Abstract
In this study, we evaluated a concatenated low pH (pH 3) and high pH (pH 10) reversed-phase liquid chromatography strategy as a first dimension for two-dimensional liquid chromatography tandem mass spectrometry (“shotgun”) proteomic analysis of trypsin-digested human MCF10A cell sample. Compared with the more traditional strong cation exchange method, the use of concatenated high pH reversed-phase liquid chromatography as a first-dimension fractionation strategy resulted in 1.8- and 1.6-fold increases in the number of peptide and protein identifications (with two or more unique peptides), respectively. In addition to broader identifications, advantages of the concatenated high pH fractionation approach include improved protein sequence coverage, simplified sample processing, and reduced sample losses. The results demonstrate that the concatenated high pH reversed-phased strategy is an attractive alternative to strong cation exchange for two-dimensional shotgun proteomic analysis.
Keywords: 2-D chromatography, Concatenation, Fractionation, High pH RP, Low pH RP, Technology
1 Introduction
Two-dimensional liquid chromatography (2-D LC) is commonly used in MS/MS proteomic approaches to improve both analytical dynamic range and proteome coverage [1–4]. The effectiveness of the 2-D LC separation depends on the chromatographic resolving power (e.g. separation peak capacity) of each separation element, as well as on the orthogonality of the two separation elements [5]. To date, low pH reversed-phase (RP) is used most often as the second LC dimension because of the ease in which it couples to MS via electrospray ionization.
With high orthogonality to RPLC, strong cation exchange (SCX) chromatography is the most widely adopted method for first-dimension fractionation [6–9]. Other fractionation methods with good orthogonality to RPLC include isoelec-tric focusing, free flow electrophoresis, and hydrophilic interaction LC, among others. Although some reports improved reproducibility and resolution [9, 10], few methods provide better overall performance than SCX–RPLC [9–12]. In terms of coverage, applied electrostatic repulsion–hydro-philic interaction chromatography (ERLIC), a mixed mode separation involving charge transfer and hydrophilic interactions is reported to increase the number of peptide and protein identified in a rat proteome relative to those obtained using SCX [13].
RPLC is another option for the first-dimension separation and affords better resolution than SCX [14]. RPLC uses low-salt or salt-free buffers that provide cleaner samples for downstream proteomic analyses than SCX and have the added potential for desalting often high salt-containing (e.g., urea or guanidine HCl) trypsin-digested proteomic samples. However, RP is not widely used as the first-dimensional separation due to the consideration of orthogonality when RP separations are used for both dimensions in 2-D LC [5]. Recently, high-pH RPLC has been shown to be semi-orthogonal to low-pH RPLC, which was explained by the dramatic change of charge distribution within the peptide chain with the RPLC mobile-phase pH values [5, 15]. RPLC–RPLC approach operated at different pH values has been shown to possess an orthogonality comparable to SCX–RPLC the most commonly applied approach in proteomic analysis [5, 15–20].
To improve the orthogonality of RPLC–RPLC separation, we explored the concatenated RPLC fractionation approach for the first dimension in a 2-D LC separation. Concatenating multiple early, middle, and late RPLC fractions that have little overlap should improve analysis coverage while maintaining high throughput. The concatenation concept was previously introduced to reduce analysis time for practical implementation of 2-D HPLC [21]. RPLC–RPLC has been demonstrated for phosphopeptide analysis by pooling two fractions with equal time interval [22]; however, the effectiveness of concatenated RPLC–RPLC compared with SCX–RPLC is unknown. To evaluate the benefit of the concatenation strategy and whether it is superior to SCX–RPLC, we applied various pH RPLC–RPLCs for MS/MS analysis of trypsin-digested human MCF10A cell proteins. We showed that concatenated high-pH RPLC–low pH RPLC is an attractive alternative to SCX–RPLC for MS/MS proteomic analysis with improvement of protein sequence coverage.
2 Materials and methods
2.1 Materials
Ammonium bicarbonate and ACN were purchased from Fisher Scientific (Fair Lawn, NJ). Sequencing grade, modi-fied trypsin was purchased from Promega (Madison, WI). Bicinchoninic acid (BCA) assay reagents and standards were obtained from Pierce (Rockford, IL). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO). Water was purified using a Barnstead Nanopure Infinity water purifi-cation system (Dubuque, IA).
2.2 Cell culture and preparation of digested lysate
Human breast epithelial cells (MCF10A) were transfected with a pQCXIH vector and were cultured in DMEM/F12 (Invitrogen, Carlsbad, CA) supplemented with 5% horse serum (Invitrogen), 20 ng/mL EGF, 0.5 μg/mL hydro-cortisone, 100 ng/mL cholera toxin, 10 μg/mL insulin, and 50 μg/mL penicillin/streptomycin until 70–80% confluence was reached. The cells were then lysed in a buffer containing 8 M urea, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 50 mM ammonium bicarbonate, one-third tablet of protease inhibitor, and 2 mM sodium ortho-vanadate. Proteins were denatured, reduced, and alkylated after which tryptic digestions were performed at an enzyme/substrate ratio of 1:50. For method comparison between SCX and RP, digested peptides were cleaned by flowing through a 1 mL solid-phase extraction C18 column (Discovery DSC-18, SUPELCO, Bellefonte, PA). Samples were concentrated using a Speed-Vac SC 250 Express (Thermo Savant, Holbrook, NY) and stored at −80°C until time for analysis. A 300.0 μg desalted peptide sample was used for each SCX, low-pH RPLC, and high-pH RPLC fractionation. A 300.0 μg nondesalted protein digest was used to evaluate the potential of high-pH approach for desalting.
2.3 SCX fractionation
SCX was performed using an Agilent 1100 HPLC System (Agilent, Palo Alto, CA) equipped with a quaternary pump, degasser, diode array detector, peltier-cooled autosampler, and fraction collector (set at 4°C for all samples). Columns included a 200 × 2.1 mm column that contained 5 μM porous (300-Å) PolySulfoethyl A stationary phase and a 10 × 2.1 mm column (PolyLC, Columbia, MD) as a guard column. The solvent consisted of 10 mM ammonium formate (pH 3.0) and 25% ACN as mobile phase (A) and 500 mM ammonium formate (pH 6.8) and 25% ACN as mobile phase (B). The separation was accomplished at a mobile phase flow rate of 0.2 mL/min using the following linear gradient: 100% A in 10 min, from 100% A to 50% B in 50 min, to 100% B in 10 min, and held at 100% B for an additional 15 min. Forty fractions were collected along with the LC separation that were subsequently pooled into 15 fractions based on collection order. Fractions were dried in a Speed-Vac and then stored at −80°C until LC-MS/MS analysis.
2.4 Low-pH RPLC fractionation and concatenation
The low-pH RPLC separation was performed with an XBridge C18, 250 × 4.6 mm analytical column containing 5 μM particles and equipped with a 20 × 4.6 mm guard column (Waters, Milford, MA); flow rate was 0.5 mL/min. The mobile-phase A consisted of 0.01% TFA and 0.585% HOAc (pH 3.0) and B 0.01% TFA, 0.585% HOAc, and 90% ACN (pH 3.0). Sample separation was accomplished using the following linear gradient: from 0 to 5% B in 10 min, from 5 to 35% B in 60 min, from 35 to 70% B in 15 min, and held at 70% B for an additional 10 min. Sixty fractions were collected along with the LC separation and were concatenated into 15 fractions by combining fractions 1, 16, 31, 46; 2, 17, 32, 47; and so on (Fig. 1A). The samples were dried in a Speed-Vac and stored at −80°C until LC-MS/MS analysis.
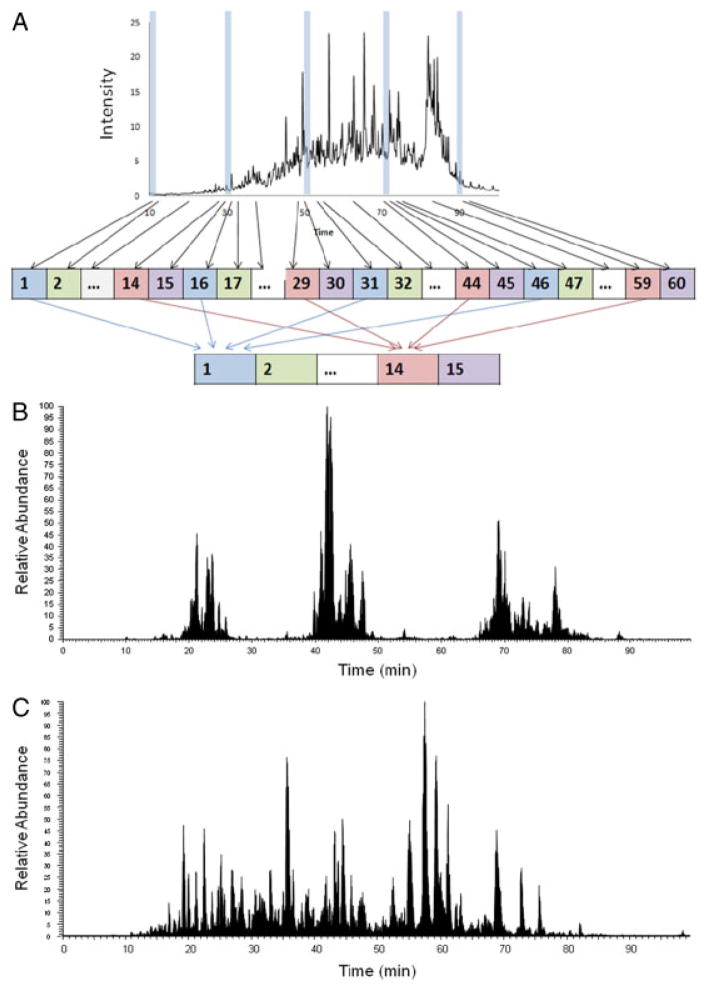
Figure 1.
Scheme of concatenation strategy applied to the first-dimensional low-pH RP and high-pH RP fractionation (A), second-dimensional LC/MS/MS base peak chromatography of low-pH RPLC (B) and high-pH RPLC (C) at postconcatenation fraction 10.
2.5 High-pH RPLC fractionation and concatenation
High-pH RPLC was performed on the same analytical and guard columns and at the same flow rate as those used for low-pH RPLC. The solvent consisted of 10 mM ammonium formate (pH 10) as mobile phase (A) and 10 mM ammonium formate and 90% ACN (pH 10) as mobile-phase B. Sample collection and concatenation were accomplished as described above for the low-pH fractions. Samples were dried in Speed-Vac and stored at −80°C until LC-MS/MS analysis.
2.6 Capillary RPLC-MS/MS analyses
A custom HPLC system was configured using 65-mL Isco Model 65D syringe pumps (Isco, Lincoln, NE), 2-position Valco valves (Valco Instruments, Houston, TX), and a PAL autosampler (Leap Technologies, Carrboro, NC) to allow for fully automated sample analysis across four separate HPLC columns. RP capillary HPLC columns were manufactured in-house by slurry packing 3 μm Jupiter C18 particles (Phenomenex, Torrance, CA) into a 70 cm × 75 μm id fused silica capillary tubing (Polymicro Technologies, Phoenix, AZ). Mobile phases A and B consisted of 0.1% formic acid in water and 0.1% formic acid in ACN, respectively. The HPLC system was equilibrated at 10 Kpsi with 100% A, and 50 min later, a near exponential gradient was achieved by switching to 100% B. A 30 cm × 15 μm id fused silica tubing was used to split ∼20 μL/min of flow before it reached the injection valve (sample loop, 5 μL). Mobile-phase flow through the capillary HPLC column was ∼400 nL/min (measured using mobile-phase A), and the split flow controlled the gradient speed under constant pressure operation (10 Kpsi). The samples were divided into three batches of 20 fractions (each batch contained five fractions from each of the four separation approaches) to reduce potential effects stemming from analytical variability. The order in which the 20 fractions within a batch were analyzed was randomly assigned after which the samples were analyzed using the same LC column in this predefined order.
Tandem mass spectra were obtained using an LTQ XL mass spectrometer (Thermo Scientific, San Jose, CA) and chemically etched electrospray emitters for electrospray ioni-zation [23]. The heated capillary temperature and spray voltage were 200°C and 2.2 kV, respectively. Data acquisition started 30 min after sample injection (i.e. 10 min into the gradient) and continued for 85 min. Full spectra (automatic gain control [AGC] 3 × 104) were collected from m/z 400 to 2000, followed by data-dependent ion-trap MS/MS spectra (automatic gain control 1 × 104) of the ten most abundant ions, using 35% collision energy. A dynamic exclusion time of 60 s was used to discriminate against the previously analyzed ions.
2.7 Data processing
Peptides were identified using SEQUEST (Thermo Finni-gan) to search the human International Protein Index (ipi.HUMAN.v3.54) and reversed human IPI protein databases with dynamic modification of methionine (+15.9949) and static modification of cysteine (+57.0215) [24]. Fragment ion tolerance was set at 0.5 Da. A false discovery rate (FDR) of 1% was estimated using the formula 100 × 2 × decoy hits/all hits % [25], and applied to all data sets at the total peptide level. To remove redundant protein entries, the software ProteinProphet [26] was applied as a clustering tool to group related proteins into a single group entry. The theoretical pI and hydrophobicity values of peptides were calculated using in-house software (Protein-DigestionSimulator). Identified proteins were categorized using STRAP tool [27].
3 Results and discussion
In this study, we explored both low-pH and high-pH RPLC concatenated fractionation approaches as an off-line first dimension in 2-D LC-MS/MS analyses of trypsin-digested human MCF10A cell proteins. Results were compared with those obtained using more traditional SCX LC for off-line fractionation. (The experimental workflow is shown in Supporting Information Fig. 1). To ensure a fair comparison, we started with the same amount of peptide sample for each approach and employed the same aliquot of peptide fractions in the second dimension. We also randomized the sample order for LC-MS/MS analysis and applied consistent database search and data analysis parameters.
3.1 Concatenation strategy
Figure 1A shows the RPLC concatenation strategy we applied in this study. To improve orthogonality of RP–RP separation, 60 fractions were collected along with the first-dimensional RPLC separation and were concatenated into 15 fractions by combining four preconcatenation RPLC fractions with equal time interval into one postconcatena-tion fraction, such as pooling preconcatenation fractions 1, 16, 31, and 46 into postconcatenation fraction 1. Figure 1B and C are examples of base peak chromatography from postconcatenation fractions of low- and high-pH RPLC, which show the benefit of concatenation approach. For the low-pH RP–low-pH RPLC, without concatenation, only one peptide cluster would be expected due to the poor orthogonality. With the concatenation, several peptide clusters were observed across a wide range of LC elution time, indicating good use of the separation space in the second-dimensional separation. However, significant separation space remained un-utilized, indicated by the gaps between the peptide clusters. For the high-pH RP–low-pH RPLC, the separation space was widely covered, as shown in Fig. 1C.
3.2 Peptide and protein identifications
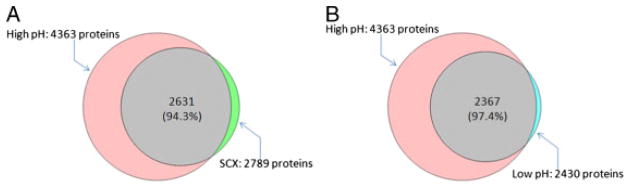
Table 1 summarizes the numbers of peptides and proteins identified from 2-D LC-MS/MS of SPE-desalted samples for each fractionation approach. The detailed information of identified proteins and peptides are provided in Supporting Information table. High-pH RPLC provided the largest number of peptide and protein identifications relative to SCX. Although low-pH RPLC is anticipated to have the least orthogonality to second-dimensional low-pH RPLC, it resulted in the comparable number of peptide and protein identifications to SCX, presumably due to the benefit of fraction concatenation that improves the orthogonality. In a previous study, concatenated pH 7.5 RPLC yielded 30% more phosphopeptide identifications than obtained using a conventional RPLC approach [22]. In our study, concatenated pH 3 RPLC provided an ~2-fold increase in the number of peptide and protein identifications compared with the nonconcatenated pH 3 RPLC approach (data not shown). High-pH RPLC fractionation prior to LC-MS/MS increased the number of protein identifications (≥2 unique peptides per protein) by n1.6- and 1.8-fold compared with SCX and low-pH RPLC, respectively, and covered 94.3 and 97.4% of proteins identified with the SCX and low-pH RPLC approaches, respectively (Fig. 2). Additionally, high-pH RPLC provided the highest average protein sequence coverage among the approaches examined in this study (Table 1), and would be expected to benefit the reliability of quantitative proteomic measurements.
Table 1.
Numbers of peptide and protein identifications (1% false discovery rate) for the different fractionation methods
| SCX | Concatenated low-pH RPLC | Concatenated high-pH RPLC | |
|---|---|---|---|
| Total peptides | 49 854 | 51 889 | 81 974 |
| Unique peptides | 20 573 | 18 750 | 37 633 |
| Unique proteins | 4286 | 3753 | 5907 |
| Proteins (≥2 unique peptides) | 2789 | 2430 | 4363 |
| Total peptides/unique peptides | 2.4 | 2.8 | 2.2 |
| Average protein sequence coverage (%) | 15.1 | 15.8 | 19.4 |
Figure 2.
Venn diagrams showing the overlap of proteins identified using high-pH RPLC with proteins identified using SCX (A) and Low-pH-RPLC (B).
The physicochemical properties of these peptides are shown in Supporting Information Fig. 2. It appears that higher number of peptide identifications of high-pH RPLC–low pH RPLC were primarily from peptides with low pI (e.g. 4–6). This may be due to the relatively low resolution of SCX separations for these low pI early fractions. For charge-based SCX separation (loading condition at pH ~3), we have collected the flow-through fractions; however, most peptides should be retained in the column and elute in early fractions. For high-pH RPLC, the peptides containing pI 4–6 widely distributed across fractions based on their hydrophobicity, and therefore the complexity of peptides with the low pI is reduced, increasing the likelihood of peptide detection and identification.
Supporting Information Fig. 3 further shows that high-pH RPLC leads to more protein identifications in cellular component and molecular function categories compared with the other two fractionation approaches, and that there is no significant percentage bias toward any category.
3.3 Effectiveness of the 2-D separation schemes
The overall efficiency of 2-D separations is determined by the resolution power of each separation element and the orthogonality of the two separation elements [28].
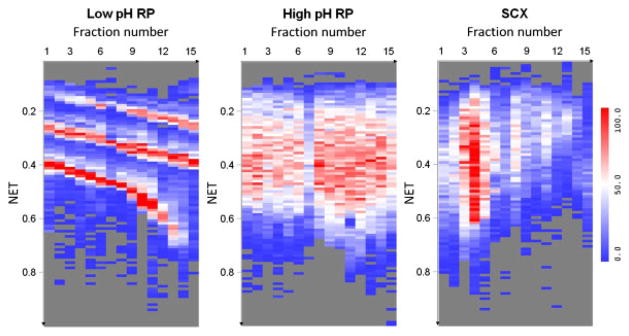
For the different fractionation methods (Supporting Information Fig. 4), low pH and high pH resulted in higher resolution separations of tryptically digested peptides than SCX, evidenced by the number of resolvable LC peaks. This observation results from the faster partitioning process on the reversed-phase surface when compared with the charged exchange surface. We have further optimized the SCX gradient for a more uniform LC elution profile using the same salt ammonium formate and similar number of peptides and proteins was identified (results not shown here). The volatile salt ammonium formate was selected for this study to facilitate the downstream MS analysis. Compared with a KCl salt gradient using a phosphate buffer, formate ions generally have weaker eluting strength than chloride ions in ion-exchange chromatography. The salt effects on the resolution of SCX separations should be limited [7]. In this study, we performed elution using pH as well as salt gradient, as we did in the previous studies [29–32]. Zhang et al. have previously reported that continuous pH gradient provides higher resolution fractionation in the first (SCX) dimension with good column recovery [33]. We therefore expect that the salt/pH gradient will have comparable or better performance compared with salt gradient alone using phosphate buffer. Figure 3 shows the number of distinct peptides identified in each LC-normalized elution time (NET) bin for each fraction for the three different approaches (NET values were obtained by regressing the elution times of the observed peptides against the predicted NET values to transform the elution times to a range of 0–1 [34]). The distribution of the number of unique peptides across fractions (first dimension) and the online low-pH RPLC elution time (second dimension) illustrates the orthogonality of the first-dimension separation (i.e. low-pH RPLC, high-pH RPLC, or SCX) with the second-dimension (low pH) RPLC separation. Using low-pH RPLC as the first dimension, the differences in both pH and selectivity relative to the second-dimension separation are small, i.e., low orthogonality with limited separation power as evidenced by the three diagonal bands shown in Fig. 3. Without concatenation, only one band would appear across the 2-D display [5, 14]. On the contrary, high-pH RPLC–low-pH RPLC uniformly covers the 2-D display and indicates improved orthogonality as a result of changes in peptide charges upon altering pH in the second dimension, as well as the concatenation strategy applied in this study. Without concatenation, high-pH RPLC is reported as being semi-orthogonal to low-pH RPLC [5, 21]. An additional advantage of concatenation is that it reduces overall analysis time by pooling several fractions into one. Dwivedi et al. showed that combining one early and one middle pH 10 RPLC fraction, reduced analysis time by 50% with only a 15–20% reduction in the number of identified proteins [21]. Theoretically, SCX has good orthogonality to RPLC; however, most tryptic peptides carry 2+, 3+, and 4+ charges that form big clusters during SCX fractionation, which leads to a nonuniform use of the 2-D space and a reduction in overall separation efficiency.
Figure 3.
Two-dimensional displays of separated species, illustrating the orthogonality of the first-dimension separation (i.e. low-pH RPLC, high-pH RPLC, or SCX) with the second-dimension (low pH) RPLC separation. The heat maps illustrate the distinct number of unique peptides identified in each bin, with the X-axis representing the postconcatenation fraction number (1-D) and the Y-axis, the NET of the second-dimensional RPLC (2-D). The number of peptides (low to high) is indicated by the blue–white–red color scale.
For all of the off-line fractionation methods investigated in this study, approximately more than 85% of the peptide identifications derived consistently from single fractions, which indicates little overlap between neighboring fractions and good separation efficiency. The average and standard deviations of the overlap are 0.14±0.02, 0.14±0.01, and 0.15±0.08 for low-pH RPLC, high-pH RPLC, and SCX, respectively. These results confirm the effective utilization of separation power provided by each separation element in the 2-D LC.
3.4 High-pH RPLC capability for desalting
Based on the encouraging results obtained using high-pH RPLC to fractionate desalted samples, we evaluated the method using samples in which salts had not been removed. Results obtained using high-pH RPLC-LC-MS/MS to analyze peptide samples that contained 1 M urea resulted in identification of 3948 unique proteins that overlapped 3580 unique proteins obtained for SPE-cleaned samples. This high degree of overlap (91%) indicates that desalting is not necessary prior to high-pH RPLC fractionation. The advantages of a combined fractionation-desalting operation are that it reduces sample preparation time and decreases sample losses, which is critical for small-sized biological/clinical samples.
4 Concluding remarks
In this study, we demonstrated the benefits of a concatenated RPLC fractionation method for 2-D LC-MS/MS proteomic analysis. Concatenated low-pH RPLC fractionation method resulted in comparable peptide and protein identifications to SCX. For the high-pH RPLC, this new method provided 1.8-and 1.6-fold increases in peptide and protein identifications, respectively, when compared with SCX and improved protein sequence coverage. Additionally, the new method eliminated the need to desalt samples prior to 2-D LC-MS, which reduces the sample loss and sample processing time. These results indicate that the concatenated high-pH RPLC–low–pH RPLC is a promising alternative to conventional SCX–low-pH RPLC-MS/MS proteomics. RPLC–RPLC with concatenation can be automated in an off-line format, which is flexible and allows multiple analyses of individual fractions to improve coverage [35]. Our next step is to evaluate a concatenated SCX approach for peptide identification, as the concatenation of SCX fractions may also afford more uniform peptide distributions.
Supplementary Material
Acknowledgments
This research was supported by NIH grants GM068487 (R. L. K.), CA097022 (R. L. K.) and RR018522 (R. D. S.). Work was performed in the NIH NCRR P41 Biomedical Technology Research Center for Proteomics located in the Environmental Molecular Sciences Laboratory (EMSL), a US Department of Energy (DOE) Office of Biological and Environmental Science national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RL01830. The authors thank Dr. Andrew Alpert from PolyLC Inc., for his helpful input on SCX chromatography.
Abbreviations
- NET
normalized elution time
- SCX
strong cation exchange
Footnotes
The authors have declared no conflict of interest.
References
- 1.Wang H, Chang-Wong T, Tang HY, Speicher DW. Comparison of extensive protein fractionation and repetitive LC-MS/MS analyses on depth of analysis for complex proteomes. J Proteome Res. 2009;9:1032–1040. doi: 10.1021/pr900927y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang Y, Robinson DP, Foster LJ. Quantitative analysis of proteome coverage and recovery rates for upstream fractionation methods in proteomics. J Proteome Res. 2010;9:1902–1912. doi: 10.1021/pr901063t. [DOI] [PubMed] [Google Scholar]
- 3.Fournier ML, Gilmore JM, Martin-Brown SA, Washburn MP. Multidimensional separations-based shotgun proteomics. Chem Rev. 2007;107:3654–3686. doi: 10.1021/cr068279a. [DOI] [PubMed] [Google Scholar]
- 4.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotech. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 5.Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of separation in two-dimensional liquid chromatography. Anal Chem. 2005;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- 6.Opiteck GJ, Lewis KC, Jorgenson JW, Anderegg RJ. Comprehensive on-line LC/LC/MS of proteins. Anal Chem. 1997;69:1518–1524. doi: 10.1021/ac961155l. [DOI] [PubMed] [Google Scholar]
- 7.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LCMS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2002;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 8.Ishihama Y, Rappsilber J, Mann M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J Proteome Res. 2006;5:988–994. doi: 10.1021/pr050385q. [DOI] [PubMed] [Google Scholar]
- 9.Slebos RJC, Brock JWC, Winters NF, Stuart SR, et al. Evaluation of strong cation exchange versus isoelectric focusing of peptides for multidimensional liquid chromatography-tandem mass spectrometry. J Proteome Res. 2008;7:5286–5294. doi: 10.1021/pr8004666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manadas B, English JA, Wynne KJ, Cotter DR, Dunn MJ. Comparative analysis of offgel, strong cation exchange with pH gradient, and RP at high pH for first-dimensional separation of peptides from a membrane-enriched protein fraction. Proteomics. 2009;9:5194–5198. doi: 10.1002/pmic.200900349. [DOI] [PubMed] [Google Scholar]
- 11.Malmström J, Lee H, Nesvizhskii AI, Shteynberg D, et al. Optimized peptide separation and identification for mass spectrometry based proteomics via free-flow electrophoresis. J Proteome Res. 2006;5:2241–2249. doi: 10.1021/pr0600632. [DOI] [PubMed] [Google Scholar]
- 12.Boersema P, Mohammed S, Heck A. Hydrophilic interaction liquid chromatography (HILIC) in proteomics. Anal Bioanal Chem. 2008;391:151–159. doi: 10.1007/s00216-008-1865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao P, Guo T, Li X, Adav SS, et al. Novel application of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) in shotgun proteomics: comprehensive profiling of rat kidney proteome. J Proteome Res. 2010;9:3520–3526. doi: 10.1021/pr100037h. [DOI] [PubMed] [Google Scholar]
- 14.Gilar M, Olivova P, Daly AE, Gebler JC. Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J Sep Sci. 2005;28:1694–1703. doi: 10.1002/jssc.200500116. [DOI] [PubMed] [Google Scholar]
- 15.Delmotte N, Lasaosa M, Tholey A, Heinzle E, Huber CG. Two-dimensional reversed-phase ×ion-pair reversed-phase HPLC: an alternative approach to high-resolution peptide separation for shotgun proteome analysis. J Proteome Res. 2007;6:4363–4373. doi: 10.1021/pr070424t. [DOI] [PubMed] [Google Scholar]
- 16.Gilar M, Olivova P, Chakraborty AB, Jaworski A, et al. Comparison of 1-D and 2-D LC MS/MS methods for proteomic analysis of human serum. Electrophoresis. 2009;30:1157–1167. doi: 10.1002/elps.200800630. [DOI] [PubMed] [Google Scholar]
- 17.Toll H, Oberacher H, Swart R, Huber CG. Separation, detection, and identification of peptides by ion-pair reversed-phase high-performance liquid chromatography-electrospray ionization mass spectrometry at high and low pH. J Chromatogr A. 2005;1079:274–286. doi: 10.1016/j.chroma.2005.03.121. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura T, Kuromitsu J, Oda Y. Evaluation of comprehensive multidimensional separations using reversed-phase, reversed-phase liquid chromatography/mass spectrometry for shotgun proteomics. J Proteome Res. 2008;7:1007–1011. doi: 10.1021/pr7005878. [DOI] [PubMed] [Google Scholar]
- 19.Gilar M, Jaworski A, Olivova P, Gebler JC. Peptide retention prediction applied to proteomic data analysis. Rapid Commun Mass Spectrom. 2007;21:2813–2821. doi: 10.1002/rcm.3150. [DOI] [PubMed] [Google Scholar]
- 20.Callipo L, Capriotti AL, Cavaliere C, Gubbiotti R, et al. Evaluation of different two-dimensional chromatographic techniques for proteomic analysis of mouse cardiac tissue. Biomed Chromatogr. 2010 doi: 10.1002/bmc.1487. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 21.Dwivedi RC, Spicer V, Harder M, Antonovici M, et al. Practical implementation of 2D HPLC scheme with accurate peptide retention prediction in both dimensions for high-throughput bottom-up proteomics. Anal Chem. 2008;80:7036–7042. doi: 10.1021/ac800984n. [DOI] [PubMed] [Google Scholar]
- 22.Song C, Ye M, Han G, Jiang X, et al. Reversed-phase-reversed-phase liquid chromatography approach with high orthogonality for multidimensional separation of phos-phopeptides. Anal Chem. 2009;82:53–56. doi: 10.1021/ac9023044. [DOI] [PubMed] [Google Scholar]
- 23.Kelly RT, Page JS, Luo Q, Moore RJ, et al. Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal Chem. 2006;78:7796–7801. doi: 10.1021/ac061133r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 25.Liu T, Qian WJ, Gritsenko MA, Camp DG, et al. Human plasma N-glycoproteome analysis by immunoaffi-nity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 27.Bhatia VN, Perlman DH, Costello CE, McComb ME. Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal Chem. 2009;81:9819–9823. doi: 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dowell JA, Frost DC, Zhang J, Li L. Comparison of two-dimensional fractionation techniques for shotgun proteomics. Anal Chem. 2008;80:6715–6723. doi: 10.1021/ac8007994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu T, Qian WJ, Chen WNU, Jacobs JM, et al. Improved proteome coverage by using high efficiency cysteinyl peptide enrichment: the human mammary epithelial cell proteome. Proteomics. 2005;5:1263–1273. doi: 10.1002/pmic.200401055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian WJ, Liu T, Monroe ME, Strittmatter EF, et al. Probability-based evaluation of peptide and protein identi-fications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2004;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 31.Liu T, Qian WJ, Strittmatter EF, Camp DG, et al. High-throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal Chem. 2004;76:5345–5353. doi: 10.1021/ac049485q. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Qian WJ, Chin MH, Petyuk VA, et al. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J Proteome Res. 2006;5:361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Lanham KA, Peterson RE, Heideman W, Li L. Characterization of the adult zebrafish cardiac proteome using online pH gradient strong cation exchange-RP 2-D LC coupled with ESI MS/MS. J Sep Sci. 2010;33:1462–1471. doi: 10.1002/jssc.200900780. [DOI] [PubMed] [Google Scholar]
- 34.Zimmer JSD, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Y, Tolić N, Masselon C, Paša-Tolić L, et al. Ultrasensitive proteomics using high-efficiency on-line micro-SPE-NanoLC-NanoESI MS and MS/MS. Anal Chem. 2003;76:144–154. doi: 10.1021/ac030096q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.