, we generally infect the cells at multiplicities of infection (MOI) between 0.1 and 0.001 pfu/cell. Viruses that do not grow well should be grown towards the higher end of that range.
-
17
On the next day, when the cells reach 75% confluence (∼3 × 107 cells), aspirate the growth medium and wash the cells with DME0 to remove any residual medium.
-
18
Infect the cells with the desired number of pfu of P1 virus diluted in 5 ml of DME2 for each flask. Rock the flask from side to side three times to distribute the virus over the cells. Continue to rock the flasks on a rocker for 60 min at room temperature.
For example, to infect at a MOI of 0.001 you will need 3 × 104 pfu per flask. For a P1 virus that has achieved a titer of 1 × 10,7 this equates to 3 µl of P1 virus in a volume of 5 ml DME2 for each 175‐cm2 flask.
-
19
Feed the cells with 20 ml DME2 and incubate at 37°C. Observe the cell culture daily and follow the development of CPE.
Typically, after 36 to 72 hr of incubation, depending upon the MOI and the strain of MHV, 95% or more of the cells are involved in syncytia and ∼25% of the cells have detached from the monolayer.
-
20
Freeze the flask at −80°C for at least 1 hr.
-
21
Thaw the cells at 37°C in a water bath taking care to remove them from the water bath before they are completely thawed. Complete the thawing at room temperature.
-
22
Transfer the cell suspension from each flask into a 50‐ml polypropylene tube and sonicate on ice in a cup sonicator at 100 W peak envelope power, three bursts of 20 sec, allowing the samples to rest on ice for 20 sec between each burst.
-
23
Clarify the lysate by centrifuging 10 min at 3000 rpm, 4°C. Pool the supernatants.
-
24
Aliquot the virus stock.
Depending upon the size of the stock and the anticipated titer, we generally make several different sized aliquots, ranging from 50 ml down to 0.5 ml portions, and freeze at −80°C. A small number of larger aliquots are often more convenient for storage than a very large number of small aliquots. These large aliquots can later be thawed once and broken down into smaller aliquots for subsequent use.
-
25
Take one 0.5‐ml aliquot and determine the titer of the P2 stock by plaque assay (Basic Protocol 1).
Support Protocol 4. Characterization of MHV Stock Virus
Viral stocks can be characterized in many different ways, ranging from simple assays such as determination of the virus titer to more lengthy procedures such as determination of growth rate or determination of the complete nucleotide sequence of the viral RNA genome. Here we provide a procedure to determine plaque size and plaque morphology. This assay is most useful for characterizing mutants using the wild‐type strain as a reference and can be carried out using the same plates that were used to carry out a plaque assay to determine the titer of the stock.
Additional Materials (also see Basic Protocol 1)
Viral stocks to be titered, including a reference “wild‐type” stock
Two rulers: a flat 6‐in (∼15‐cm)' plastic ruler that has a centimeter/millimeter scale and a 12‐in. (∼30‐cm) ruler with a similar scale
Either an overhead projector or an imaging system allowing you to photograph the plates, saving the image to a tiff file, combined with a computer projection system allowing you to subsequently project the recorded images
-
1
Determine the titer of the stocks to be compared by performing a plaque assay as described in Basic Protocol 1, steps 1 to 9.
-
2
Let the plates dry overnight after staining.
-
3
Set up the overhead projector with the 6‐in. ruler next to the plate being measured and project on to a wall. Alternatively photograph the plates and the ruler with a computerized imaging system, capture the data as tiff files, and subsequently project the image with an LCD projector.
-
4
Determine the enlargement factor of the projected image of the 6‐in. ruler by measuring the size of the projected image of the millimeter/centimeter scale on the ruler.
-
5
Determine the diameter of 25 to 50 well isolated plaques by measuring the diameter of their projected images. Determine the mean value and using the enlargement factor calculated in step 4, converting back to the actual size of the plaques.
It is important to keep the same degree of enlargement for all of the stocks being compared. This means that it is possible to measure plaque diameter to an accuracy of greater than 0.1 mm.
-
6
Note the morphology of the plaques.
Most strains of MHV make clear plaques on L2 cells, but other coronaviruses or MHV mutants that do not produce as much cell fusion may make “turbid” or “cloudy” plaques.
Plaque diameter for a given virus can vary considerably from day to day when repeating this assay. Thus, meaningful comparisons are only achieved by comparing the diameter of a particular virus to a reference “wild‐type” strain.
Basic Protocol 3. Purification of MHV by Equilibrium Ultracentrifugation Through Sucrose Gradients
MHV is easily purified from infected cell supernatants by ultracentrifugation. To produce virus for purification, cells are normally infected at a relatively low multiplicity of infection, 0.1 to 0.001 pfu per cell (Basic Protocol 2). When CPE is apparent throughout the monolayer, the virus‐containing medium is removed to 50‐ml polypropylene conical centrifuge tubes and chilled on ice, and any detached cells or cellular debris are pelleted by low‐speed centrifugation at 5000 × g for 1 hr at 4°C. The clarified supernatant medium can then be frozen at −80°C for purification at a later time. The most commonly used medium for ultracentrifugation is sucrose, and a protocol for purifying MHV using two cycles of equilibrium ultracentrifugation through sucrose gradients is presented below. An alternative method that gives slightly cleaner virus uses velocity ultracentrifugation followed by an equilibrium ultracentrifugation step, and is described in Alternate Protocol 2. These protocols are easily adapted to the purification of other coronaviruses. For both protocols, virus is first concentrated by centrifugation through a sucrose or potassium tartrate pad prior to further purification by density‐gradient ultracentrifugation. Please note that all manipulations with virus should be done in a bio‐safety cabinet, and waste should be disposed of appropriately.
Materials
Virus‐infected cell supernatants (e.g., Basic Protocol 2)
Sucrose gradient solutions (see recipe): 20%, 30%, and 60% sucrose, chilled before use
MOPS‐saline‐EDTA (MSE) buffer (see recipe)
Disinfectant
-
Ultraclear ultracentrifuge tubes:
SW28 tubes, 3.5‐in. high × 1‐in. diameter (Beckman, part number 344058)
SW41 tubes, 3.5‐in. high × 9/16‐in. diameter (Beckman, part number 344059)
Precooled Beckman SW28 and SW41 rotors or equivalent. For radiolabeled samples when only relatively small volumes of virus will be purified, only an SW41 rotor is required.
Ultracentrifuge rated to accept the SW28 and SW41 rotors
Cup sonicator
Gradient maker, available from Hoefer Scientific and GE Healthcare (we have found the 30‐ml size, i.e., the Hoefer SG30, to be the most useful
No.‐00 rubber stopper pierced through the center with a 21‐G 1.5‐in. needle
Ring stand with small three‐finger clamp
22‐G needle and 3‐ml syringe
Refractometer
Metal probe (16‐ to 18‐G, 4‐ to 6‐in. length)
Thin Tygon tubing (∼0.045‐ to 0.065‐in. inside diameter; must fit snugly over the probe)
Peristaltic pump
-
1
Keep samples cold (on ice whenever possible) at all times. Pool the clarified infected cell culture fluids and measure the volume to be purified.
The SW28 rotor can spin a maximum of six buckets at one time, and each bucket can hold a maximum volume of 36 ml of virus‐containing culture fluids. Thus, the largest volume of culture fluids that can be concentrated in one spin is 216 ml.
-
2aIf a large prep is being purified:
- Pipet 1.5 ml of 30%(w/w) sucrose gradient solution into a sterile microcentrifuge tube for each ultracentrifuge tube that will be used to pellet the virus.
- Pipet 15 ml of the virus prep into each of the SW28 tubes. Place a Pasteur pipet into the SW28 tube holding it almost vertically, placing it down to the bottom of the tube.
-
Using a second Pasteur pipet, carefully pipet 1.5 ml of 30% sucrose gradient solution from the 1.5‐ml microcentrifuge tube into the first Pasteur pipet, running the sucrose slowly down the side, being careful not form an airlock that prevents the sucrose solution from flowing down through the vertical Pasteur pipet.If an airlock forms it can usually be disrupted by moving the Pasteur pipet up and down through the air bubble. The sucrose should flow through the Pasteur pipet to form a 1.5 ml pad at the bottom of the ultracentrifuge tube.
- Slowly remove the Pasteur pipet from the ultracentrifuge tube, being careful not to disturb the interface between the virus‐containing culture medium and the sucrose solution.
-
Slowly pipet 21 ml of virus‐containing culture medium into the ultracentrifuge tubes, carefully running the liquid down the side of the tube, disturbing the sucrose‐culture medium interface as little as possible.After all of the tubes to be centrifuged have been filled, they should be put into the SW28 buckets and weighed in a small beaker cushioned with a tissue, recording the weight of each. MSE can then be added to each tube to ensure that the weight of each bucket is equal, with a tolerance of 0.1 g or less. Failure to balance the rotor can result in catastrophic failure of the rotor.
- Put the caps on the buckets and tighten. Load the buckets into the ultracentrifuge rotor and place into the ultracentrifuge.
-
2bIf a small prep of radiolabeled virus is being purified:
- Pipet 0.5 ml of 30% sucrose gradient solution into a sterile microcentrifuge tube for each ultracentrifuge tube that will be used to pellet the virus.
- Pipet 5 ml of the virus prep into each of the SW41 tubes and underlay with 0.5 ml of 30% sucrose gradient solution using Pasteur pipets as described in step 2a.
-
Fill the tubes with 6 ml of additional virus‐containing culture fluids and balance as described in step 2a.Failure to balance the rotor can result in catastrophic failure of the rotor.
- Put the caps on the buckets and tighten. Load the buckets into the ultracentrifuge rotor and place into the ultracentrifuge.
-
3
For the SW28 rotor (large preps), pellet the virus by centrifuging for 2.5 hr at 112,500 × g (25,000 rpm in SW28 rotor), 4°C, to pellet the virus. If purifying a small prep in the SW41 rotor, centrifuge 1 hr at 210,000 × g (35,000 rpm in SW41 rotor), 4°C.
-
4
Resuspend pelleted virus for further purification:
- Pour off the supernatant liquids into a beaker containing disinfectant. Keep the ultracentrifuge tubes inverted and remove residual liquid by blotting the inside (top) of the ultracentrifuge tube with a cotton swab or by inverting on paper towels for about a minute (if using the SW28 rotor, a small pellet of virus is often visible).
-
Put the ultracentrifuge tubes on ice.The pelleted virus must be resuspended in a small volume of MSE buffer for further purification. This is most easily accomplished in a cup sonicator as described in the following.
- Pipet 0.5 ml MSE into the first ultracentrifuge tube and seal with Parafilm. Fill the cup of the cup sonicator with chipped ice plus sufficient water to make slush. Sonicate for three bursts of 20 sec each at 100 W peak envelope power, pausing for 1 min between bursts. Replenish ice as needed.
-
Transfer the resuspended virus to the next ultracentrifuge tube and sonicate as described above to resuspend the pelleted virus in the second tube.Virus from up to three tubes can be resuspended in 0.5 ml of MSE buffer by this method.
-
Use an additional 0.5 ml of MSE buffer to resuspend virus from the remaining three ultracentrifuge tubes if necessary.If a cup sonicator is not available, you can resuspend the virus pellets by adding 0.5 ml of MSE buffer to the virus pellet and letting the pellet soften for 5 to 10 min on ice. Break up the pellet by pipetting the MSE buffer up and down in a 100‐µl pipet tip; if necessary, you can use the pipet tip to break up the pellet at the bottom of the tube. Once the pellet is broken up, transfer the virus suspension to a second tube of pelleted virus, and repeat the process. Virus from up to three tubes can be resuspended in 0.5 ml of MSE buffer by this method. Keep the resuspended virus on ice until needed.
-
5
Further purify virus:
-
Prepare two 11‐ml 20% to 60% sucrose gradients in SW41 Ultraclear ultracentrifuge tubes using a gradient maker (see Fig. 1):These gradients may be prepared in advance while the virus is being pelleted in step 4.
- Carefully overlay the resuspended virus onto the top of the gradient. Overlay the second centrifuge tube with an equal volume of MSE to serve as a balance. Load the tubes into the SW41 buckets and weigh on a balance. If necessary, adjust the weight of the bucket containing the balance tube by adding or removing small amounts of MSE until they are equal in weight.
-
Centrifuge 4 hr at 210,000 × g (35,000 rpm in SW41 rotor), 4°C.Alternatively if it is more convenient, you can centrifuge at 112,500 × g (25,000 rpm in SW41 rotor), 4oC, overnight.
Figure 1.
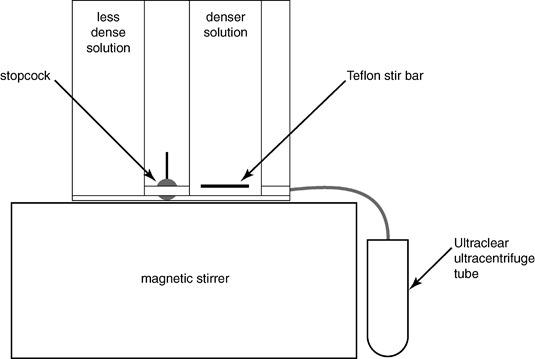 A schematic drawing of the preparation of density gradients for virus purification.
A schematic drawing of the preparation of density gradients for virus purification.
-
-
6aFor large preps of virus (generally from 100 ml or more of a high titered virus such as MHV‐A59):
-
Remove the tubes from the ultracentrifuge buckets.For these large preps, a visible band of virus can be seen at approximately the midpoint of the gradient when the tube is illuminated from above against a black background (see Fig. 2A).
-
To remove the virus from the ultracentrifuge tube, carefully remove about 3 ml of liquid from the top of the sucrose gradient without disturbing the purified virus band.You must remove a sufficient volume of liquid to allow a no.‐00 rubber stopper to be placed into the tube without disturbing the gradient.
-
Carefully and slowly pierce the tube just below the visible virus band by rotating a 22‐G needle attached to a syringe while applying steady pressure. Once the tube is penetrated, move the needle tip to just below the virus band and slowly aspirate the virus.You typically will collect approximately 1 ml of virus. Prior to withdrawing your needle from the tube, place a finger over the hub of the needle in the rubber stopper to keep the remaining sucrose from pouring out through the hole in the side of the tube.
- Withdraw the needle and syringe, transfer the virus to a sterile polypropylene tube, and place on ice. Place the pierced ultracentrifuge tube in a beaker to allow the liquid in the tube to drain. Sterilize liquid in beaker with disinfectant prior to discarding.
- Discard ultracentrifuge tube as biohazard waste and autoclave the rubber stopper pierced with a needle prior to storage for subsequent use.
-
Determine the refractive index of the virus‐containing sucrose solution in a refractometer.The buoyant density of the purified virus can then be looked up from the International Critical Tables and should be between 1.17‐1.19 g/cm3.
Figure 2.
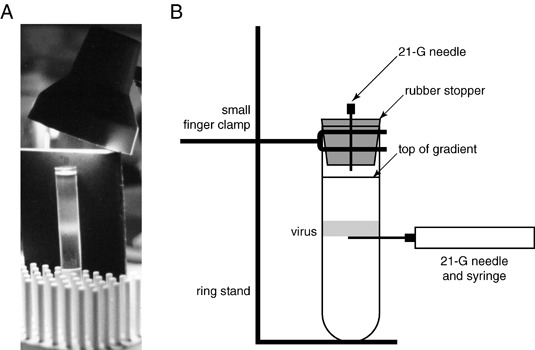 Purification of MHV. (A) A photograph of a potassium tartrate density gradient containing purified by MHV‐A59. (B) A schematic diagram of the setup for collecting density–gradient‐purified coronavirus.
Purification of MHV. (A) A photograph of a potassium tartrate density gradient containing purified by MHV‐A59. (B) A schematic diagram of the setup for collecting density–gradient‐purified coronavirus.
-
-
6bFor small radiolabeled virus preps:
-
Place a metal probe attached to thin Tygon tubing and attached to a peristaltic pump into the bottom of the ultracentrifuge tube.A visible virus band may not be present.
-
Pump the gradient out at a flow rate of ∼1 ml/min and collect 0.5‐ml fractions.Radiolabeled virus can then be detected by counting aliquots of each fraction.
- Pool the virus containing peak of radioactivity, generally around fractions 9 to 11, and determine the buoyant density of as described in step 6a.
-
To prepare highly purified virus
To prepare highly purified virus, a second cycle of equilibrium centrifugation is often necessary.
-
7
Dilute virus with sufficient volume of cold MSE buffer to bring the buoyant density below that of 20% sucrose, generally 2 to 2.5 ml. Prepare two 8‐ml 20% to 60% sucrose gradients in SW41 Ultraclear ultracentrifuge tubes using a gradient maker. Carefully overlay the diluted virus onto the sucrose gradient.
-
8
Centrifuge 4 hr at 210,000 × g (35,000 rpm in SW41 rotor), 4°C.
Alternatively if it is more convenient, you can centrifuge at 112,500 × g (25,000 rpm in SW41 rotor), 4oC, overnight.
-
9
Collect purified virus as described above in step 6. To further concentrate virus for downstream applications, dilute with 12 ml MSE and pellet virus by centrifugation for 1 hr at 210,000 × g (35,000 rpm in SW41 rotor), 4°C.
The pelleted virus can be resuspended in a small volume of buffer or medium as described in step 4, or dissolved in SDS sample buffer or guanidium‐HCl buffers for subsequent protein and RNA analyses, respectively.
Alternate Protocol 2. Purification of MHV by Velocity and Equilibrium Ultracentrifugation Through Potassium Tartrate Gradients
While the most commonly used medium for ultracentrifugation is sucrose (see Basic Protocol 3), the alternative method described below gives slightly cleaner virus and uses velocity ultracentrifugation followed by an equilibrium ultracentrifugation step. Potassium tartrate is less viscous than sucrose; thus, preparing the gradients is somewhat faster with this medium. Potassium tartrate does have the disadvantage that trace amounts of potassium carried over with purified virus can interfere with downstream analysis of purified virus, particularly if sodium dodecyl sulfate (SDS) is involved. Either Basic Protocol 3 or this protocol is easily adapted to the purification of other coronaviruses. For both protocols, virus is first concentrated by centrifugation through a sucrose or potassium tartrate pad prior to further purification by density gradient ultracentrifugation. Please note that all manipulations with virus should be done in a biosafety cabinet, and waste should be disposed of appropriately.
Additional Materials (also see Basic Protocol 3)
Potassium tartrate gradient solutions (see recipe): 5%, 10%, 15%, 25%, and 40% potassium tartrate
-
1
Keep samples cold (on ice whenever possible) at all times. Pool the clarified infected cell culture fluids and measure the volume to be purified.
The SW28 rotor can spin a maximum of six buckets at one time, and each bucket can hold a maximum volume of 36 ml of virus‐containing culture fluids. Thus, the largest volume of culture fluids that can be concentrated in one spin is 216 ml.
-
2aIf a large prep is being purified:
- Pipet 1.5 ml of 15% (w/w) potassium tartrate gradient solution into a sterile microcentrifuge tube for each ultracentrifuge tube that will be used to pellet the virus.
- Pipet 15 ml of the virus prep into each of the SW28 tubes. Place a Pasteur pipet into the SW28 tube holding it almost vertically, placing it down to the bottom of the tube.
-
Using a second Pasteur pipet, carefully pipet 1.5 ml of 15% (w/w) potassium tartrate gradient solution from the 1.5‐ml microcentrifuge tube into the first Pasteur pipet, running the potassium tartrate solution slowly down the side, being careful not form an airlock that prevents the solution from flowing down through the vertical Pasteur pipet.If an airlock forms it can usually be disrupted by moving the Pasteur pipet up and down through the air bubble. The potassium tartrate solution should flow through the Pasteur pipet to form a 1.5‐ml pad at the bottom of the ultracentrifuge tube.
- Slowly remove the Pasteur pipet from the ultracentrifuge tube, being careful not to disturb the interface between the virus‐containing culture medium and the potassium tartrate solution.
-
Slowly pipet 21 ml of virus‐containing culture medium into the ultracentrifuge tubes, slowly running the liquid down the side of the tube, disturbing the potassium tartrate solution–culture medium interface as little as possible.After all of the tubes to be centrifuges have been filled they should be put into the SW28 buckets and weighed in a small beaker cushioned with a tissue, recording the weight of each. MSE can then be added to each tube to ensure that the weight of each bucket is equal with a tolerance of 0.1 grams or less. Failure to balance the rotor can result in catastrophic failure of the rotor.
- Put the caps on the buckets and tighten. Load the buckets into the ultracentrifuge rotor and place into the ultracentrifuge.
-
2b
If a small prep of radiolabeled virus is being purified:
- Pipet 0.5 ml of 15% potassium tartrate gradient solution into a sterile microcentrifuge tube for each ultracentrifuge tube that will be used to pellet the virus.
- Pipet 5 ml of the virus prep into each of the SW41 tubes and underlay with 0.5 ml of 15% potassium tartrate gradient solution using Pasteur pipets as described in step 2a.
-
Fill the tubes with 6 ml of additional virus‐containing culture fluids and balance as described in step 2a.Failure to balance the rotor can result in catastrophic failure of the rotor.
- Put the caps on the buckets and tighten. Load the buckets into the ultracentrifuge rotor and place into the ultracentrifuge.
-
3
For the SW28 rotor (large preps), pellet the virus by centrifuging for 2.5 hr at 112,500 × g (25,000 rpm in SW28 rotor), 4°C, to pellet the virus. If purifying a small prep in the SW41 rotor, centrifuge 1 hr at 210,000 × g (35,000 rpm in SW41 rotor), 4°C.
-
4
Resuspend pelleted virus for further purification:
- Pour off the supernatant liquids into a beaker containing disinfectant. Keep the ultracentrifuge tubes inverted and remove residual liquid by blotting the inside (top) of the ultracentrifuge tube with a cotton swab or by inverting on paper towels for about a minute (if using the SW28 rotor, a small pellet of virus is often visible).
-
Put the ultracentrifuge tubes on ice.The pelleted virus must be resuspended in a small volume of MSE buffer for further purification. This is most easily accomplished in a cup sonicator as described in the following.
- Pipet 0.5 ml MSE into the first ultracentrifuge tube and seal with Parafilm. Fill the cup of the cup sonicator with chipped ice plus sufficient water to make slush. Sonicate for three bursts of 20 sec each at 100 W peak envelope power, pausing for 1 min between bursts. Replenish ice as needed.
-
Transfer the resuspended virus to the next ultracentrifuge tube and sonicate as described above to resuspend the pelleted virus in the second tube.Virus from up to three tubes can be resuspended in 0.5 ml of MSE buffer by this method.
-
Use an additional 0.5 ml of MSE buffer to resuspend virus from the remaining three ultracentrifuge tubes if necessary.If a cup sonicator is not available, you can resuspend the virus pellets by adding 0.5 ml of MSE buffer to the virus pellet and letting the pellet soften for 5 to 10 min on ice. Break up the pellet by pipetting the MSE buffer up and down in a 1000‐µl pipet tip; if necessary, you can use the pipet tip to break up the pellet at the bottom of the tube. Once the pellet is broken up, transfer the virus suspension to a second tube of pelleted virus, and repeat the process. Virus from up to three tubes can be resuspended in 0.5 ml of MSE buffer by this method. Keep the resuspended virus on ice until needed.
-
5
Further purify virus:
-
Prepare two 11 ml 5% to 25% potassium tartrate gradients in SW41 Ultraclear ultracentrifuge tubes using a gradient maker (see Fig. 1).These gradients may be prepared in advance while the virus is being pelleted in step 4.
- Carefully overlay the resuspended virus onto the top of the gradient. Overlay the second centrifuge tube with an equal volume of MSE to serve as a balance. Load the tubes into the SW41 buckets and weigh on a balance. If necessary, adjust the weight of the bucket containing the balance tube by adding or removing small amounts of MSE until they are equal in weight.
- Centrifuge 45 min 210,000 × g (35,000 rpm in SW41 rotor), 4°C.
-
-
6a
For large preps of virus (generally from 100 ml or more of a high titered virus such as MHV‐A59):
-
Remove the tubes from the ultracentrifuge buckets.For these large preps, a visible band of virus can be seen at approximately the midpoint of the gradient when the tube is illuminated from above against a black background (see Fig. 2A).
-
To remove the virus from the ultracentrifuge tube, carefully remove about 3 ml of liquid from the top of the potassium tartrate gradient without disturbing the purified virus band.You must remove a sufficient volume of liquid to allow a no.‐00 rubber stopper to be placed into the tube without disturbing the gradient.
-
Carefully and slowly pierce the tube just below the visible virus band by rotating a 22‐G needle attached to a syringe while applying steady pressure. Once the tube is penetrated, move the needle tip to just below the virus band and slowly aspirate the virus.You typically will collect approximately 1 ml of virus. Prior to withdrawing your needle from the tube, place a finger over the hub of the needle in the rubber stopper to keep the remaining potassium tartrate gradient solution from pouring out through the hole in the side of the tube.
- Withdraw the needle and syringe, transfer the virus to a sterile polypropylene tube, and place on ice. Place the pierced ultracentrifuge tube in a beaker to allow the liquid in the tube to drain. Sterilize liquid in beaker with disinfectant prior to discarding.
- Discard ultracentrifuge tube as biohazard waste and autoclave the rubber stopper pierced with a needle prior to storage for subsequent use.
-
-
6b
For small radiolabeled virus preps:
-
Place a metal probe attached to thin Tygon tubing and attached to a peristaltic pump into the bottom of the ultracentrifuge tube.A visible virus band may not be present.
-
Pump the gradient out at a flow rate of ∼1 ml/min and collect 0.5‐ml fractions.Radiolabeled virus can then be detected by counting aliquots of each fraction.
- Pool the virus‐containing peak of radioactivity, generally around fractions 9 to 11.
-
To prepare highly purified virus
To prepare highly purified virus, a second cycle of equilibrium centrifugation is often necessary.
-
7
Dilute virus with at least an equal volume of cold MSE buffer to bring the buoyant density below that of 10% (w/w) potassium tartrate and the final volume of diluted virus to 3 to 3.5 ml. Prepare two 8‐ml 10% to 40% potassium tartrate gradients in SW41 Ultraclear ultracentrifuge tubes using a gradient maker. Carefully overlay the diluted virus onto the potassium tartrate gradient.
-
8
Centrifuge 4 hr at 210,000 × g (35,000 rpm in SW41 rotor), 4°C.
Alternatively if it is more convenient, you can centrifuge at 112,500 × g (25,000 rpm in SW41 rotor), 4oC, overnight.
-
9
Collect purified virus as described above in step 6. Determine the refractive index of the virus‐containing potassium tartrate solution in a refractometer.
The buoyant density of the purified virus can then be looked up in the table provided (Table 3) and should be between 1.17 and 1.19 g/cm3.
Table 3.
Relationships of Refractive Index, Percent Potassium Tartrate (w/w), and Buoyant Densitya
|
Potassium tartrate concentration (w/w) |
Refractive index |
Buoyant density |
Potassium tartrate concentration (w/w) |
Refractive index |
Buoyant density |
|---|---|---|---|---|---|
|
1.340 |
1.004 |
1.366 |
1.152 |
||
|
1.341 |
1.010 |
1.367 |
1.158 |
||
|
1.342 |
1.015 |
1.368 |
1.164 |
||
|
1.343 |
1.021 |
1.369 |
1.169 |
||
|
1.344 |
1.027 |
1.370 |
1.175 |
||
|
1.345 |
1.033 |
1.371 |
1.180 |
||
|
5% |
1.346 |
1.038 |
25% |
1.372 |
1.186 |
|
1.347 |
1.044 |
1.373 |
1.192 |
||
|
1.348 |
1.050 |
1.374 |
1.198 |
||
|
7.5% |
1.349 |
1.055 |
1.375 |
1.204 |
|
|
1.350 |
1.061 |
1.376 |
1.209 |
||
|
1.351 |
1.067 |
1.377 |
1.215 |
||
|
10% |
1.352 |
1.072 |
30% |
1.378 |
1.220 |
|
1.353 |
1.078 |
1.379 |
1.226 |
||
|
1.354 |
1.084 |
1.380 |
1.232 |
||
|
1.355 |
1.090 |
1.381 |
1.238 |
||
|
1.356 |
1.095 |
1.382 |
1.243 |
||
|
1.357 |
1.101 |
1.383 |
1.249 |
||
|
1.358 |
1.107 |
35% |
1.384 |
1.255 |
|
|
15% |
1.359 |
1.112 |
1.385 |
1.261 |
|
|
1.360 |
1.118 |
1.386 |
1.266 |
||
|
1.361 |
1.124 |
1.387 |
1.272 |
||
|
1.362 |
1.129 |
1.388 |
1.278 |
||
|
1.363 |
1.135 |
1.389 |
1.283 |
||
|
1.364 |
1.141 |
1.390 |
1.289 |
||
|
20% |
1.365 |
1.147 |
40% |
1.391 |
1.295 |
Table is calculated by the following formula: density (at 25°C) = aN D (at 25°C) – b. For potassium tartrate, a = 5.700 and b = 6.634.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
-
10
To further concentrate virus for downstream applications, dilute with 12 ml MSE and pellet virus by centrifugation for 1 hr at 210,000 × g (35,000 rpm in SW41 rotor), 4°C.
The pelleted virus can then be resuspended in a small volume of buffer or medium as described in step 4, or dissolved in guanidium‐HCl buffers for subsequent RNA analysis. Potassium dodecyl sulfate is poorly soluble in aqueous buffers.
-
11
If it is necessary to analyze purified virus by SDS‐PAGE, gently rinse the pellet once with 0.5 ml of MSE buffer, pour off the wash solution, and, keeping the centrifuge tube inverted, blot any residual MSE buffer that adheres to centrifuge tube.
The pellet can then be solubilized in SDS‐PAGE sample buffer.
Basic Protocol 4. Generation of Recombinant MHV from cDNA
Coronaviruses have the largest genome known for an RNA virus. Direct genetic modification of an RNA‐virus genome is not feasible. In the past decade, coronavirus reverse genetics was performed using defective interference (DI) genomes, extensively deleted genomic remnants that replicate by using the RNA synthesis machinery of a helper virus. Using the DI system, a number of cis‐acting elements important for viral transcription and replication have been defined. However, by the nature of the competition assay in the DI system, phenotype changes observed in the DI system do not strictly reflect the behavior of the virus when the mutations were introduced into the whole virus genome (Johnson et al., 2005; Dong et al., manuscript in preparation). Recently, reverse genetic systems for a number of coronaviruses have been established using nontraditional approaches based on bacterial artificial chromosomes (Almazan et al., 2000), the use of vaccinia virus as a vector for the propagation of coronavirus genomic cDNAs (Thiel and Siddell, 2005), or the in vitro ligation of coronavirus cDNA fragments (Yount et al., 2000, 2002, 2003; Donaldson et al., 2008). These reverse genetic systems enable us to genetically modify the coronavirus genome at any position and provide a powerful tool to investigate viral transcription, replication, and virus‐host interactions in the whole genome system. Here, we introduce in vitro cDNA assembly protocol developed by Yount et al. (2002, 2003).
The seven plasmids, named A to G, contain cDNAs that represent the entire MHV‐A59 genome. These plasmids are propagated in E. coli (see Support Protocol 5), then plasmid DNA is extracted and restriction enzyme digested to generate the seven cDNA fragments. These fragments are sequentially ligated to generate a full‐length cDNA copy of the MHV genome under the control of a T7 promoter. In vitro transcription with T7 RNA polymerase is used to generate full‐length genomic RNA, which is electroporated into BHK‐R cells to regenerate infectious virus that can then be characterized further. This procedure has been successfully adapted to many other coronaviruses including TGEV (Yount, Curtis, and Baric, 2000), SARS‐CoV (Yount et al., 2003), and IBV (Youn et al., 2005).
NOTE: The nomenclature for the different cDNA fragments and the restriction enzymes used will vary from virus to virus.
Materials
Plasmids A to G containing cDNAs that represent the entire MHV‐A59 genome (Support Protocol 5)
10 U/µl restriction enzymes AhdI, BglI, BsmBI, MluI, and SfiI (New England Biolabs)
10× NEBuffer 2 and 3 (New England Biolabs)
100× (10 mg/ml) bovine serum albumin (BSA; New England Biolabs, cat. no. B9001S)
0.8% agarose gel (Voytas, 2000)
10 U/µl calf intestinal alkaline phosphatase (CIAP; New England Biolabs) or 1 U/µl shrimp alkaline phosphatase
500 mM EDTA (see recipe)
3 M sodium acetate, pH 5.2 (appendix mca02a)
Chloroform
Isopropanol
70% ethanol
95% ethanol
Nuclease‐free water (e.g., DEPC‐treated; appendix mca02a)
3 U/µl T4 DNA ligase and 10× ligation buffer (Promega, cat. no. M180B)
QiaQuick gel extraction kit (Qiagen, cat. no. 28704)
Lambda HindIII markers (Invitrogen, cat. no. 15612‐013)
10 µM ATP
100 mM dithiothreitol (DTT)
-
Primers for N gene:
A59SP6 Ng(+):
5′TCGGCCTCGATGGCCATTTAGGTGACACTATAGATGTCTTTTGTTCCTGGGCAAG3′
A59Ng3′ (‐): 5′TCCGGA(TTT)8TTACACATTAGAGTCATCTTCTAACC3′
100% ethanol
BHK‐R cells (not commercially available; cells can be obtained from Dr. Kathryn Holmes, University of Colorado Medical School; kathryn.holmes@ucdenver.edu)
DBT cells (not commercially available; cells can be obtained from most investigators working with MHV, including the authors)
Phosphate‐buffered saline (PBS; appendix mca02a)
RNAeasy kit (Qiagen)
16°, 50°, 55°, and 65°C water baths
SpeedVac evaporator (Savant Instruments; or equivalent centrifugal vacuum evaporator)
Dark Reader light box (Clare Chemical Research, http://www.clarechemical.com/)
Orange viewing glasses for Dark Reader (Clare Chemical Research, http://www.clarechemical.com/)
Razor blade
NanoDrop spectrophotometer (http://www.nanodrop.com)
Ambion mMESSAGE mMACHINE High Yield Capped DNA Kit [Applied Biosystems; use T7 kit (cat. no. AM1344) for transcription of the ligated full‐length cDNA and the SP6 kit (cat. no. AM1340) for transcription of the N gene RNA]
25‐, 75‐ and 175‐cm2 tissue culture flasks
Electroporator with 4‐mm‐gap cuvettes
50‐ and 15‐ml conical polypropylene centrifuge tubes (e.g., BD Falcon)
Cup sonicator
Tabletop centrifuge (e.g., IEC Clinical), 4°C
Additional reagents and equipment for agarose gel electrophoresis (Voytas, 2000), the polymerase chain reaction (PCR; Kramer and Coen, 2001), culturing/passaging cell lines (Support Protocol 1), and plaque purification of viruses (Basic Protocol 2)
Prepare plasmids
Plasmids A though G are restriction digested sequentially, and fragments are gel extracted. Purified cDNA fragments can be stored at −70°C for up to 4 months. Fragments A and G need to be treated with calf intestinal alkaline phosphatase (CIAP) between the first and second digestion steps. As a general rule, these two plasmids are done in parallel and before the other plasmids. All digestions will be monitored by removing an aliquot and analyzing the progress of the digestion by agarose gel electrophoresis to ensure that the digestion is complete prior to running a preparative gel.
Plasmids A and G
A minimum of 50 µg of DNA is required. We generally start with 100 µg DNA. This should yield sufficient fragments A or G for 10 ligations.
-
1
Set up digestion in 100 to 500 µl final volume, depending upon DNA concentration.
- For plasmid A, add 100 µg plasmid DNA, 100 U MluI, and 1/10 of final volume of 10× NEBuffer 3, and incubate at 37°C overnight.
- For plasmid G, add 100 µg DNA, 100 U SfiI, 1/10 of final volume of 10× NEBuffer 2, and 1/100 of final volume of 100× BSA, and incubate at 50°C overnight.
-
2
Remove 1 µl and analyze by electrophoresis in a 0.8% agarose gel (Voytas, 2000).
If digestion is complete (all plasmids should be converted to a single fragment of ∼8.4 kpb for A, or 13 kpb for G; Table 4), proceed to step 5.
Table 4.
Seven Plasmids for MHV‐A59‐1000, Their Backbones, Antibiotic Resistance, and Restriction Enzymes Used
|
Plasmid name |
Vector backbone |
Antibiotic resistance |
Vector fragment (kpb) |
Insert size (kpb) |
RE to excise fragment |
|---|---|---|---|---|---|
|
A |
PCR‐XL‐TOPO (Invitrogen) |
Kanamycin 30 µg/ml |
3.5 |
4.9 |
MluI × BsmBI |
|
B |
pSMART‐LC (Lucigen) |
Ampicillin 50 µg/ml |
1.2 + 0.8 |
4.7 |
BglI × BsmBI |
|
C |
pSMART‐LC (Lucigen) |
Ampicillin 50 µg/ml |
1.2 + 1.5 |
2.0 |
BglI × BsmBI |
|
D |
pSMART‐LC (Lucigen) |
Ampicillin 50 µg/ml |
1.0 + 1.0 |
1.45 |
AhdI × BsmBI |
|
E |
pSMART‐LC (Lucigen) |
Ampicillin 50 µg/ml |
1.2 + 0.8 |
2.8 |
BglI × BsmBI |
|
F |
PCR‐XL‐TOPO (Invitrogen) |
Kanamycin 30 µg/ml |
3.5 |
7.0 |
BsmBI |
|
G |
pBR322 |
Ampicillin 50 µg/ml |
2.2 + 2.1 |
8.7 |
SfiI × BsmBI |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
-
3
If digestion is not complete, redigest with 10 µl of fresh 10 U/µl restriction enzyme and monitor digestion by gel electrophoresis as in step 3.
-
4
Perform alkaline phosphatase digestion.
This step is crucial to eliminate self‐ligation of the A and G fragments at the MluI and SfiI sites during subsequent ligation steps.
- Add 1 µl of 10 U/µl CIAP or 10 µl of 1 U/µl SAP to the digestion reaction. Incubate at 37°C for at least 4 hr.
- Add 2 µl of 500 mM EDTA to stop the reaction and heat kill CIAP at 65°C for 30 min.
- Add 10 µl of 3 M sodium acetate, pH 5.2, per 100 µl digestion. Add an equal volume of chloroform to the reaction and mix, then microcentrifuge at maximum speed for 2 min. Remove the aqueous phase and transfer into a fresh tube.
- Precipitate the DNA by the addition of an equal volume of isopropanol and let stand at room temperature for 10 min. Pellet DNA by microcentrifuging for 10 min at full speed.
- Discard supernatant and add 1 ml 70% ethanol. Mix well and pellet DNA for 5 min. Discard supernatant and add 1 ml 95% ethanol. Mix well and pellet DNA for 5 min. Pour off supernatant and let the pellet air dry. Dissolve the dried DNA in 50 µl of nuclease‐free water.
-
5
Test adequacy of CIAP treatment by taking 1 µl of dissolved DNA, 1 µl of 10× ligation buffer, 7 µl of nuclease‐free water, and 1 µl of 3 U/µl T4 DNA ligase. Incubate the ligation reaction at 16°C overnight. Run the ligation reaction on 0.8% agarose gel (Voytas, 2000); if no dimer forms, the CIAP treatment is adequate. If dimers are observed, repeat step 5 and retest for adequacy of CIAP treatment.
-
6
Set up the second restriction digestion reactions by adding 40 µl digested and CIAP‐treated plasmid A or G, 5 µl NEBuffer 3, and 5 µl BsmBI (10 U/µl). Incubate at 55°C overnight.
-
7
Monitor digestion by electrophoresing 1 µl of the reaction on a 0.8% agarose gel (Voytas, 2000).
If digestion is complete, all of the plasmid A DNA should be converted from the single 8.4‐kbp fragment to two fragments: a 4.9‐kbp A fragment and the 3.5‐kbp vector; all the plasmid G DNA should be converted from the single 13‐kbp fragment to three fragments of 8.7‐kbp (G fragment) and 2.2‐kpb plus 2.1‐kbp vector fragments (Table 4). If BsmBI digestion is not complete, add 2 µl of BsmBI and continue digestion at 55oC for 4 additional hours.
-
8
Concentrate the digested DNA in a centrifugal vacuum evaporator such as a SpeedVac to approximately 100 µl. Prepare a 0.8% agarose gel (you can load ∼50 µl of sample into each well). Leave an empty well between each set of samples. Put a cardboard box over the gel to keep out light and electrophorese at ∼100 V for 2 hr or when adequate resolution of the fragments has been achieved. DO NOT PUT UNDER UV LIGHT TO SEE BANDS. Visualize gel in a dark room with the Claire Research Dark Reader light box by wearing orange viewing glasses. Cut out bands with a clean razor blade and transfer to labeled 1.5‐ml microcentrifuge tubes.
The gel purification is generally done in parallel with several other fragments.
-
9
Extract the purified DNA fragments from the gel slice using the QiaQuick gel extraction kit.
About four QiaQuick spin columns are needed per band.
-
10
Quantitate DNA with a NanoDrop spectrophotometer.
You will need at least 570 ng of purified fragment A and 1000 ng of fragment G for each ligation. The usual yield from 100 µg of plasmid A is greater than 6 µg of purified fragment A and 10 µg of purified fragment G from 100 µg of plasmid G.
Plasmid B to E
A minimum of 50 µg of DNA is required. We generally start with 100 µg DNA. This should yield sufficient fragments B to E for 10 ligations.
-
11
Set up the following first digestions in 100 to 500 µl final volume, depending upon DNA concentration (the reaction is written for a 100‐µl reaction with 50 to 100 µg of plasmid DNA).
- Restriction digestion reaction: Plasmid B
- 50 to 100 µg Plasmid B DNA
- 10 µl 10× NEBuffer 3
- Adjust volume with nuclease‐free water to 90 µl, then add:
- 10 µl 10 U/µl BglI
- Restriction digestion reaction: Plasmid C
- 50 to 100 µg Plasmid C DNA
- 10 µl 10× NEBuffer 3
- Adjust volume with nuclease‐free water to 90 µl, then add:
- 10 µl 10 U/µl BglI
- Restriction digestion reaction: Plasmid E
- 50 to 100 µg Plasmid E DNA
- 10 µl 10× NEBuffer 3
- Adjust volume with nuclease‐free water to 90 µl, then add:
- 10 µl 10 U/ µl BglI
- Restriction digestion reaction: Plasmid D
- 50 to 100 µg Plasmid D DNA
- 10 µl 10× NEBuffer 4
- 1 µl 10 mg/ml acetylated BSA
- Adjust volume with nuclease‐free water to 90 µl, then add:
- 10 µl 10 U/µl AhdI
-
12
Incubate BglI and AhdI digestions at 37°C overnight.
-
13
Remove 1 µl and electrophorese in a 0.8% agarose gel (Voytas, 2000). If digestion is complete (all plasmid is converted to a single fragment of 6.7 kpb for B, 4.7 kpb for C, 3.45 kpb for D, and 4.8 kpb for E; Table 4), go to step 15.
-
14
If digestion is not complete, redigest with 10 µl of fresh enzyme and monitor digestion by gel electrophoresis as in step 4.
-
15
Set up the second digestions by adding 42 µl BglI‐ or AhdI digested plasmids B to E, 5 µl NEBuffer 3, and 3 µl 10 U/µl BsmBI (10 U/µl). Incubate at 55°C overnight.
Note: For plasmid D, you need to precipitate the first digestion and use NEBuffer 3 for the second digestion.
-
16
Monitor digestion by electrophoresing 1 µl of each reaction on a 0.8% agarose gel (Voytas, 2000); if digestion is complete all the plasmid A DNA should be converted from linearized molecules to the size fragments listed in Table 4. If BsmBI digestion is not complete, add 2 µl 10 U/µl BsmBI and continue digestion at 55°C for 4 additional hours.
-
17
Perform gel purification the same way as in step 8 for plasmids A and G.
-
18
Extract DNA from the excised gel slices using the QiaQuick gel extraction kit.
You need about four QiaQuick spin columns per band.
-
19
Quantitate DNA with NanoDrop.
-
Based on 1000 ng of fragment G for each ligation, the following quantities of plasmid DNA will be needed:
- Purified fragment B: 540 ng
- Purified fragment C: 230 ng
- Purified fragment D: 170 ng
- Purified fragment E: 330 ng
-
The usual yield from 100 µg of plasmids B to E is:
- Purified fragment B: 5 to 10 µg
- Purified fragment C: 5 to 10 µg
- Purified fragment D: 5 to 10 µg
- Purified fragment E: 5 to 10 µg
-
Ligate purified restriction fragments
Ligations are performed stepwise. A schematic diagram depicting the order of ligations is provided in Figure 3. Each ligation is checked for completion by agarose gel electrophoresis and then religated if needed to go to completion.
Figure 3.
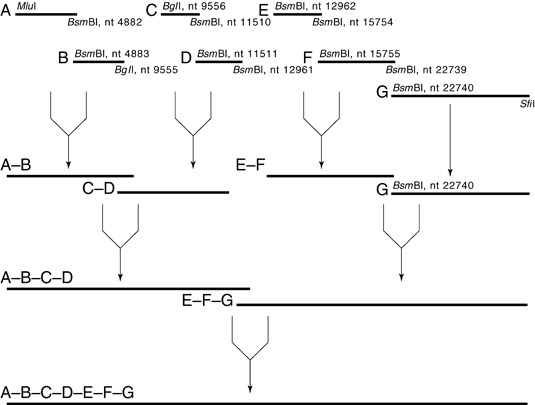
Stepwise ligation to create full‐length MHV A59‐1000 cDNA.
PROCEDURE 1: Ligate A+B, C+D, and E+F
Ligations are set up such that the molar ratios of the various fragments is 1:1 with two exceptions: fragments A and G can be molar excess. The magic ratios in micrograms for the various fragments are:
A = 570 ng
B = 540 ng
C = 230 ng
D = 170 ng
E = 330 ng
F = 810 ng
G = 1000 ng
As noted above, we aim to have sufficient DNA to do ligations based on a minimum of 1000 ng of G fragment.
-
20
Assemble the ligation reactions (A+B, C+D, and E+F) in the smallest volume possible using each fragment the ratios described (generally, this will be 50 µl).
Each ligation reaction will contain 2.5 µl of T4 DNA ligase or 5% of the ligation reaction (the ligase should be relatively fresh).
-
21
Ligate at 16°C overnight.
-
22
Check ligation results by removing 1 µl from each ligation reaction and running in a 0.8% agarose gel (Voytas, 2000) in parallel to Lambda HindIII markers.
- Ligation A+B is complete when fragment B has all been converted to high‐molecular‐weight form. If needed, add additional A fragment and continue ligation as described in step 3d. If some A and B fragments remain unligated in approximately equal amounts, then continue ligation as described in 22d, below.
- Ligation C+D is complete if both C and D have been converted to high‐molecular‐weight product. If some C and D fragments remain unligated in approximately equal amounts, then continue ligation as described in step 22d, below. If one fragment has disappeared and another remains, the quantification of the fragment is off or a pipetting error has been made. Adjust the ligation reaction by adding a small amount of the missing fragment and continue ligation as described in step 22d.
- Ligation E+F is complete if both E and F have been converted to high‐molecular‐weight product. If some E and F fragments remain unligated in approximately equal amounts, then continue ligation as described in step 22d. If one fragment has disappeared and one remains, the quantification of the fragment is off or a pipetting error has been made. Adjust the ligation reaction by adding a small amount of the missing fragment and continue ligation as described in step 22d.
- Add 2 µl of 3 U/µl T4 DNA ligase, 1 µl of 10 µM ATP, and 0.5 µl of 100 mM DTT. Incubate at 16°C a minimum 6 hr. Monitor completion of ligation by analyzing on gel as described above.
PROCEDURE 2: Ligate A‐B + C‐D and E‐F + G
-
23
Take the E‐F ligation product from Procedure 1 and set up a ligation with fragment G. Again keep the volume as small as possible. Use 2.5 µl of 3 U/µl ligase, 1 µl of 10 mM ATP, and 0.5 µl of 100 mM DTT. Add sufficient 10× ligation buffer to make the final ligation reaction 1× with respect to buffer, remembering that the E‐F ligation reaction already is at 1× buffer concentration. Incubate ligation at 16°C overnight.
-
24
Take the A‐B and C‐D ligation products from Procedure 1 and set up ligation for A‐B‐C‐D. Use 2.5 µl of 3 U/µl ligase, 1 µl of 10 mM ATP, 0.5 µl of 100 mM DTT, and 0.45 µl of 10× ligation buffer. Incubate ligation at 16°C overnight.
PROCEDURE 3 Ligate A‐B‐C‐D and E‐F + G
-
25
Take the A‐B‐C‐D and E‐F‐G ligation products and set up ligation for A‐B‐C‐D‐E‐F‐G. Use 2.5 µl of ligase, 1 µl of 10 mM ATP, 0.5 µl of 100 mM DTT, and 0.45 µl of 10× ligation buffer. Incubate ligation at 16°C overnight.
Prepare template for N gene transcription
The N gene transcription template contains the N gene under the control of a SP6 promoter. The coronavirus nucleocapsid (N) protein is a multifunctional protein which encapsidates the RNA genome in the virion, and has a functional role in RNA replication and transcription. Co‐electroporation of N‐gene transcripts with genome RNA increases the number of infectious centers and the speed at which virus replication initiates.
-
26
Synthesize N gene transcription template by PCR (Kramer and Coen, 2001) from plasmid G using A59SP6 Ng(+) and A59Ng3′ (‐) primers.
- Use 10 ng of G plasmid per 50 µl of PCR reaction as template and set up 10 PCR reactions.
- Use the following PCR conditions:
1 cycle:1 min95°C(initial denaturation)5 cycles:50 sec95°C(denaturation)1 min52°C(annealing)1 min, 35 sec72°C(extension)5 cycles:50 sec95°C(denaturation)1 min55°C(annealing)1 min, 35 sec72°C(extension)55 cycles:50 sec95°C(denaturation)1 min61°C(annealing)1 min, 35 sec72°C(extension)1 cycle:5 min72°C(final extension).This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.- Combine all of the PCR reactions, concentrate them down to 1/5 volume, and purify the PCR product by electrophoresis on a 0.8% agarose gel (Voytas, 2000).
- Cut out the band containing the PCR‐amplified N gene cDNA under the control of the SP6 promoter and purify using the QiaQuick gel extraction kit. Dilute the purified N gene PCR product to 100 ng/µl using nuclease‐free water.
The N gene yield from 10 PCR reactions is generally enough for 15 transcription reactions.
In vitro transcription reaction
-
27
Chloroform extract the ligated‐full length cDNA from step 25 by adding 1/10 volume of 3 M sodium acetate, pH 5.2, and an equal volume of chloroform; mix by brief vortexing, then spin at full speed in a microcentrifuge for 30 sec. Transfer the aqueous phase into a new microcentrifuge tube and add 2× volume of 100% ethanol. Mix and let stand at room temperature for 10 min, spin for 10 min, take out the supernatant, add another 200 µl of 100% ethanol, and spin for 5 min. Discard liquid, invert tube to air dry the pellet for 10 min (the pellet should have a white appearance). Then, dissolve the pellet with 9 µl of nuclease‐free water.
-
28
Set up the following transcription reactions using the Ambion mMESSAGE mMACHINE kit for both in vitro transcription reactions. Use the T7 kit (cat. no. AM1344) for transcription of the ligated full‐length cDNA and the SP6 kit (cat. no. AM1340) for transcription of the N gene RNA.
- Full‐length cDNA transcription reaction:
- 7 µl template
- 5 µl buffer
- 25 µl 2× NTP/Cap
- 7 µl GTP
- 5 µl enzyme mix
- MHV N gene transcription reactions (sufficient for 4 electroporations):
- 20 µl template
- 8 µl buffer
- 40 µl 2× NTP/Cap
- 7 µl GTP
- 5 µl enzyme mix
-
29
Place both reactions in a thermal cycler set for 40.5°C for 30 min, 37°C for 50 min, and 40.5°C for 30 min. Hold at 4°C.
Electroporation
-
30a
For BHK‐R cells: Starting (normally) with a confluent 75‐cm2 flask on the day before electroporation, set up two 175‐cm2 flasks, seeding with 7–8 × 106 cells (on the next day, the BHK‐R cells should be 50% to 80% confluent). Trypsinize the cells (see Support Protocol 1), wash three times with cold PBS, resuspend to a concentration of 107 cells per ml, aliquot 800 µl into a 4‐mm‐gap cuvette, and place the cuvette on ice.
0.8 ml of cell suspension will be needed for each electroporation reaction, including a negative control.
-
30b
For DBT cells:, Starting (normally) with a confluent 75‐cm2 flask on the day before electroporation, trypsinize (Support Protocol 1), and seed 106 cells into a 75‐cm2 flask for each mutant plus an additional 75‐cm2 flask for the negative control, which will not receive in vitro–transcribed RNA.
-
31
Add 45 µl of the in vitro–transcribed full‐length RNA and 20 µl of the transcribed N gene to your washed BHK‐R cells with nuclease‐free pipet tips, and pipet up and down three or four times to mix. Use electroporation settings of 0.85 kV, 200 Ω, 25 µF. Electroporate the cells three times (the time constant is usually 0.44 to 0.62 msec). Perform one electroporation reaction without added in vitro–transcribed RNA to serve as a negative control.
-
32
Let the cells rest in the cuvette for 5 to 10 min, then transfer each electroporation into separate 75‐cm2 flasks containing DBT cells.
However, for viruses that have been shown to be nonviable where the electroporation is going to be used to detect negative‐strand RNA synthesis, transfer your electroporated cells directly into growth medium, as the extra DBT cells will only increase your background in RT‐PCR assays.
-
33
Incubate at 37°C overnight.
Cells electroporated with wild‐type genomic RNA usually develop widespread CPE 24 to 48 hr after electroporation, a sign of virus infection. Mutants with decreased rates of growth can be incubated as long as 72 hr.
-
34
If CPE has developed, remove the medium to a 15‐ml conical tube labeled as P0 and freeze at −80°C. If no CPE is observed, continue to step 35. If virus is recovered at this stage (CPE is widespread in the culture), skip to step 36.
-
35
If no CPE develops by the third day after electroporation, freeze the culture at –80°C for at least 1 hr. Thaw and transfer to a 50‐ml conical centrifuge tube. Sonicate on ice in a cup sonicator with 3 bursts of 20 sec each at 100 W peak envelope power. Centrifuge 10 min at 3000 rpm, 4°C. Infect a fresh 75‐cm2 flask of 50% confluent DBT cells with 3 ml of the lysate and observe for CPE (blind passage 1). If no CPE is observed by 3 days, freeze cells as before and perform a second blind passage, and if necessary a third.
We do not consider a mutation to be lethal until we have performed three independent electroporations, and serially blind passed each electroporation three times without seeing any CPE. Recombinant viruses are labeled as P0 and frozen at −80°C.
-
36
Recombinant viruses are subsequently plaque purified from the P0 pool as described in Basic Protocol 2. Pick at least 8 well separated plaques.
-
37
Expand four plaques in DBT cells.
Purified plaques are inoculated into 25‐cm2 flasks of DBT cells to prepare P1 stocks as described in Basic Protocol 2, with one exception. When extensive CPE has developed in the culture, rather than freezing the 25‐cm2 flask, remove the supernatant medium, and freeze as the P1 stock.
-
38
Take the infected cell monolayer and extract total RNA using an RNAeasy kit according to the vendor's instructions.
The RNA is subsequently used as a PCR template to sequence regions of the MHV genome to confirm the genotype of the virus recovered.
Support Protocol 5. Plasmid Propagation and Maintenance
The seven plasmids named A to G contain the cDNAs required to create a cDNA representing the entire MHV‐A59 genome by in vitro ligation. Several of these plasmids are metastable. This means that the plasmid sometimes deletes all or a portion of their MHV cDNA insert. These deleted plasmids have a growth advantage and grow out in plasmid preps. It also means that in plate cultures, the bacteria that contain the desired cDNA produce smaller colonies than those that have deletions. This is a particular problem with the F clone. A small amount of plasmid DNA from a construct that worked must be maintained at −70°C in addition to maintaining a glycerol stock of bacteria containing these plasmids. Although many of the plasmid vectors used to propagate these cDNAs were chosen to increase stability (see Table 4), to minimize problems it is necessary to grow the plasmids at 30°C in Top 10 cells (Invitrogen).
Materials
Bacteria containing the desired plasmids (either glycerol stocks or plates <6 weeks old); plasmids can be obtained from Ralph Baric, University of North Carolina (rbaric@sph.unc.edu), and initially should be transformed into Top 10 cells (Invitrogen, cat. no. C44040‐03) as described below
LB plates containing 50 µg/ml ampicillin or 30 µg/ml kanamycin (also see Table 4)
2× YT broth (see recipe) containing appropriate selection antibiotic
Plasmid Midiprep kit (BioRad, cat. no. 732‐6120; alternatively use Promega Wizard kit)
Restriction enzymes AdhI, BglI, BsmBI, MluI, SfiI (New England Biolabs; see Table 4)
0.8% and 1% agarose gels prepared with TAE buffer (Voytas, 2000)
1 mg/ml ethidium bromide (see recipe)
30°C incubator
Tabletop centrifuge, 4°C
Additional reagents and equipment for agarose gel electrophoresis (Voytas, 2000)
Procedure
-
1
Streak bacteria containing the desired plasmids, from either glycerol stocks or from prior plates less than 6 weeks old, on an LB agar plate containing the appropriate antibiotic (see Table 4; 50 µg/ml ampicillin or 30 µg/ml kanamycin, depending upon the plasmid). Incubate at 30°C for 24 to 36 hr. Store plates at 4°C after wrapping in Parafilm.
Alternatively, plasmids can be transformed into competent Top 10 cells (Invitrogen, cat. no. C4040‐03). In recovering the transformants, it is important to do the 1‐hr incubation step prior to plating at 30oC. The transformation reaction is spread on LB agar plates containing the appropriate antibiotic and incubated at 30oC for 24 to 36 hr. Save plates at 4oC after wrapping in Parafilm.
-
2
Inoculate a single colony into 300 ml of 2×YT with appropriate antibiotic at 30°C and incubate 20 to 24 hr. Divide into 50‐ml aliquots, pellet in a cold tabletop centrifuge for 10 min at 5000 rpm, 4°C, then freeze at −70°C.
-
3
Defrost two aliquots, each corresponding to 50 ml of bacterial culture, and process for plasmid midiprep, 100 ml/midiprep, using midiprep kit from BioRad or Promega (Wizard).
Both of the abovementioned kits work; it has been a matter of personal preference which one to use.
-
4
Confirm that the plasmid has not undergone rearrangement or deletion by digestion of 2 µl (approximately 500 ng) of plasmid DNA in a 10‐µl reaction with 0.5 µl of the appropriate restriction enzymes (see Table 4). Resolve the restriction digest is by electrophoresis in a TAE agarose gel (0.8% agarose for A, B, F, and G; 1% agarose for C, D, and E) containing 1 drop (50 to 100 µl) of 1 mg/ml ethidium bromide per small gel, 2 drops per medium gel, or 3 drops per large gel. Photograph gel.
If restriction digests yield the predicted size restriction fragments, the plasmids are fine. If not, discard all aliquots and start back at step 1 or step 2.
Basic Protocol 5. Generation of Recombinant Viruses by Targeted Recombination
Targeted RNA recombination is a reverse genetic system developed by the Masters lab to introduce site‐directed mutations into the 3′ one‐third of the coronavirus genome that encodes all the viral structural proteins (Kuo et al., 2000). The method given below for MHV‐A59 is based upon the use of a chimeric coronavirus in which the ectodomain of the MHV‐A59 spike glycoprotein, which determines host range, has been replaced by that of feline peritonitis virus (FIPV), and thus can only infect feline cells. The chimeric virus is used for recombination with a synthetic donor RNA (transcribed from a plasmid, pMH54, that is available from Paul Masters, Wadsworth Laboratory, Albany, New York) that contains a portion of the MHV‐A59 HE gene, the original spike (S) gene, and extends to the 3′ end of the genome (see Fig. 4). The recombinant viruses are selected by their regained ability to infect murine cells. Mutations inserted into the donor RNA will be transferred to the recombinant virus and generate new mutant viruses (Kuo et al., 2000; Ontiveros et al., 2001; Fu et al., 2004). Similar systems have been constructed for other strains of MHV, and in principle similar systems can be created for any other coronavirus.
Figure 4.
A schematic representation of the targeted recombination procedure. In the first step, fMHV is created by homologous recombination of the felinized S gene (light gray) into the genomic RNA of the MHV‐A59 virus to generate a chimeric virus containing the felinized S gene, fMHV. In the second step, a mutant MHV‐A59 virus is formed by homologous recombination of the donor RNA containing additional mutations (denoted by the asterisk) in the original S gene with the genomic RNA of the felinized MHV. The dark gray region indicates the region of the genome into which a mutation may be inserted into MHV by this methodology.
The procedure for targeted RNA recombination starts with the generation of a chimeric acceptor virus in which the ectodomain of the MHV‐A59 spike glycoprotein has been replaced by that of FIPV. A donor RNA is prepared that contains a chimeric felinized S gene that contains the ectodomain of FIPV, which allows infection of feline cells, fused to the transmembrane and C‐terminal domain of MHV, allowing incorporation into MHV virions. Murine cells are infected with the original MHV virus and transfected with the donor RNA containing the felinized S gene. Homologous recombination occurs producing a felinized MHV strain (fMHV). fMHV is then selected by its ability to grow in feline cells.
For targeted recombination with MHV‐A59, the acceptor virus contains the ectodomain of feline infectious peritonitis virus and is designated fMHV. fMHV can be obtained from Paul Masters, Wadsworth Laboratory, Albany, New York. fMHV can only infect feline cells, and we routinely use FCWF cells for propagating fMHV and for targeted recombination. A detailed description of the isolation of fMHV is described in Kuo et al. (2000), and we and others have developed similar felinized acceptor viruses for additional strains of MHV (Ontiveros et al., 2001; McGruder et al., unpub. observ.). fMHV functions as a recipient for an in vitro–synthesized donor RNA that contains the original wild‐type MHV S gene and additional targeted mutations. The cells are infected with fMHV and subsequently electroporated with the donor RNA. Homologous recombination events between the donor RNA and the viral genomic RNA that occur upstream of the S gene produce mutant viruses that can grow in murine cells such as DBT cells and can thus be selected.
NOTE: All culture incubations should be performed in a humidified 37°C, 5% CO2 incubator unless otherwise specified. L2 cells should be incubated at 3% CO2.
NOTE: All cell cultures are cultured in DME10 except for FCWF cells, which are cultured in DME10 prepared with 10% fetal bovine serum in place of the usual calf serum.
NOTE: The incubation time with the felinized MHV can be change according to the MHV type. Usually the incubation period takes 4 to 6 hr; fMHV/A59 requires a 4‐hr incubation, fMHV/JHM requires a slightly longer incubation, and fMHV/MHV1 requires at least 6 hr of incubation.
Materials
FCWF cells (ATCC, cat. no. CRL‐2787)
DME10/FBS (for FCWF cells; see recipe for DME10 but use FBS where calf serum is called for)
DME0 (see recipe)
DME2 (see recipe)
PacI restriction enzyme (New England Biolabs)
0.8% agarose gel in TAE buffer (Voytas, 2000)
3 M ammonium acetate, pH 5.2 (appendix mca02a)
70% and 96% (v/v) ethanol
QuickClean 5M PCR Purification Kit (Genscript, cat. no. L00198; optional)
Turbo DNase kit (Applied Biosystems)1% agarose gel in TBE buffer (Voytas, 2000)
DBT cells (not commercially available; cells can be obtained from most investigators working with MHV, including the authors)
Recipient virus fMHV (Dr. Paul Masters, Wadsworth Laboratory, Albany, N.Y.; masters@wadsworth.org)
Dulbecco's phosphate buffered saline (DPBS; e.g., Invitrogen) without calcium and magnesium
RNeasy mini kit (Qiagen)
SuperScript III Reverse Transcriptase kit (Invitrogen)
25‐, 75‐, and 175‐cm2 tissue culture flasks with filter cap
Platform rocker
15‐ and 50‐ml conical polypropylene tubes with screw caps
Cup sonicator
Tabletop centrifuges (e.g., IEC Clinical), 4°C and room temperature
Cryogenic tubes
mMESSAGE mMACHINE High Yield Capped RNA T7 Transcription Kit (Applied Biosystems)
Cell Line Nucleofector Kit V (Amaxa Biosystems; http://www.lonzabio.com)
Nucleofector (Amaxa Biosystems; http://www.lonzabio.com)
Cup sonicator
Cryogenic tubes
Additional reagents and equipment for preparation/passaging of cell cultures, including trypsinization (Support Protocol 1), agarose gel electrophoresis (Voytas, 2000), counting cells using a hemacytometer (appendix mca04a), plaque purification (Basic Protocol 1), and expansion of plaques to produce stocks (Basic Protocol 2)
Propagate recipient fMHV
-
1
Maintain FCWF cells in DME10/FBS by trypsinizing slightly subconfluent cultures, passaging and seeding 5 × 106 cells into 175‐cm2 flasks.
This procedure provides consistent maintenance of FCWF cells as well as cultures for the generation of the recipient virus stock.
A 1:4 split of FCWF cells will usually provide cultures that approach confluence after 72 to 96 hr.
-
2
Wash FCWF cells once with DME0 and inoculate with fMHV in 5 ml DME2 at an MOI of 0.01 to 0.1.
-
3
Incubate by rocking for 60 min at room temperature, then add 15 ml of DME2 and incubate at 37°C.
-
4
After 24 to 36 hr or when the monolayer exhibits extensive CPE and 25% or more of the cells have detached from the flask surface, freeze the culture at −80°C for at least 1 hr. Partially thaw in a 37°C water bath, and then place into an ice bucket to complete thawing.
-
5
Transfer the medium with the cells to a 50‐ml conical polypropylene tube. Sonicate in a cup sonicator, three bursts of 20 sec each at 100 W peak envelope power, resting 20 sec on ice between each cycle.
-
6
Clarify the medium by centrifugation 10 min at 2500 rpm, 4°C, in a prechilled tabletop centrifuge.
-
7
Store the supernatant in aliquots of 1 ml in cryogenic tubes at −80°C until use.
Typical titers obtained for fMHV are around 5 × 106 to 1 × 107 pfu/ml.
Preparation of synthetic donor RNA
The donor RNA is synthesized by in vitro transcription from vector pMH54. pMH54 contains a cDNA corresponding to the 5′ end of the genome (467 nt) fused to the hemagglutinin gene at codon number 28 and extends to the 3′ end of the genome, including the poly(A) tail. There is a PacI restriction site just after the poly(A) tail. The viral cDNA is under the control of a T7 RNA polymerase promoter and targeted mutations can be introduced into the cDNA construct, which is then transcribed and transferred into the recombinant virus by homologous recombination.
-
8
Linearize 10 µg of pMH54 with 25 U PacI restriction enzyme in a reaction volume of 125 µl for 2 hr at 37°C.
-
9
Check the linearization of the plasmid by analyzing a 2‐µl aliquot of the reaction mixture by 0.8% (w/v) agarose gel electrophoresis in 1 × TAE buffer (Voytas, 2000).
-
10
Purify the linearized plasmid by either of two different methods:
- Precipitation with salt and ethanol: Add one‐tenth volume of 3 M ammonium acetate and 2.5 volumes of 96% (v/v) ethanol to the linearized plasmid, and incubation at −20°C for 30 min. Pellet the DNA precipitates by microcentrifugation at 4°C for 20 min at 14,000 rpm. Wash the pellet with 70% (v/v) ethanol, dry the pellet, and dilute it in 10 µl of water.
- Using PCR purification kit: Perform the purification according to the manufacturer's instruction. Briefly, add 2.5 volumes of binding buffer to the enzymatic reaction, transfer the solution to a column, spin, wash the linearized plasmid with washing buffer containing 96% ethanol, dry the columns, and elute with 65°C pre‐heated nuclease‐free water.
-
11
Transcribe the donor RNA in a 20‐µl reaction volume with the T7 mMESSAGE mMACHINE kit according to the manufacturer's instructions, with small modifications as follows. Briefly, add, in this order: nuclease‐free water to complete the reaction to 20 µl, then add 10 µl of 2 × NTP/CAP, 2 µl of 10 × reaction buffer, 1 µl of 30 mM GTP to enhance the transcription reaction, at least 1 µg of the purified linear plasmid, and finally 2 µl of the enzyme. Mix the reaction by pipetting up and down 5 times and incubate at 37C for 2 hr.
-
12
Treat the transcript with 2 µl of Turbo DNase for additional 30 min at 37°C to eliminate the DNA template.
-
13
Check RNA synthesis by analyzing one‐tenth of the reaction volume (2 µl) by electrophoresis on a 1% (w/v) agarose gel in 1× TBE buffer.
-
14
Store the RNA at −80°C until use. Do not use transcripts stored more than 48 hr.
Infection and nucleofection
The generation of recombinant MHV is accomplished by the infection of feline FCWF cells with fMHV and the transfer of the transcribed donor RNA into the cells by nucleofection or electroporation. The recombinant MHV is selected on DBT cells and subsequently plaque purified on L2 cells. The synthetic RNA can be transferred into cells by either electroporation or nucleofection. Our lab has found that the donor RNA is transferred more efficiently by the Nucleofector Technology (Amaxa), and this method will be described here in detail.
-
15
Seed 1.4 × 107 FCWF cells into a 175‐cm2 flask 16 to 24 hr prior to the infection and incubate at 37°C.
Typically, one 175‐cm2 flask containing confluent cells will be sufficient for 3 to 4 nucleofection reactions.
-
16
On the next day, before you begin the infection, seed 2 × 106 DBT cells into 25‐cm2 flasks; the number of flasks will be determined according to the number of the nucleofection reactions.
To be confident that any phenotype that you observe in a recovered virus mutant, it is best to have two or more independent isolates from separate nucleofection reactions to characterize.
-
17
Wash the FCWF cells with 10 ml of DME0 and infect the cells with fMHV at MOI of 0.5 to 1 in a volume of 5 ml DME0, incubate at room temperature with rocking for 60 min, than add 15 ml of DME10/FBS and incubate at 37°C.
-
18
After 4 hr of incubation wash the cells with 10 ml of DPBS.
-
19
Detach the cells from the flask surface using trypsin/EDTA solution (see Support Protocol 1).
-
20
Add 10 ml of DME10/FBS to the cell suspension, then count the cells using a hemacytometer under the microscope (appendix mca04a) and adjust the cell density to at least 1.5 × 106 cells per reaction. Add an additional control reaction with cells nucleofected without the donor RNA.
-
21
Pellet the cells in 15‐ml tubes by centrifugation 10 min at 720 rpm, room temperature, in a tabletop centrifuge, remove the supernatant, and wash the cell pellet with 5 ml of DPBS (without calcium and magnesium).
-
22
Prepare fresh Nucleofection solution by mixing Nucleofection solution V and Supplement 1 solution in a ratio of 9:2 according to the manufacturer's instruction. Gently resuspend the cells in 100 µl of Nucleofection solution.
-
23
Add 6 µl of the donor RNA to the cell suspension, mix, and transfer the mixture into the Amaxa‐certified cuvette. Insert the cuvette into the holder of the Nucleofector and start program T‐0200.
-
24
Remove the cells into a new 1.5‐ml microcentrifuge tube, add 0.5 ml of fresh DME10/FBS and mix gently.
-
25
Transfer the cell suspension into the 25‐cm2 flasks containing a subconfluent monolayer culture of DBT cells in order to allow the propagation of the recombinant virus.
-
26
Follow the development of cytopathic effect in the DBT cells by microscopic observation and remove 1 ml of culture medium after the first and second days of incubation. Freeze the remaining culture with the medium samples at −80°C on day 3 or when all the cells have developed CPE, whichever occurs first.
-
27
To expand any recombinant viruses that may have been generated, passage the lysate and supernatants from days 1 and 2 in murine cells (DBT cells). After 24 hr, seed three new 25‐cm2 flasks with 2 × 106 DBT cells per flask, and also defrost the cultures frozen on the third day after nucleofection and the medium samples from the first and second days of incubation. Transfer the defrosted cells to a 15‐ml conical tube and sonicate in a cup sonicator, using three bursts of 20 sec each, resting 20 sec on ice between each cycle. Centrifuge the lysate 10 min at 2500 rpm, 4°C, in a tabletop centrifuge, and collect the supernatant. Use the clarified lysate and the day‐1 and ‐2 tissue culture supernatants to infect the fresh DBT 25‐cm2 culture flasks.
-
28
Incubate and follow the flasks until the monolayer exhibits extensive cytopathic effect or up to 3 days of incubation.
-
29
When cytopathic effects are clearly visible in the DBT cell cultures, freeze the cultures at −80°C for at least 1 hr. Defrost and sonicate the cell suspension as described previously, and clarify by centrifugation for 10 min at 2500 rpm, 4°C. Freeze the clarified stock at −80°C in 1‐ml aliquots.
Plaque purify and expand recombinant viruses
Recombinant MHV are grown on DBT or 17Cl‐1 cells and plaque‐purified on L2 cells.
-
30
Plaque purify recombinant viruses on L2 cell monolayers as described in Basic Protocol 1.
-
31
Pick at least eight plaques and store at −80°C.
-
32
Expand four plaques in DBT cells. Inoculate purified plaques into 25‐cm2 flasks of DBT cells to prepare P1 stocks as described in Basic Protocol 2, with one exception. When extensive CPE has developed in the culture, rather than freezing the 25‐cm2 flask, remove the supernatant medium, and freeze as the P1 stock. Take the infected cell monolayer and extract total RNA using the RNAeasy kit according to the vendor's instructions.
The RNA is subsequently used as a PCR template to sequence regions of the MHV genome to confirm the genotype of the virus recovered.
Verify the recombinant genotype
-
33
Analyze the recombinant viruses by reverse transcription–PCR using SuperScript III and the RNA that was yielded in step 2 (above) as a template. Amplify the target region on the cDNA product with gene‐specific, sense, and antisense, primers.
-
34
Analyze the RT‐PCR product by cutting with unique restriction enzymes and direct sequencing.
Reagents and Solutions
Use tissue–culture grade water in all recipes and protocol steps. For common stock solutions, see appendix mca02a; for suppliers, see mcaspl.
Agarose, 1.6%
Dilute agarose type I (FMC, medium endosmosis; Sigma, cat. no. A‐6013) to 1.6% (w/v) with tissue culture‐grade water. Autoclave to sterilize. Store at room temperature up to 1 year.
DME0
Powdered medium can be purchased from many vendors. We typically purchase Dulbecco's Modified Eagle Medium powder, high glucose, with l‐glutamine, with pyridoxine hydrochloride and 110 mg/liter sodium pyruvate, without sodium bicarbonate, from Invitrogen in 5‐liter packets (cat. no. 12800‐082). Dissolve the powdered medium in 4.5 liters of Milli‐Q quality water. Add 18.5 g sodium bicarbonate. Add 75 ml of 1 M HEPES/MOPS/TES buffer, pH 7.3 (see recipe). Bring to a final volume of 5 liters. Filter sterilize through a 0.22‐µm filter, dispensing 450 ml into 500‐ml sterile bottles. Seal tightly and label DME0. Store up to 3 months at room temperature.
DME10
To a bottle containing 450 ml of Dulbecco's Modified Eagle Medium, add: 50 ml calf serum (or FBS; see below), 10 ml 200 mM glutamine, and 5 ml 100× penicillin‐streptomycin solution (10,000 U penicillin base and 10,000 µg streptomycin sulfate per ml; available from Invitrogen as well as other vendors). Label as DME10 and store up to 4 weeks at 4°C.
Our lab has found it unnecessary to use more expensive fetal bovine serum (FBS) for the support of most of the murine cell lines we routinely utilize. We have evaluated and used many different less expensive supplemented and characterized calf sera over the years and have always found lots that were satisfactory for 17Cl‐1, L2, DBT, and BHK‐R cells. However, we utilize FBS for FCWF cells, since these cells seem to require FBS for long‐term maintenance.
DME2
To a bottle containing 450 ml of Dulbecco's Modified Eagle Medium, add: 10 ml calf serum, 10 ml 200 mM glutamine, 5 ml 100× penicillin‐streptomycin solution (10,000 U penicillin base and 10,000 µg streptomycin sulfate per ml; available from Invitrogen as well as other vendors). Label as DME2 and store up to 4 weeks at 4°C.
DME2, 2×
Prepare 2× DME0:
Dissolve powdered Dulbecco's Modified Eagle Medium in half the normal volume of water recommended by the manufacturer
Add 15 ml of 1 M HEPES/MOPS/TES buffer, pH 7.3, per 500 ml final volume
Sterilize by filtration through a 0.22‐µm filter and label as 2× DME0
2×DME0 can be stored at room temperature in tightly closed bottles.
Prior to use, add (per 500 ml):
20 ml calf serum
20 ml 200 mM glutamine
10 ml 100× penicillin‐streptomycin solution (10,000 U penicillin base and 10,000 µg streptomycin sulfate per ml; available from Invitrogen as well as other vendors)
Label as 2×DME2 and store up to 4 weeks at 4°C
EDTA, 0.5 M
Add 16.8 g disodium EDTA to 100 ml tissue‐culture‐grade water; adjust pH to 8.0 with NaOH. Autoclave 20 min to sterilize. Store up to 1 year at room temperature.
Ethidium bromide, 1 mg/ml
Add 100 mg ethidium bromide powder to 100 ml sterile water and stir for several hours to dissolve; transfer to dark bottle or wrap container with foil. Store up to 1 year at room temperature.
Glutamine, 0.2 M
Add 29.2 g glutamine to 1 liter Milli‐Q water. Filter sterilize and freeze in 50‐ml aliquots (store up to 1 year at −20°C).
HEPES/MOPS/TES buffer, pH 7.3
Combine the following:
125.58 g MOPS
142.98 g HEPES
137.52 g TES
15 g NaOH
Milli‐Q water to a final volume of 600 ml
Adjust to pH 7.3 with NaOH
Autoclave and store up to 1 year at 4°C
LB plates
Combine the following:
10 g Bacto tryptone (BD Difco)
5 g Bacto yeast extract (BD Difco)
10 g NaCl
15 g Bacto agar (BD Difco)
Milli‐Q water to 1 liter
Autoclave for 45 min
Cool down to ∼45°C
Add 50 mg/ml ampicillin or 30 mg/ml kanamycin (depending on plasmid)
Pour into 100‐mm‐diameter plates
Store up to 2 weeks at 4°C
MOPS‐saline‐EDTA (MSE) buffer
10 mM MOPS, pH 6.8
150 mM NaCl
1 mM EDTA
Store up to 1 year at room temperature
Sucrose gradient solutions
Potassium tartrate gradient solutions
5% (w/w) potassium tartrate in MSE
5 g potassium tartrate
95 ml MSE buffer
10% (w/w) potassium tartrate in MSE
10 g potassium tartrate
90 ml MSE buffer
15% (w/w) potassium tartrate in MSE
15 g potassium tartrate
85 ml MSE buffer
25% (w/w) potassium tartrate in MSE
25 g potassium tartrate
75 ml MSE buffer
40% (w/w) potassium tartrate in MSE
40 g potassium tartrate
60 ml MSE buffer
Store potassium tartrate solutions up to 1 year at 4°C and chill down prior to use.
Trypsin‐EDTA solution
Add 100 mg phenol red, 175 mg NaHCO3, 4 g NaCl, 200 mg KCl, 500 mg glucose, 500 mg EGTA, and 600 mg Tris base to 500 ml Milli‐Q water. Adjust pH to 8.1 and keep on ice‐cold water until all ingredients dissolved. Add 500 mg trypsin (Sigma, cat. no. T4799) to the solution. Filter sterilize and aliquot into 50‐ml conical tubes, store up to 1 year at −20°C.
YT broth, 2×
Combine the following:
16 g Bacto tryptone
10 g Bacto yeast extract
5 g NaCl
Milli‐Q water to 1 liter
Mix until dissolved
Autoclave 30 min
Store up to 1 year at room temperature.
Commentary
Background Information
The first coronaviruses were isolated in the 1930s (infectious avian bronchitis, IBV) and 1940s [mouse hepatitis virus (MHV) and transmissible gastroenteritis virus (TGEV)], but were not recognized as a related group of viruses until the late 1960s when it was noted that these viruses had a unique appearance in transmission electron micrographs. The negative‐stained EM appearance of these viruses is that of round enveloped particles, approximately 100 to 160 nm in diameter, bearing bulbous spike proteins on the lipid envelope that give the virus particle an appearance somewhat reminiscent of a crown, thus giving this group of virus its name. Further EM studies of infected cells revealed that coronaviruses bud through intracellular membranes in the ERGIC‐Golgi and are subsequently released from the infected cell by exocytosis. Coronavirus particles contain four or five proteins: a nucleocapsid protein (N); a triple membrane‐spanning protein, M; a small membrane protein with ion channel activity, E (Wilson et al., 2004); and a spike protein (S) that makes up the characteristic bulbous peplomer and functions to bind to host cell receptors and mediate virus entry to the cytoplasm. In addition, most but not all members of the Group IIa coronaviruses contain a hemagglutinin‐esterase (HE) protein, which makes up a second, somewhat shorter spike protein on these viruses. The virus particles contain a helical nucleocapsid made up of the N protein and a large single‐stranded, positive‐sense, nonsegmented RNA genome, between 27 and 32 kbp in length.
The genus Coronavirus is one of two genera in the Coronaviridae family, the other being Torovirus. Both of these genera are members of the order Nidovirales that also contains the Arterivirus family. The coronaviruses were originally further classified as belonging to one of three subgroups, subgroup I, II, or III, based on serologic relatedness. Viruses belonging to subgroups I and II infect mammals; subgroup III viruses infect birds. This classification is supported by phylogenetic analyses based on sequence similarity and genetic organization common to each of the subgroups.
A large number of molecular biologic and sequencing studies established that the coronaviruses share a common genetic organization and mode of replication. Infected cells contain a set of 6 to 9 virus‐specific polyadenylated RNAs that make up a 3′ co‐terminal nested set, with the largest member of this set being the genome RNA and the remaining RNAs representing subgenomic mRNAs. The subgenomic RNAs encode the structural proteins enumerated above in a characteristic order: HE (if present)‐S‐E‐M‐N. In addition to these structural genes, coronavirus subgenomic RNAs encode accessory proteins that are not essential for viral replication. They generally modulate cellular responses to infection and are specific for each coronavirus subgroup. The replicase proteins are encoded in the 5′ two‐thirds of the genome. This region contains two very long partially overlapping open reading frames that are translated by a ribosomal frame shifting mechanism. The ∼750‐kDa primary translation product is cotranslationally autocatalytically cleaved into 16 nonstructural proteins (nsps) that encode the machinery needed for RNA replication and transcription.
Many of the coronaviruses are associated with respiratory or enteric diseases, although some viruses replicate and cause disease at other sites, including the liver, spleen, and central nervous system. Most of the coronaviruses are relatively species specific, only infecting their natural host and cell cultures derived from that host or a closely related host species. However, there are several examples of coronaviruses jumping species, with SARS‐CoV being the most well known example. SARS‐CoV is thought to have originated in bats, where SARS‐like CoVs have been identified, and then transmitted to civets and raccoon dogs in wild animal markets in Southern China, from which they were subsequently transmitted to humans (Lau et al., 2010). During this chain of transmission, several key adaptations to the new host occurred (Li et al., 2005a,b). CCoV and FeCoV provide another exception to the high degree of species specificity of coronaviruses, in that both viruses are able to grow in feline cells. Phylogenetic analysis of the group Ia coronaviruses, CCoV, FeCoV, and TGEV, suggest a complex set of recombination events has given rise to these viruses from a common ancestor, likely explaining the ability of both CCoV and FeCoV to infect feline cells (Wesseling et al., 1994; Lorusso et al., 2008). The species specificity of coronaviruses is controlled by the ectodomain of the S protein, and can be altered by creating chimeric S proteins that contain the ectodomain of a distantly related CoV (Kuo et al., 2000).
Many of the newly identified coronaviruses have not been successfully propagated in cell culture as yet, and are only known through RT‐PCR sequencing. This is particularly true for the bat coronaviruses, since suitable bat cell lines that might assist in the recovery of these viruses are not commonly available. The barrier to their successful cultivation is likely due to the species specificity of their spike proteins.
The development of reverse genetic systems for CoVs has had a major impact on research into these agents, enabling investigations into basic mechanisms both in viral replication and in pathogenesis. Two approaches, targeted recombination (Kuo et al., 2000) and in vitro cDNA assembly (Yount et al., 2002; Youn et al., 2005), which have been employed to perform reverse genetics in coronaviruses, are described here. Other reverse genetic systems have been developed for various coronaviruses. These employ recombinant vaccinia viruses, essentially as cloning vectors carrying a coronavirus genome (Casais et al., 2001; Thiel et al., 2001), or use a bacterial artificial chromosome (BAC) as a cloning vector (Almazan et al., 2000). Each of these systems has advantages and disadvantages. The in vitro cDNA assembly system has the advantage of ease of manipulation in terms of introducing mutations, since the genome has been cloned in seven separate cDNA fragments. The corresponding disadvantage is that the plasmids carrying these cDNAs must be maintained and ligated and subsequently transcribed in vitro to regenerate infectious virus. Vaccinia virus–based systems are easy to maintain, but it is slightly more cumbersome to generate mutant recombinant viruses using this system, and it exposes personnel to the added risk of working with vaccinia virus. The BAC‐based system established for TGEV contains a single cDNA representing the entire TGEV genome, and has the advantage that the genome is under the control of a CMV promoter, allowing virus to be regenerated by simple transfection into permissive cells rather than relying on in vitro transcription and electroporation of the resulting RNA into permissive cells. This is balanced by the awkwardness of introducing mutations into the 31‐kbp CoV insert in the BAC. All of these systems have been successfully utilized by various labs.
Critical Parameters and Troubleshooting
Infectivity assays
Both the plaque assay and the end‐point dilution assays are quite reliable, with the largest variable being the physiologic state of the cells used for the assays. We generally do not use cells that have been carried in cell culture more than 20 passages after defrosting, to try to minimize variability. For plaque assays, it is important to seed cells at a consistent concentration and to allow them to reach confluence. Once cells have reached confluence they should be used that day. Holding them an additional day will depress the titer and result in smaller plaques. A second critical parameter for the plaque assay is avoiding drying out the monolayer, either during washing the monolayers or viral adsorption, steps 3 and 4 respectively in Basic Protocol 1. For this reason, we generally will wash the monolayers only in groups of four 6‐well plates, and similarly start the viral adsorption four plates at a time. Also, we also do not aspirate all of the medium at these two steps, but leave ∼0.2 ml behind to avoid drying out the plates. It is better to leave too much medium behind than too little. If the plates are dried out, there will be a crescent of dead cells in the plates upon staining. Another critical step is, when adding the agarose overlay medium to the plates (Basic Protocol 1, step 6), to be sure to mix the virus inoculum with the overlay medium before the overlay hardens. Failure to do so will result in residual liquid between the overlay medium and the agarose, which will produce comet‐shaped plaques that are hard to count. The critical parameter for the end‐point dilution assay is the density of the cells at the time of infection. We have found that seeding the cells at 50,000 cells per well the evening prior to the assay has worked well in our hands. It is also possible to seed the cells first thing in the morning and perform the assay 6 to 8 hr after seeding, although this is rarely as convenient. The cell monolayer should be 75% confluent to just barely confluent at the time the assay is performed. Cell monolayers that are crowded will be hard to read, since some of the cells may come off the substrate by the end of the assay.
Growth and characterization of MHV stock virus
The most important variables for growing high‐titer stocks are the density and physiologic status of the cells, the particular strain of virus used, and the input multiplicity of infection. One strain of MHV, MHV‐A59, grows to very high titer (108 to 109 plaque‐forming units/ml); most other strains commonly used (MHV‐3, MHV‐2, MHV‐1, and MHV‐JHM) achieve titers between 106 and 108 pfu/ml), and some strains, such as MHV‐Yale, grow to still lower titers (105 to 106 pfu/ml). Especially for viruses that grow relatively poorly, it is important that the cell monolayers not be confluent when they are infected. We generally infect culture flasks that are 50% to 75% confluent, depending upon the strain of MHV we are growing. In Basic Protocol 2, we give a range of multiplicities of infection for growing stock virus, from 0.1 to 0.0001. The multiplicity of infection, the confluence of the monolayer, and the speed at which virus infection spreads through the cell monolayer are related variables. Generally, viruses that reach high titer replicate more quickly, and thus the infection spreads through the monolayer more rapidly. Hence, a lower multiplicity of infection can be used for these viruses. For viruses that spread somewhat more slowly, it is best to use multiplicities of infection toward the higher portion of this range. The density of the cells used to grow the stock should be adjusted for the estimated amount of time it will take for 90% or more of the cells to develop cytopathogenic effect, with cells infected at relatively low densities (∼50% confluence) if the infection is expected to take 3 days; cell monolayers that are ∼75% confluent are generally used for infections that are expected to take 48 hr or less. It often takes some experience with an individual virus to determine the optimal conditions for growing a virus stock, since these differ somewhat from strain to strain.
Mouse hepatitis virus purification
The most critical parameter for purifying unlabeled virus is to start with sufficient virus to be able to see a band after gradient centrifugation. It is often advisable, therefore, to titer the virus to be purified to be certain it has grown reasonably well. For MHV‐A59, a virus that grows to high titer, we generally start the purification with a minimum of 108 ml of clarified infected tissue culture supernatant. For viruses that do not grow as vigorously, we generally start with a minimum of 216 ml of clarified infected tissue culture supernatant, and preferably with 432 ml or more. For volumes larger than 216 ml, it is necessary to pellet the virus through a pad (Basic Protocol 3, steps 1 to 3), and after pouring off the supernatant liquid, including the pad, a second 216 ml can pelleted through a fresh pad in the same tube. The pellet from both centrifugations can then be resuspended in buffer and virus purified by density‐gradient centrifugation. It is important to be careful when preparing density gradients and overlaying the sample onto the gradients. It is important to disturb the gradient as little as possible in moving centrifuge tubes into and out of the centrifuge buckets.
Generation of recombinant MHV from cDNA
There are several critical points in utilizing the cDNA assembly reverse genetic system for coronaviruses. First, several of the large cDNAs have a tendency to undergo deletion when grown in E. coli if care is not taken in making plasmid preps. For this reason, we always grow the bacteria containing these plasmids at 30°C (Support Protocol 2) and check that the inserts are the correct size prior to use. We maintain DNA stocks of each plasmid that worked successfully, as well as glycerol stocks of the bacteria, so that we can retransform with a plasmid that we know to be correct. A second critical parameter relates to the ligation of the gel‐purified cDNAs. We always monitor each ligation step by agarose electrophoresis. If a ligation has not gone to completion, we add additional ligase and continue the incubation for an additional 6 to 24 hr. Gel‐purified cDNA fragments should be stored at −70°C for no longer than 8 weeks for best results at the ligation step. It is also important to use relatively fresh ligase. We recommend that you avoid using ligase that has been on hand for greater than 4 weeks. In vitro transcription kits can lose activity during storage in the freezer. We keep our T7 polymerase in a Stratacooler (Stratagene) and take care that it is kept cold when taken out of the freezer. It is essential to have a good transcription reaction and to avoid RNA degradation; therefore, good technique is called for. We monitor the transcription reaction by gel electrophoresis to be sure that we have synthesized very large RNA transcripts. Finally, when making recombinant viruses containing mutations, it is possible to recover undesired wild‐type contaminants. To minimize the likelihood of this occurring, we keep all of the reagents and buffers that we use for transcription and electroporation segregated and in small aliquots. The electroporations are performed in a hood that is only used for uninfected cells, and we always perform a negative control electroporation that has not received any RNA to monitor for contamination. It is essential to partially sequence any viruses recovered to ascertain that you have recovered the desired virus and not a contaminant. The sequencing should cover any mutations that you have introduced and regions containing marker mutations. For efficient electroporation, BHK‐R cells need to be growing well, and the density of the BHK‐R cells should be 50% to 80% of confluency prior to harvesting. This is readily achieved by setting up 175‐cm2 flasks with 7–8 × 107 cells 24 hr prior to harvesting, as we describe in Basic Protocol 4.
Targeted recombination
Many of the key parameters are generally similar for targeted recombination experiments and for the cDNA assembly system. Plasmids are maintained as described above and grown at 30°C. Good in vitro transcription reactions and good‐quality FCWF cells for nucleofection are key to obtaining recombinants. The FCWF cells are more fastidious than the other cells that we use, in that they require FBS and sometimes grow more slowly without apparent cause. The cells must be growing well to have reliable results with targeted recombination (Basic Protocol 5). In vitro–synthesized RNA transcripts must be stored at −80°C until thawed for nucleofection, and then kept briefly at 4°C until the nucleofection reaction takes place.
Anticipated Results
Plaque assay
Plaques are generally visible after 48 hr of incubation. The plaque size varies considerably among the different strains of MHV, and there is a lesser degree of variation from assay to assay. Although the plaques are typically clear for viruses that produce cell fusion of infected cells (most strains of MHV with the exception of MHV‐2), viruses that do not fuse cells may produce cloudy plaques. Plaque sizes for the strains of MHV that are used in our laboratory used are given in Table 5.
Table 5.
Plaque Sizes and Titers of Various Strains of MHV
|
Virus |
Plaque size (mm)a |
Titer of stocks |
|---|---|---|
|
MHV‐A59 |
2.0‐2.7 |
1 × 108‐1 × 109 |
|
MHV‐JHM |
1‐1.5 |
1 × 106‐1 × 107 |
|
MHV‐1 |
1.2‐1.6 |
3 × 106‐3 × 107 |
|
MHV‐3 |
1.5‐2.0 |
5 × 106‐5 × 107 |
|
MHV‐Yale |
0.4‐0.8 |
2 × 105‐8 × 105 |
Plaque assays performed in L2 cells and stained after 48‐hr incubation.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Growth of MHV stock virus
Different strains of MHV grow to different titers, with the MHV‐A59 strain achieving the highest titers. Most strains grow to titers that are approximately 107 pfu/ml. The ranges of titers that we achieve in growing working stocks of some different strains of MHV are given in Table 5.
Mouse hepatitis virus purification
When purifying virus by density‐gradient centrifugation, the virus is generally visible as an opalescent band in the middle of the gradient. This band can be visualized more easily in a darkened room with a light shined onto the tube against a black or dark gray background, as shown in Figure 2A. They buoyant densities of purified coronaviruses are 1.17 to 1.19 g/cm3.
Generation of recombinant MHV from cDNA
Cells electroporated with wild‐type genomic RNA usually develop widespread CPE 24 to 48 hr after electroporation, a sign of virus infection. Mutants with decreased rates of growth can be incubated as long as 72 hr. We consider a mutant to be lethal if no cytopathic effect develops in at least three totally independent electroporations on different days and three sequential blind passages of each electroporation in DBT cells fail to recover infectious virus. We always include at least one experiment in which electroporated cells were incubated at 34°C and 39.5°C to recover temperature‐sensitive virus. Also, we always verify that there are no problems with the cDNA fragments that we are using by performing at least one positive‐control electroporation to generate wild‐type virus in parallel to the mutant.
Generation of recombinant viruses by targeted recombination
It may take up to 3 days for CPE to spread to the DBT cells after electroporation of the infected FCWF cells (Basic Protocol 5, step 12). On infrequent occasions, we have observed CPE throughout the monolayer but have failed to recover recombinant virus that grows in murine cells. We also recover recombinants when it is not clear that spread of the infection to DBT cells has been achieved. Therefore, we now always pass supernatants from the nucleofection or electroporation through murine DBT cells at least one time to select for recombinant virus (Basic Protocol 5, steps 25 to 26). In successful experiments where recombination has taken place, we typically see cytopathic effect at the first passage in DBT cells.
Time Considerations
Infectivity assays
Both the plaque assay and the end‐point dilution assay typically take 2 days to perform once the cell monolayers are ready. It generally takes about 2 hr to perform either assay on a small number (<10) of samples. Larger assays, up to 140 samples, can be performed in single day by one person who is experienced in the assay, and scored 2 days later. To accomplish this, it is necessary the day prior to the assay to label all of the tubes needed for serial dilutions and the plates needed for the assays. Scoring very large assays may take the better part of a day.
Growth of MHV stock virus
Preparation of working virus stocks (P2 or above) from a single plaque (Basic Protocol 2) generally takes from 2 to 3 weeks, depending on which strain of MHV you are using. Less vigorous strains tend to take longer, particularly because, to obtain a large working stock with a poorly growing virus, it may be necessary to grow a P3 stock. It generally takes 3 days or less to grow P1 stocks from a single plaque and another 2 days to determine the titer. The same timetable holds true for the growth of P2 working stocks. Characterization of the stocks can take much longer, depending on how it is characterized. If the characterization of the virus stock is limited to determining the titer of the virus stock and the plaque morphology, these two determinations can be done in the same plaque assay.
Mouse hepatitis virus purification
Once clarified supernatants are obtained, it typically takes 2 days to purify virus through two cycles of density‐gradient centrifugation. Most of the time is centrifugation time, freeing the investigator to do other things. Preparing the first density gradients (Basic Protocol 3, step 5) generally takes about an hour and should be done during the 3‐hr initial pelleting step (Basic Protocol 3, step 3). It is generally more convenient to centrifuge the second density gradient overnight and collect the virus band the following day.
Generation of recombinant MHV from cDNA
Recovery of recombinant virus from cDNA clones generally takes 2 to 3 weeks. This time includes almost 2 weeks to propagate all seven plasmids and perform the gel purifications of the cDNAs. Once the cDNA fragments have been gel purified, it takes approximately 3 days for the ligation steps to be completed, and another day for the transcription and electroporation steps, followed by up to 3 days incubation. Preparation of plaque‐purified virus stocks from the lysates obtained from a successful electroporation may take another 2 to 3 weeks.
Generation of recombinant viruses by targeted recombination
Generation of a recombinant strain of MHV by targeted recombination takes approximately 3 to 4 weeks. We typically prepare enough linearized plasmid DNA for several experiments 1 to 2 days in advance of a targeted recombination experiment. Once the linearized plasmid DNA has been transcribed in vitro, we will either use the RNA product for nucleofection the same day (preferred) or store it at −80°C for no more than 48 hr prior to nucleofection of the FCWF cells. If we are going to use the nucleofected RNA on the same day as the transcription reaction, we do the experiment in the following order. On the day that we are going to perform the experiment, we first set up the DBT cells (infection and nucleofection, Basic Protocol 5, step 16), then set up the in vitro translation reaction as described. While the transcription reaction is incubating we then infect the FCWF cells with fMHV (infection and nucleofection, Basic Protocol 5, step 17). During the 4‐hr incubation of the fMHV‐infected FCWF cells, the in vitro transcription reaction is completed through Basic Protocol 5, step 12, and the analysis of the transcription product is started (step 13). It generally takes 1 to 2 hr to prepare the cells and perform the nucleofection procedure (infection and nucleofection, steps 19 to 25). Once the nucleofection reaction has been performed, there are up to 3 days of incubation time, followed by another 3 days for blind passaging the virus supernatant through DBT cells. Preparation of plaque‐purified virus working stocks from the lysates obtained in a successful targeted recombination may take an additional 2 to 3 weeks.
Literature Cited
- Almazan, F. , Gonzalez, J.M. , Penzes, Z. , Izeta, A. , Calvo, E. , Plana‐Duran, J. , and Enjuanes, L. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U.S.A. 97:5516‐5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais, R. , Thiel, V. , Siddell, S.G. , Cavanagh, D. , and Britton, P. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359–12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, S.R. , O'Shea, T.J. , Oko, L.M. , and Holmes, K.V. 2007. Detection of group 1 coronaviruses in bats in North America. Emerg. Infect. Dis. 13:1295‐1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, E.F. , Yount, B. , Sims, A.C. , Burkett, S. , Pickles, R.J. , and Baric, R.S. 2008. Systematic assembly of a full‐length infectious clone of human coronavirus NL63. J. Virol. 82:11948‐11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dveksler, G.S. , Pensiero, M.N. , Cardellichio, C.B. , Williams, R.K. , Jiang, G. , Holmes, K.V. , and Dieffenbach, C.W. 1991. Cloning of the mouse hepatitis virus (MHV) receptor: Expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 65:6881‐6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, L. , Gonzales, D.M. , Sarma, J.D. , and Lavi, E. 2004. A combination of mutations in the S1 part of the spike glycoprotein gene of coronavirus MHV‐A59 abolishes demyelination. J. Neurovirol. 10:41‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.F. , Feng, M. , Liu, P. , Millership, J.J. , Yount, B. , Baric, R.S. , and Leibowitz, J.L. 2005. Effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J. Virol. 79:14570‐14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, M.F. and Coen, D.M. 2001. Enzymatic amplification of DNA by PCR: Standard procedures and optimization. Curr. Protoc. Mol. Biol. 56:15.1.1–15.1.14. [DOI] [PubMed] [Google Scholar]
- Kuo, L. , Godeke, G.J. , Raamsman, M.J. , Masters, P.S. , and Rottier, P.J. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: Crossing the host cell species barrier. J. Virol. 74:1393‐1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, S.K. , Woo, P.C. , Li, K.S. , Huang, Y. , Tsoi, H.W. , Wong, B.H. , Wong, S.S. , Leung, S.Y. , Chan, K.H. , and Yuen, K.Y. 2005. Severe acute respiratory syndrome coronavirus‐like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 102:14040‐14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, S.K. , Li, K.S. , Huang, Y. , Shek, C.T. , Tse, H. , Wang, M. , Choi, G.K. , Xu, H. , Lam, C.S. , Guo, R. , Chan, K.H. , Zheng, B.J. , Woo, P.C. , and Yuen, K.Y. 2010. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome‐related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self‐limiting infection that allows recombination events. J. Virol. 84:2808‐2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Shi, Z. , Yu, M. , Ren, W. , Smith, C. , Epstein, J.H. , Wang, H. , Crameri, G. , Hu, Z. , Zhang, H. , Zhang, J. , McEachern, J. , Field, H. , Daszak, P. , Eaton, B.T. , Zhang, S. , and Wang, L.F. 2005a. Bats are natural reservoirs of SARS‐like coronaviruses. Science 310:676‐679. [DOI] [PubMed] [Google Scholar]
- Li, W. , Zhang, C. , Sui, J. , Kuhn, J.H. , Moore, M.J. , Luo, S. , Wong, S.K. , Huang, I.C. , Xu, K. , Vasilieva, N. , Murakami, A. , He, Y. , Marasco, W.A. , Guan, Y. , Choe, H. , and Farzan, M. 2005b. Receptor and viral determinants of SARS‐coronavirus adaptation to human ACE2. EMBO J. 24:1634‐1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso, A. , Decaro, N. , Schellen, P. , Rottier, P.J. , Buonavoglia, C. , Haijema, B.J. , and de Groot, R.J. 2008. Gain, preservation, and loss of a group 1a coronavirus accessory glycoprotein. J. Virol. 82:10312‐10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontiveros, E. , Kuo, L. , Masters, P.S. , and Perlman, S. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230‐238. [DOI] [PubMed] [Google Scholar]
- Reed, L.J. and Muench, H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hygiene 27:493‐497. [Google Scholar]
- Robb, J.A. and Bond, C.W. 1979. Pathogenic murine coronaviruses I. Characterization and biologic behavior in vitro and virus‐specific intracellular RNA of strongly neurotopic JHMV and weakly neurotopic A59V viruses. Virology 94:352‐370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephensen, C.B. , Casebolt, D.B. , and Gangopadhyay, N.N. 1999. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 60:181‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, V. and Siddell, S.G. 2005. Reverse genetics of coronaviruses using vaccinia virus vectors In Coronavirus Genome Structure and Replication, Vol. 287 (Brian D. and Baric R., eds.) pp. 199‐227. Springer, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, V. , Herold, J. , Schelle, B. , and Siddell, S.G. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273‐1281. [DOI] [PubMed] [Google Scholar]
- Vijaykrishna, D. , Smith, G.J. , Zhang, J.X. , Peiris, J.S. , Chen, H. , and Guan, Y. 2007. Evolutionary insights into the ecology of coronaviruses. J. Virol. 81:4012‐4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas, D. 2000. Agarose gel electrophoresis. Curr. Protoc. Mol. Biol. 51:10.4.1‐10.4.8. [DOI] [PubMed] [Google Scholar]
- Wesseling, J.G. , Vennema, H. , Godeke, G.J. , Horzinek, M.C. , and Rottier, P.J. 1994. Nucleotide sequence and expression of the spike (S) gene of canine coronavirus and comparison with the S proteins of feline and porcine coronaviruses. J. Gen. Virol. 75:1789‐1794. [DOI] [PubMed] [Google Scholar]
- Wilson, L. , McKinlay, C. , Gage, P. , and Ewart, G. 2004. SARS coronavirus E protein forms cation‐selective ion channels. Virology 330:322‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn, S. , Leibowitz, J.L. , and Collisson, E.W. 2005. In vitro assembled, recombinant infectious bronchitis viruses demonstrate that the 5a open reading frame is not essential for replication. Virology 332:206‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount, B. , Curtis, K.M. , and Baric, R.S. 2000. Strategy for systematic assembly of large RNA and DNA genomes: Transmissible gastroenteritis virus model. J. Virol. 74:10600‐10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount, B. , Denison, M.R. , Weiss, S.R. , and Baric, R.S. 2002. Systematic assembly of a full‐length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065‐11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount, B. , Curtis, K.M. , Fritz, E.A. , Hensley, L.E. , Jahrling, P.B. , Prentice, E. , Denison, M.R. , Geisbert, T.W. , and Baric, R.S. 2003. Reverse genetics with a full‐length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 100:12995‐13000. [DOI] [PMC free article] [PubMed] [Google Scholar]


