Abstract
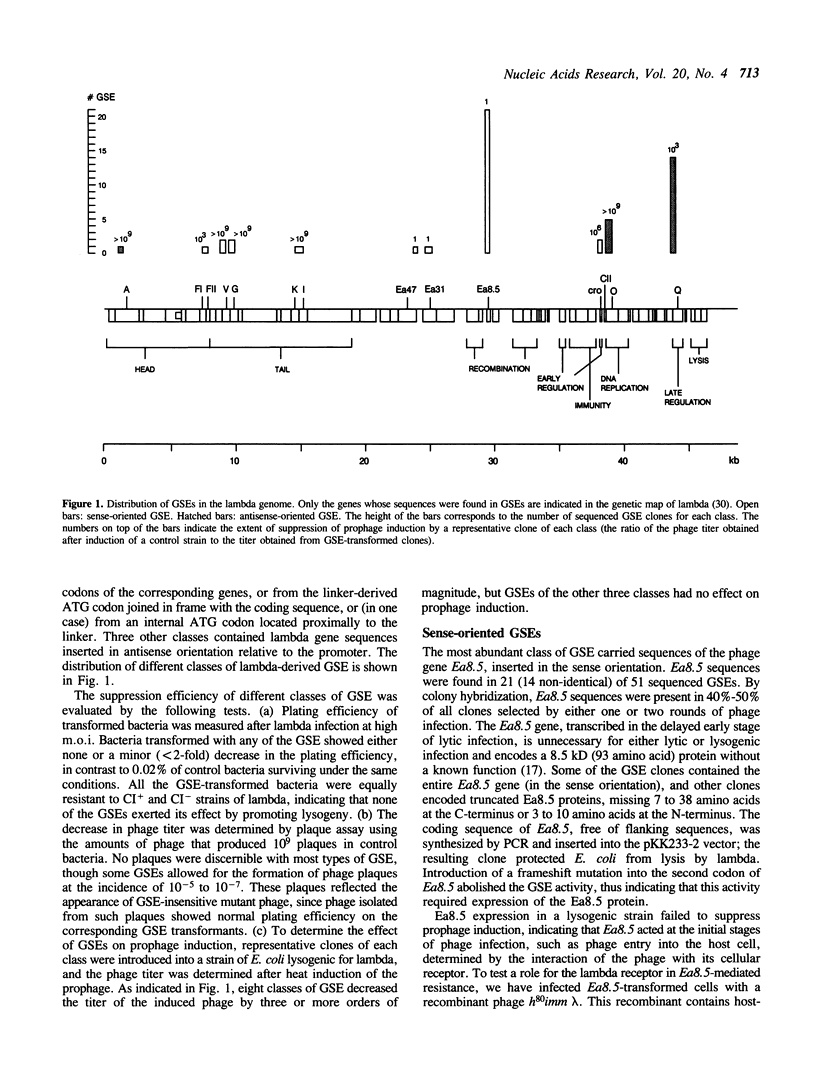
Selective inhibition of specific genes can be accomplished using genetic suppressor elements (GSEs) that encode antisense RNA, dominant negative mutant proteins, or other regulatory products. GSEs may correspond to partial sequences of target genes, usually identified by trial and error. We have used bacteriophage lambda as a model system to test a concept that biologically active GSEs may be generated by random DNA fragmentation and identified by expression selection. Fragments from eleven different regions of lambda genome, encoding specific peptides or antisense RNA sequences, rendered E. coli resistant to the phage. Analysis of these GSEs revealed some previously unknown functions of phage lambda, including suppression of the cellular lambda receptor by an 'accessory' gene of the phage. The random fragment selection strategy provides a general approach to the generation of efficient GSEs and elucidation of novel gene functions.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann E., Brosius J. "ATG vectors' for regulated high-level expression of cloned genes in Escherichia coli. Gene. 1985;40(2-3):183–190. doi: 10.1016/0378-1119(85)90041-1. [DOI] [PubMed] [Google Scholar]
- Baltimore D. Gene therapy. Intracellular immunization. Nature. 1988 Sep 29;335(6189):395–396. doi: 10.1038/335395a0. [DOI] [PubMed] [Google Scholar]
- Boulain J. C., Charbit A., Hofnung M. Mutagenesis by random linker insertion into the lamB gene of Escherichia coli K12. Mol Gen Genet. 1986 Nov;205(2):339–348. doi: 10.1007/BF00430448. [DOI] [PubMed] [Google Scholar]
- Cameron F. H., Jennings P. A. Specific gene suppression by engineered ribozymes in monkey cells. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9139–9143. doi: 10.1073/pnas.86.23.9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty B. L., Hotta K., Kumar C., Ahn Y. H., Zhu J. D., Pestka S. Antisense RNA: effect of ribosome binding sites, target location, size, and concentration on the translation of specific mRNA molecules. Gene Anal Tech. 1989 Jan-Feb;6(1):1–16. doi: 10.1016/0735-0651(89)90007-1. [DOI] [PubMed] [Google Scholar]
- Debarbouille M., Cossart P., Raibaud O. A DNA sequence containing the control sites for gene malT and for the malPQ operon. Mol Gen Genet. 1982;185(1):88–92. doi: 10.1007/BF00333795. [DOI] [PubMed] [Google Scholar]
- Deiss L. P., Kimchi A. A genetic tool used to identify thioredoxin as a mediator of a growth inhibitory signal. Science. 1991 Apr 5;252(5002):117–120. doi: 10.1126/science.1901424. [DOI] [PubMed] [Google Scholar]
- Franklin N. C. Deletions and functions of the center of the phi80 -lambda phage genome. Evidence for a phage function promoting genetic recombination. Genetics. 1967 Oct;57(2):301–318. doi: 10.1093/genetics/57.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J. V., Miller A. D. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991 Apr 11;350(6318):508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- Hasan N., Somasekhar G., Szybalski W. Antisense RNA does not significantly affect expression of the galK gene of Escherichia coli or the N gene of coliphage lambda. Gene. 1988 Dec 10;72(1-2):247–252. doi: 10.1016/0378-1119(88)90150-3. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987 Sep 17;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Hoopes B. C., McClure W. R. A cII-dependent promoter is located within the Q gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1985 May;82(10):3134–3138. doi: 10.1073/pnas.82.10.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J., Wodak S. J. Structural domains in proteins and their role in the dynamics of protein function. Prog Biophys Mol Biol. 1983;42(1):21–78. doi: 10.1016/0079-6107(83)90003-2. [DOI] [PubMed] [Google Scholar]
- Kerr S. M., Stark G. R., Kerr I. M. Excess antisense RNA from infectious recombinant SV40 fails to inhibit expression of a transfected, interferon-inducible gene. Eur J Biochem. 1988 Jul 15;175(1):65–73. doi: 10.1111/j.1432-1033.1988.tb14167.x. [DOI] [PubMed] [Google Scholar]
- Krinke L., Wulff D. L. OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependent mechanism. Genes Dev. 1987 Nov;1(9):1005–1013. doi: 10.1101/gad.1.9.1005. [DOI] [PubMed] [Google Scholar]
- Leiter J. M., Krystal M., Palese P. Expression of antisense RNA fails to inhibit influenza virus replication. Virus Res. 1989 Oct;14(2):141–159. doi: 10.1016/0168-1702(89)90035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. I., Schjeide O. A. Micro estimation of RNA by the cupric ion catalyzed orcinol reaction. Anal Biochem. 1969 Mar;27(3):473–483. doi: 10.1016/0003-2697(69)90061-x. [DOI] [PubMed] [Google Scholar]
- Randall-Hazelbauer L., Schwartz M. Isolation of the bacteriophage lambda receptor from Escherichia coli. J Bacteriol. 1973 Dec;116(3):1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes A., James W. Inhibition of human immunodeficiency virus replication in cell culture by endogenously synthesized antisense RNA. J Gen Virol. 1990 Sep;71(Pt 9):1965–1974. doi: 10.1099/0022-1317-71-9-1965. [DOI] [PubMed] [Google Scholar]
- Simon K. O., Cardamone J. J., Jr, Whitaker-Dowling P. A., Youngner J. S., Widnell C. C. Cellular mechanisms in the superinfection exclusion of vesicular stomatitis virus. Virology. 1990 Jul;177(1):375–379. doi: 10.1016/0042-6822(90)90494-c. [DOI] [PubMed] [Google Scholar]
- Sullenger B. A., Gallardo H. F., Ungers G. E., Gilboa E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell. 1990 Nov 2;63(3):601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- Takayama K. M., Houba-Herin N., Inouye M. Overproduction of an antisense RNA containing the oop RNA sequence of bacteriophage lambda induces clear plaque formation. Mol Gen Genet. 1987 Nov;210(1):184–186. doi: 10.1007/BF00337778. [DOI] [PubMed] [Google Scholar]
- Takayama K. M., Inouye M. Antisense RNA. Crit Rev Biochem Mol Biol. 1990;25(3):155–184. doi: 10.3109/10409239009090608. [DOI] [PubMed] [Google Scholar]
- van der Krol A. R., Mol J. N., Stuitje A. R. Modulation of eukaryotic gene expression by complementary RNA or DNA sequences. Biotechniques. 1988 Nov-Dec;6(10):958–976. [PubMed] [Google Scholar]