Summary
Multiple mechanisms have been proposed to explain how Rb and p53 tumor suppressor loss lead to chromosome instability (CIN). It was recently shown that Rb pathway inhibition causes overexpression of the mitotic checkpoint gene Mad2 but whether Mad2 overexpression is required to generate CIN in this context is unknown. Here we show that CIN in cultured cells lacking Rb family proteins requires Mad2 upregulation and that this upregulation is also necessary for CIN and tumor progression in vivo. Mad2 is also repressed by p53 and its upregulation is required for CIN in a p53 mutant tumor model. These results demonstrate that Mad2 overexpression is a critical mediator of the CIN observed upon inactivation of two major tumor suppressor pathways.
Significance
Chromosome instability (CIN) is thought to be the major evolutionary driving force for tumor progression. Here we show that overexpression of the mitotic checkpoint gene Mad2 is required for the CIN observed upon inhibition of the Rb and p53 pathways, two pathways frequently inactivated in human cancer. Our results demonstrate that acquisition of CIN is hard-wired into loss of the major tumor suppressor pathways via hyperactivation of the mitotic checkpoint pathway. The inhibition of these tumor suppressor pathways in the early stages of cancer progression may account for the widespread presence of CIN in tumors in the absence of other genetic events and underscores the therapeutic value of targeting aneuploid cells.
Introduction
Tumor initiation and progression are multistep processes associated with multiple genetic and epigenetic alterations (Hanahan and Weinberg, 2011). One common alteration tightly associated with human cancer is abnormal chromosome numbers or aneuploidy. Since the 1800s, researchers have observed that cancer cells missegregate their chromosomes during mitosis and that such missegregation events are more prevalent in advanced stages of the disease. It is not until recently, however, that the causative role of aneuploidy in tumor initiation has been established in animal models: at least 24 independent genetic lesions which lead to aneuploidy also initiate or enhance the progression of tumorigenesis in mice (Schvartzman et al., 2010). Aneuploidy likely leads to the loss of tumor suppressors or gain of oncogenic signals and indeed in one mouse model, enhancing aneuploidy leads to accelerated loss of heterozygosity of the p53 tumor suppressor locus (Baker et al., 2009).
How aneuploidy arises in tumors is still a matter of debate. It has been postulated that this occurs by mitotic checkpoint loss or weakness. In fact, loss of a number of key mitotic checkpoint genes leads to aneuploidy and tumorigenesis in mice (Schvartzman et al., 2010). However, data from human tumors does not support this hypothesis as loss or weakness of the mitotic checkpoint is rare in clinical samples (Rhodes et al., 2007).
An unsuspected connection between a tumor suppressor pathway and the acquisition of aneuploidy is the fact that inhibition of the Rb tumor suppressor pathway and E2F activation directly lead to enhanced expression of the mitotic checkpoint gene Mad2 (Hernando et al., 2004). Mad2 overexpression is sufficient to cause non-disjunction, aneuploidy and tumor initiation in mice (Sotillo et al., 2007). Thus, loss of at least one of the major tumor suppressor pathways is hard-wired to a pathway which directly leads to CIN during mitosis. Three recent reports propose an alternative mechanism by which Rb pathway inhibition leads to CIN via centromeric chromatin decondensation but whether this is necessary for establishing CIN remains unknown (Coschi et al., 2010; Manning et al., 2010; van Harn et al., 2010) (See also Discussion).
The p53 pathway is a second pathway that has long been recognized as a primary suppressor of CIN. In vitro, p53 loss or mutation results in loss of cell cycle control and aneuploidy (Negrini et al., 2010). Genetic disruption of p53 function in the mouse has been shown to cause CIN and accelerate tumor development in models of various types of cancer (Donehower and Lozano, 2009). Additionally, strong correlations between abnormal p53 status and aneuploidy have been observed in human tumors (Junttila and Evan, 2009).
The dissection of the mechanism by which p53 prevents genomic instability is complicated by the fact that p53 executes other cellular functions that are intricately linked, including the induction of cell cycle arrest, apoptosis and senescence (Junttila and Evan, 2009). Despite this, evidence for a crucial role of p21Waf1/Cip1/Sdi1 (p21), whose expression is directly induced by p53, has accumulated. For example, mice carrying mutant p53R172P knock-in alleles, which maintain the ability to induce p21 yet are unable to trigger apoptosis, develop tumors with stable, diploid genomes; however, tumors that develop in mice carrying the same mutation in a p21−/− background are invariably aneuploid (Barboza et al., 2006). This suggests that part, if not all, of p53’s function to maintain genomic stability is executed by p21.
While the p53 and Rb pathways are often studied independently, the relevance of crosstalk between these major tumor suppressor pathways is increasingly recognized (Polager and Ginsberg, 2009). p21 represents an obvious connection between the two pathways as its expression is directly induced by p53 and, as a cyclin-dependent kinase inhibitor, p21 retains the Rb family of pocket proteins in a hypophosphorylated state, thus preventing unscheduled E2F activation and cell cycle entry. The notion that p53 maintains genomic stability largely, if not exclusively, by p21 induction suggests that p53 may prevent CIN through a p21-cyclin/Cdk-Rb-E2F repression pathway. Because increased Mad2 expression, a primary cause of CIN, is a direct result of aberrant Rb pathway signaling, disruption of p53 function may also cause CIN through elevated levels of Mad2.
While Mad2 overexpression is sufficient to initiate tumorigenesis, whether it is necessary for the tumorigenic effects of Rb or p53 pathway inhibition and the nature of these effects have not been previously explored. The experiments presented here address this hypothesis.
Results
p53 Represses Mad2 Expression by Induction of p21
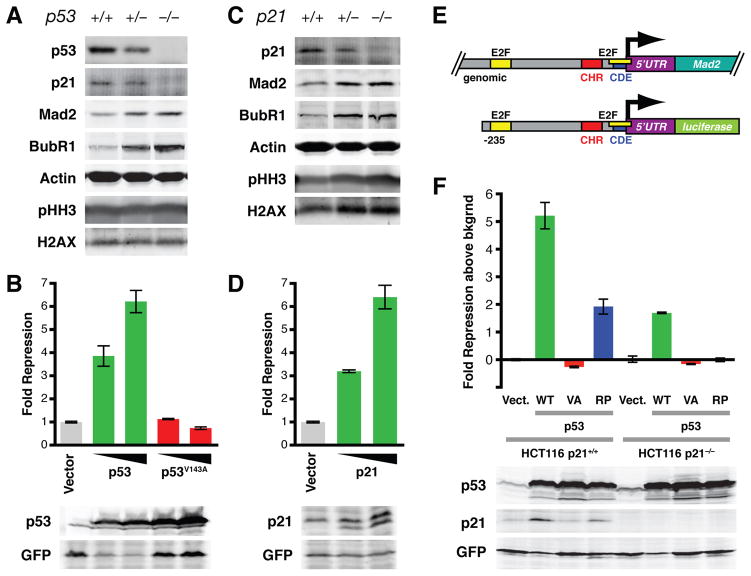
Rb inactivation results in upregulation of Mad2 (Hernando et al., 2004), a direct E2F target, and Mad2 overexpression leads to CIN both in vitro and in vivo (Sotillo et al., 2007). Disruption of the p53 pathway, directly or indirectly, leads to and maintains CIN (Donehower et al., 1995; Thompson and Compton, 2010; Tomasini et al., 2008). To determine whether Mad2 overexpression plays a role in the acquisition of CIN observed upon p53 inactivation, we first analyzed Mad2 protein levels in primary mouse embryonic fibroblasts (MEFs) derived from wild-type, p53+/− and p53−/− mice. p53+/− and p53−/− MEFs expressed higher levels of Mad2 than wild-type cells (Figure 1A). Consistent with previous studies showing that p53 or p21 loss does not lead to changes in the distribution of cells in G1, S, G2 or M phase of the cell cycle (Deng et al., 1995), levels of phospho-histone H3 (Ser10), a mitotic marker, were similar for the three genotypes. Additionally, Mad2 levels were elevated in mitotic as well as in non-mitotic cells (Figure S1A). This indicates that the increase in Mad2 levels is not merely a result of an increase in the fraction of mitotic cells. The levels of BubR1, another mitotic checkpoint protein, were equally elevated, implying that BubR1 expression is controlled by p53 in a similar manner (see also Discussion).
Figure 1. P53 Represses Mad2 Expression by Induction of P21.
(A) Western blot of primary mouse embryonic fibroblasts (MEFs) derived from wild-type, p53+/− or p53−/− embryos. H2AX serves as loading control for phospho-histone H3 (Ser10; pHH3).
(B) Reporter assays showing repressive activity of wild-type and mutant p53 on the Mad2 promoter. HCT116 cells were co-transfected with a firefly luciferase vector in which luciferase expression is controlled by 235 bp of the human Mad2 promoter and an expression vector containing either wild-type p53 or the DNA-binding deficient mutant p53V143A (see also (E)). Renilla luciferase and GFP expressing vectors were co-transfected for normalization of transfection efficiency (top) and exogenous p53 protein expression (Western blots below). Triangles indicate increasing amounts of transfected p53. Means ± SD are shown for assays performed in triplicate.
(C) Western blot of MEFs derived from wild-type, p21+/− or p21−/− embryos. H2AX serves as loading control for phospho-histone H3 (pHH3).
(D) Reporter assays as in (B) using a p21 expression vector. Means ± SD are shown.
(E) Genomic structure of the Mad2 gene and the luciferase reporter construct containing 235 base pairs of the Mad2 promoter. E2F, putative E2F binding site; CHR, cell cycle homology region; CDE, cell cycle-dependent element (see also text for details).
(F) Reporter assays as in (B) using HCT116-p21+/+ or HCT116-p21−/− cells and expression vectors containing no insert (Vect.), wild-type p53 (WT), DNA-binding deficient p53V143A (VA) or the p53R175P mutant (RP), which can induce p21 expression but not apoptosis (Liu et al., 2004 and Western blots below). Means ± SD are shown.
See also Figure S1.
In order to test whether p53 is able to directly repress Mad2 promoter activity, a p53 expression vector and a vector in which luciferase expression is driven by 235 base pairs of the Mad2 promoter were co-transfected into HCT116 cells. Compared to the empty vector control, p53 effectively repressed promoter activity in a dose-dependent manner. In contrast, mutant p53V143A, which does not bind to p53 DNA binding elements (Zambetti and Levine, 1993), was unable to do so (Figure 1B). Similar results were obtained using Saos2 cells (data not shown). Sequence analysis of the Mad2 promoter revealed the presence of a putative p53 binding site approximately 160 base pairs upstream of the transcription start site. However, mutation of this site did not affect p53-mediated repression (data not shown). These results suggest that p53-mediated Mad2 repression is dependent on its ability to bind to DNA but that this effect is mediated through other transcriptional targets of p53.
Because p21 is a primary transcriptional target of p53, we asked whether Mad2 protein levels were also elevated in MEFs lacking one or both copies of the CDKN1A gene, which encodes p21. Indeed, both p21+/− and p21−/− cells expressed higher levels of Mad2 than wild-type MEFs (Figure 1C). Similar to our observations in p53+/− and p53−/− MEFs, BubR1 levels were also elevated, whereas phospho-histone H3 levels did not change in p21+/− or p21−/− cells, again confirming that Mad2 was not elevated as a consequence of an increase in the fraction of mitotic cells (Deng et al., 1995). In addition, as seen with wild-type p53, p21 repressed Mad2 promoter activity in reporter assays that contained 235 base pairs of the Mad2 promoter (Figure 1D, 1E). Loss of either both p53 alleles or both p21 alleles by means of homologous recombination in human HCT116 cells leads to a 2.5 to 3-fold increase in Mad2 promoter activity (Figure S1B) and a 1.5-fold increase in endogenous Mad2 mRNA levels (Figure S1C). Since p21 induction by p53 is dependent on its DNA binding activity, these results suggest that p53 represses Mad2 expression through induction of p21. Consistent with this, p53’s ability to repress Mad2 promoter activity in HCT116-p21+/+ cells was significantly diminished in a p21−/− background (Figure 1F). The residual repressive activity was the result of the ability of p53 to induce apoptosis as a p53 mutant that is able to induce p21 expression but unable to induce apoptosis (p53R175P) (Liu et al., 2004) completely lost repressive activity in the absence of p21 (Figure 1F). Thus, we conclude that p53 represses Mad2 expression by induction of p21.
p53 Represses Mad2 Expression through Crosstalk to the Rb Pathway
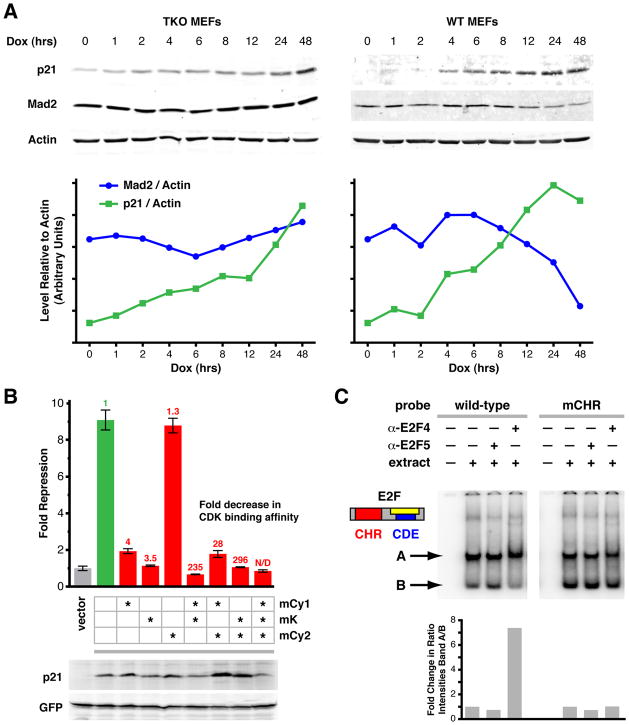
Since p21 functions as a Cdk inhibitor, we next asked whether its effect on the Mad2 promoter was dependent on pocket proteins by overexpressing p21 in Rb triple knockout (TKO) MEFs, in which all three pocket protein genes are deleted (Dannenberg et al., 2000; Sage et al., 2003). TKO MEFs were transduced with a lentiviral vector containing the reverse tetracycline transactivator (rtTA) cDNA under control of a ubiquitous promoter and a p21 cDNA whose expression is controlled by a tetracycline-responsive promoter. Following selection for transduced cells, doxycycline was added to the culture medium and cells were harvested at different time points thereafter. While p21 expression increased over time, Mad2 protein levels did not significantly change in TKO cells (Figure 2A and S2A). As expected, Mad2 levels decreased following induction of exogenous p21 expression in wild-type control MEFs (Figure 2A and S2A). Flow cytometry analysis of the TKO cells at each time point did not show significant changes in the cell cycle profiles, indicating that the TKO cells had not arrested or entered senescence (Figure S2B). Thus, p21-mediated repression of Mad2 is pocket protein-dependent.
Figure 2. P53 Represses Mad2 Expression through Crosstalk to the Rb Pathway.
(A) Top: Western blot of Rb triple knockout (p107−/−, p130−/−, pRb−/−; TKO) and wild-type (WT) MEFs overexpressing p21 (Dox (hrs) indicates time after induction). Bottom: Quantification of Western blots shown in upper panels. Relative levels of Mad2 and p21 proteins are each normalized to actin on the same blot. See also Figure S2A.
(B) Reporter assays as in Figure 1B using HCT116-p21−/− cells and mutant p21 expression vectors. Wild-type p21 (green) and p21 containing mutations in the cyclin binding domains Cy1 and/or Cy2 (mCy1, mCy2) and/or the kinase binding domain K (mK) (red) were assayed for their ability to repress Mad2 promoter activity. Means ± SD are shown. Numbers above each bar indicate the fold decrease in Cdk binding activity (green, red) as a result of the respective mutation(s), as determined by Chen et al., 1996. Western blots below show relative expression of exogenous wild-type and mutant p21 with respect to GFP.
(C) Electrophoretic mobility shift assay (EMSA) of CHR-CDE/E2F elements in the Mad2 promoter. Top: Wild-type or mutant CHR (mCHR) labeled probes encompassing the CHR-CDE/E2F elements in the Mad2 promoter were incubated with HeLa nuclear extract (extract) and antibodies specific for E2F-4 or E2F-5 (α-E2F4, α-E2F5), as indicated. Bottom: fold change in ratio of the intensities of band A/B.
See also Figure S2.
To further assess the contribution of pocket protein-independent effects of p21, we made use of a series of p21 mutants. Three domains in p21 have been implicated in its binding to cyclin/Cdk complexes, a requirement for p21-mediated inhibition of Cdk activity. Cy1 and Cy2 are cyclin binding domains and the K domain mediates Cdk binding. Mutations in each of these domains individually or combinations thereof lead to varying degrees of decrease in cyclin/Cdk binding activity toward different cyclin/Cdk complexes (cyclin A/Cdk2, cyclin D1/Cdk4, cyclin E/Cdk2) (Chen et al., 1996). However, if such p21 mutants are able to repress Mad2 promoter activity, this would suggest that p21 is able to repress in a manner independent of the Rb pathway. Reporter assays using HCT116-p21−/− and HCT116-p21+/+ cells and p21 variants containing mutations in the Cy1, K or Cy2 domains, individually or in combination, showed that six different mutants in which the Cdk binding affinity was reduced 3.5 to greater than 300-fold showed no capacity to repress the Mad2 promoter (Figure 2B, S2C). One mutant, p21mCy2, retained its capacity to repress Mad2. However, this mutant only showed a 29% reduction in Cdk binding affinity (Chen et al., 1996). While it is possible that any given mutation which disrupts the ability of p21 to bind to or inhibit Cdks might also affect its ability to signal through an Rb-independent pathway, this seems remote given the fact that multiple p21 mutations show the same phenotype.
Together, the above results suggest that p21 represses Mad2 expression through canonical signaling, i.e. inhibition of the Cdk-mediated phosphorylation of Rb and consequent stabilization of the Rb/E2F complex. In addition to multiple E2F binding sites, analysis of the Mad2 promoter revealed the presence of a cell cycle gene homology region (CHR) and a cell cycle-dependent element (CDE) close to the transcription start site (Figure S2F, G). CHR and CDE elements are also present and conserved in a number of cell cycle-regulated genes. A protein complex referred to as CDF1 (cell cycle-dependent factor 1) is recruited to CHR and CDE elements and, at least for one promoter, p21 has been shown to be recruited to these elements where it mediates transcriptional repression of the downstream gene (Fung and Poon, 2005; Liu et al., 1997; Vigneron et al., 2006). We refer to this as the non-canonical p21 repression pathway.
Disruption of either CHR or CDE elements led to derepression of the Mad2 promoter (Figure S2D upper) (Liu et al., 1997) suggesting that these elements also play an inhibitory role. However, this derepression was dependent on an upstream E2F binding site, since a smaller reporter construct that lacked this upstream element failed to show derepression when the CHR or CDE sites were mutated (Figure S2D lower).
Supporting the hypothesis that p21 repressed Mad2 expression through the CHR and CDE elements, p21-mediated repression was completely lost when either site was mutated (Figure S2E). However, the CHR element adjacent to the CDE/E2F site might be required to stabilize E2F/Rb inhibitory complexes. We therefore asked whether mutation of the CHR site affected E2F binding, whose binding site overlaps only with the CDE site. First, using an unbiased protein affinity purification protocol followed by mass spectrometric protein identification, we found that none of the activating E2Fs (E2F1, 2, 3a) and only repressor E2Fs E2F4 and E2F5 bound to repeats of wild-type CHR-CDE DNA sequences. In addition, binding of these E2Fs was enriched for the wild-type CHR-E2F/CDE bait sequence as compared to the mCHR-E2F/CDE sequence in which the CHR site was mutated (Figure S2H). We also identified E2F binding partner DP-1, Rb family member p107, which is known to preferentially bind to E2F4 and E2F5 (Ikeda et al., 1996; Moberg et al., 1996), cyclin B1, cyclin-dependent kinase-1 (Cdk1), Cdk2 and the transcriptional repressor HDAC1 (Figure S2H). These observations were confirmed for E2F4 and p107 by ChIP analysis (Figure S2I). In contrast to the mass spectrometry analysis, this assay is not quantitative and also likely includes E2F4, p107 and/or p130 precipitated from more upstream E2F sites. Cyclin B1 binding to the CHR-CDE/E2F site was also confirmed by electrophoretic mobility shift assay (EMSA) using a super-shifting antibody (data not shown). EMSAs using a wild-type and a mutant CHR (mCHR) probe also showed that a high molecular weight protein complex containing E2F4 bound to the wild-type but not to the mCHR probe (Figure 2C). These data suggest that an intact CHR site is required for the stable formation of repressor E2F- and p107-containing complexes that can inhibit expression of the upstream activatory sites. This provides further support for our observations indicating that Mad2 expression is repressed through canonical Rb pathway signaling.
Mad2 Upregulation Is Necessary for the CIN Seen upon Rb Pathway Inhibition
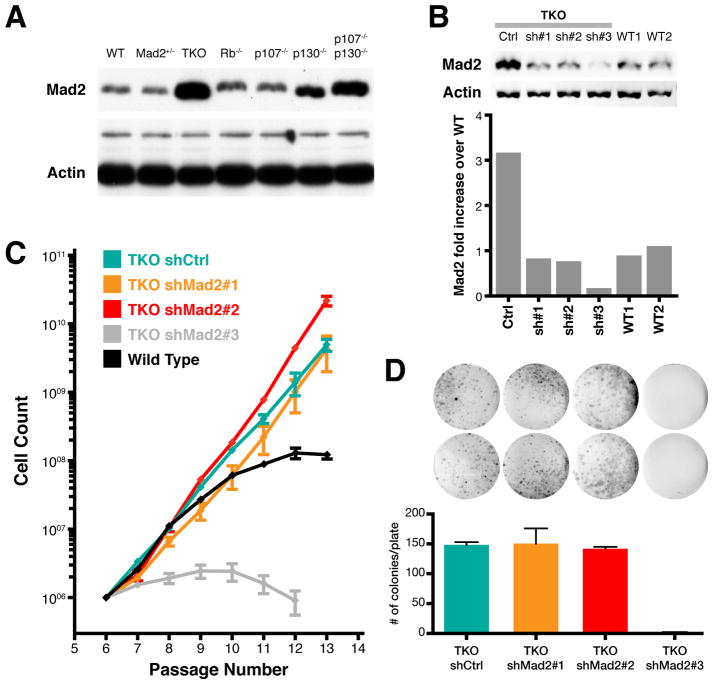
The above results suggest that p53 maintains adequate repression of the Mad2 gene through the Rb pathway. All three Rb family members (Rb, p107, p130) play a role in the inhibition of Mad2 expression, since none of the double or single knockouts showed as high a level as seen in TKO cells (Figure 3A). Mad2 protein levels in TKO MEFs were higher both in mitotic and in non-mitotic cells (Figure S3A) indicating that increased Mad2 levels were not simply the result of a higher mitotic fraction. To determine whether this observed upregulation of Mad2 was necessary for the CIN previously seen in TKO MEFs (Gonzalo et al., 2005), we sought to reduce the levels of Mad2 to those approaching wild-type. Importantly, partial downregulation of Mad2 is itself conducive to aneuploidy (Michel et al., 2001) and complete knockdown is cell lethal, so our intent was to achieve levels close to those seen in wild-type cells. Using a panel of lentiviral vectors expressing short-hairpin RNAs, two constructs (shMad2#1 and shMad2#2) were found to normalize Mad2 levels to the desired wild-type range, as determined by quantitative Western blot analysis (Figure 3B).
Figure 3. Normalization of Mad2 levels in TKO MEFs.
(A) Western blot showing Mad2 levels in wild-type (WT), Mad2 heterozygote (Mad2+/−), and Rb, p107 and p130 triple (TKO), single and double p107 and p130 knockout MEFs. Actin and a larger molecular weight non-specific band serve as a loading control.
(B) Western blot and quantification of Mad2 levels in wild-type (WT1 and WT2) and TKO MEFs transduced with lentiviral vectors expressing sh-scrambled control (Ctrl), sh-Mad2#1, sh-Mad2#2 and sh-Mad2#3. Levels are normalized to Actin.
(C) 3T3 proliferation assay for TKO MEFs transduced with corresponding short hairpin lentiviral vectors. Averages ± SD of two independent experiments are shown.
(D) Colony formation assay and quantification of colonies for TKO Control, shMad2#1, shMad2#2 and shMad2#3 vectors. Averages ± SEM of two independent experiments are shown (p < 0.002 for shMad2#3, all others p > 0.4; unpaired t-test).
See also Figure S3.
Normalization of Mad2 had no effect on the high levels of BubR1 already present in TKO MEFs (Figure S3B), again arguing against marked decreases in the number of mitotic cells. As a functional readout of the effect of Mad2 normalization on mitotic checkpoint function, we challenged the different TKO MEF lines and WT MEFs with nocodazole, a microtubule depolymerizing agent. As expected, TKO MEFs transduced with scrambled short hairpin vectors arrested in mitosis for a prolonged period of time (median 500 minutes) and normalization of Mad2 led to a decreased median arrest time (Figure S3C). These cells were still SAC competent, however, as previous studies have shown that Mad2 null MEFs fail to maintain a nocodazole arrest for more than 60 minutes (Burds et al., 2005), well below the 310 or 260 minutes seen in the Mad2 normalized TKO MEFs.
Normalization of Mad2 in TKO MEFs had no significant effect on proliferation compared with TKO MEFs transduced with a scrambled control vector as assessed by a 3T3 immortalization assay (Figure 3C). In addition, a standard proliferation assay (Figure S3D) and a low density seeding efficiency assay (Figure 3D) failed to show significant effects of Mad2 normalization on TKO MEF growth rates or viability. In agreement with previous reports, near complete knockdown of Mad2 (shMad2#3) (Figure 3B) resulted in marked decreases in both viability and seeding efficiency (Figures 3C and 3D) (Michel et al., 2004).
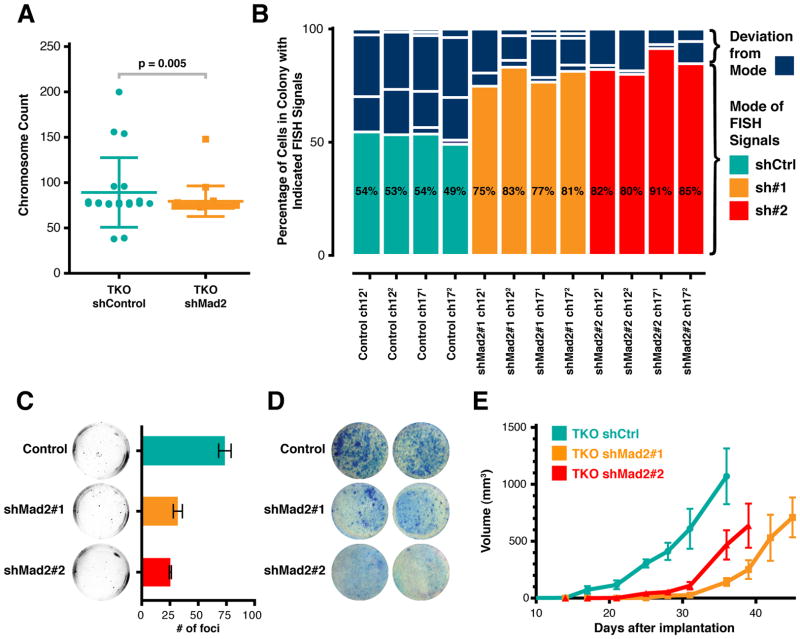
If Mad2 upregulation was responsible for the CIN in TKO MEFs, its normalization would be expected to result in a rescue of this phenotype. Karyotypes of passage 25 TKO cells with normalized levels of Mad2 were significantly less variable than those of control TKO cells (Figure 4A). Of twenty metaphase spreads analyzed in each condition, the distribution of chromosome counts in scrambled-sh control TKO cells was significantly broader than that of Mad2-normalized TKO cells. Furthermore, none of the scrambled-sh control TKO cells had the same chromosome complement while more than half of normalized Mad2 TKO cells did (Table S1). Nevertheless, this decrease in variability could be the result of subtle differences in proliferation rate or the outgrowth of a clone of cells with a fixed karyotype. To control for these possibilities, we directly measured the rate of CIN by monitoring chromosome 12 and 17 specific interphase FISH (fluorescent in situ hybridization) in colonies of 150–500 cells derived from a single cell. In this assay, the modal number of FISH signals in a colony reflects the karyotype of the initiating cell and deviation from the mode is a measure of CIN rates. The frequency distribution analysis of FISH signals for each colony (0 to 6 or more signals for each chromosome probe) revealed a significant decrease in the deviation from the mode in TKO cells with normalized Mad2 levels compared to scrambled-sh TKO controls (Figure 4B); 15.2% for shMad2 TKO colonies (SEM=1.630) and 39.5% for the shCtrl TKO colonies (SEM=3.079) (p value < 0.005). This difference was observed in three independent experiments with both short hairpin vectors (shMad2#1 and shMad2#2) that had been shown to normalize Mad2 levels. It is unlikely that we are selecting for genomically stable clones upon Mad2 normalization since colony-forming efficiency was unchanged in the normalized Mad2 TKO colonies (Figure 3D) and all scrambled-sh TKO colonies analyzed showed high deviations from the mode. These results provide direct evidence that Mad2 upregulation is necessary for the CIN observed in cells lacking Rb protein family function.
Figure 4. Mad2 Upregulation is Required for Rb Loss-Induced CIN and Accelerates Transformation of TKO MEFs.
(A) Chromosome counts for sh-scrambled control and shMad2#1 TKO MEFs. Error bars indicate SD. p value as determined by non-parametric Mann-Whitney test.
(B) CIN Assay. Each column corresponds to one chromosome analyzed in one colony by FISH. Scrambled shCtrl colonies in teal, shMad2#1 in orange and shMad2#2 in red. Percentages indicate mode. Non-modal groups that comprise deviation from the mode are in dark blue. p value for control versus individual shMad2 and combined shMad2 assays < 0.005 as determined by Mann-Whitney non-parametric test.
(C) Anchorage Independent Growth Assay. Averages ± SEM of two independent experiments are shown (p < 0.0037; unpaired t-test).
(D) Contact Inhibition Assay. Confluent plates were transfected with an HrasV12 vector and colonies stained after 3 weeks.
(E) Intradermal Allograft Growth. Each point is the average size of 6 tumors. Averages ± SEM of two independent experiments are shown. All p values between scrambled sh control and shMad2 curves < 0.05 (unpaired t-test).
Mad2-Mediated CIN Accelerates Transformation of Fibroblasts without Affecting Proliferation Rate
We next sought to investigate whether normalization of Mad2 levels in TKO cells had any effects on cellular transformation. Transformed murine fibroblasts readily form colonies when seeded at low density, show anchorage independent growth in soft agar and form tumor allografts when implanted intradermically in nude mice (Flint, 2000). In addition to showing impaired seeding efficiency, normalization of Mad2 in TKO MEFs transduced with a retroviral vector expressing the HrasV12 oncogene led to significant decreases in growth in soft agar and focus formation (Figures 4C and 4D), standard assays for anchorage independent growth and escape from contact inhibition, respectively. Moreover, HrasV12 transformed TKO MEFs implanted intradermically into immunodeficient animals form rapidly proliferating fibrosarcomas (Sage et al., 2003) and Mad2 normalization led to a significant delay in the growth of these tumors as compared with scrambled hairpin controls (Figure 4E). This difference likely reflects an underestimation of the effect of CIN on allograft tumor growth since the selective pressure to maintain short hairpin expression (puromycin resistance) is no longer present in vivo. In fact, Mad2 levels in the normalized allograft tumor samples were more similar to controls than in the pre-implanted parental lines (Figure S4A). This decrease in proliferation in vivo was not observed in vitro after HrasV12 transduction (Figure S4B), arguing that CIN induced by Mad2 upregulation plays a role in accelerating progression through tumor-suppressive events that are independent of proliferation per se and are only manifest in assays of anchorage independence, contact inhibition or implantation into recipient animals.
Mad2-Mediated CIN Is Required for the Development of Anaplastic Tumors in a Mouse Model of Mammary Adenocarcinoma
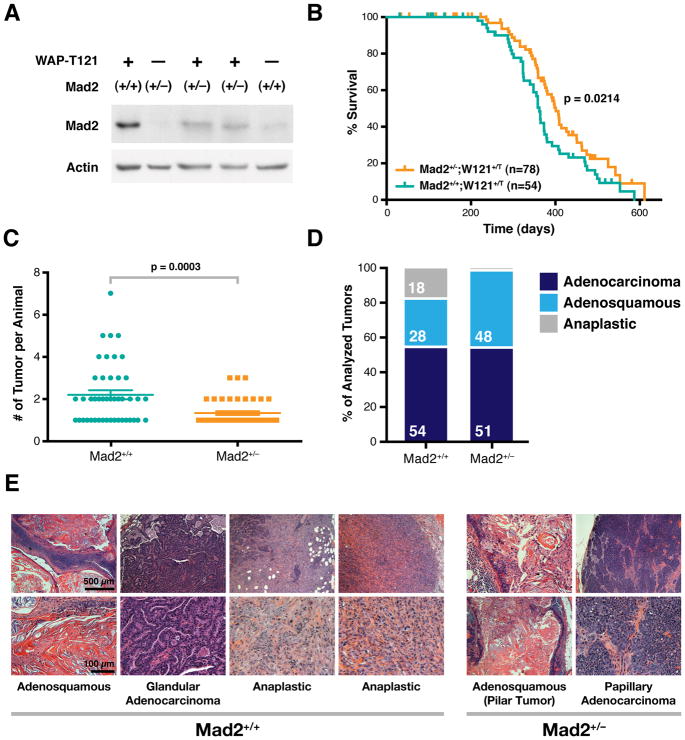
In order to determine what role Mad2 upregulation (as a result of Rb pathway inhibition) plays in a setting more comparable to common human malignancies, we focused on the previously described WAP-T121 mammary tumor model (Simin et al., 2004). These animals express T121, a fragment of SV40 Large T antigen that binds to and inhibits Rb family members, under the control of the mammary-specific Whey-Acidic-Protein promoter. WAP-T121 females develop mammary adenocarcinomas with a median latency of 12 months. Pretumorigenic mammary glands of WAP-T121 female mice showed high levels of Mad2 relative to their wild-type counterparts (Figure 5A) and we sought to determine if normalization of Mad2 levels via Mad2 heterozygosity would have an effect on the characteristics and CIN in these tumors. Pretumorigenic mammary glands of Mad2+/−;WAP-T121 females showed Mad2 levels slightly higher than those of wild-type animals but significantly lower than those of Mad2+/+;WAP-T121 mice (Figure 5A). Mad2 heterozygosity in WAP-T121 females resulted in a delay in tumor onset (Figure 5B; median latency of 362 days for Mad2+/+ versus 407 days for Mad2+/−; p = 0.0214) and a decrease in tumor burden compared to wild-type controls (Figure 5C; 2.2 versus 1.3 tumors per animal; p = 0.0003), both of which were statistically significant. The most striking phenotype was observed upon analysis of tumor histology. Mad2+/+;WAP-T121 females developed three histological types of tumors (Figure 5D and 5E): well differentiated, adenosquamous tumors (28% of all tumors), less differentiated, more basophillic glandular or papillary adenocarcinomas (54% of all tumors) and poorly differentiated anaplastic tumors (18% of all tumors) with spindle shaped cells reminiscent of an epithelial to mesenchymal transition (EMT) (Figure 5E). The anaplastic tumors had a similar appearance to the anaplastic mammary tumors seen in humans, which are associated with triple negative status (estrogen, progesterone and EGF receptor negative) and carry a particularly poor clinical prognosis (Reis-Filho and Tutt, 2008).
Figure 5. Mad2 Upregulation Is Required for the Development of Anaplastic Mammary Tumors in WAPT121 Transgenic Mice.
(A) Western blot of adult pretumorigenic mammary glands from female Mad2+/+ and Mad2+/− transgenic (WAPT121) and non-transgenic mice. Lanes 3 and 4 are lysates from 2 different animals.
(B) Tumor-free survival of Mad2+/+ and Mad2+/− in WAPT121 mice. p value determined from Gehan-Breslow-Wilcoxon Test. Vertical lines indicate censored subjects.
(C) Tumor burden (# of tumors per animal) as determined by macroscopic analysis in Mad2+/+ and Mad2+/− in WAPT121 mice. Averages ± SEM are shown.
(D) Mammary tumor type distribution in Mad2+/+ (N=65) and Mad2+/− (N=68) WAPT121 mice. p value (Fisher’s test) for Adenosquamous/Anaplastic = 0.0003; Adenocarcinoma/Anaplastic = 0.0086.
(E) Representative H&E examples of tumor types in Mad2+/+ and Mad2+/− WAPT121 mice. The two anaplastic tumors shown are examples from two different animals.
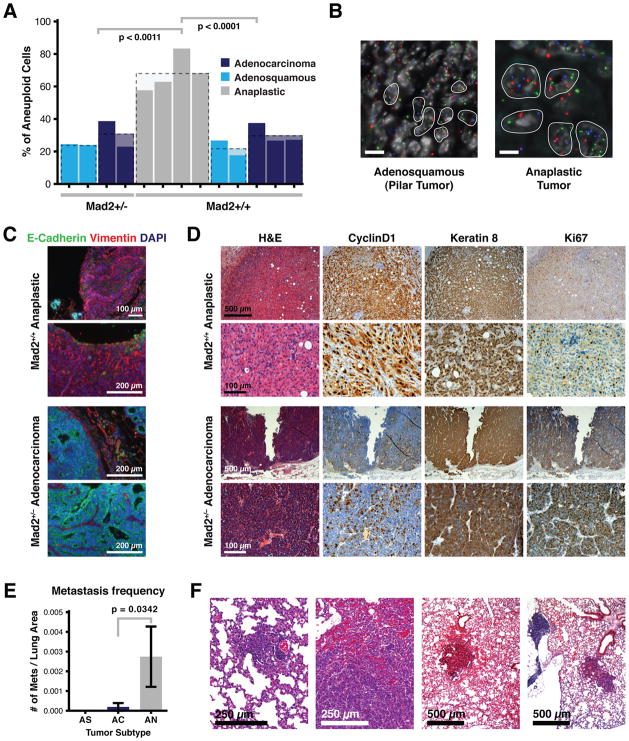
The nature of our study precludes determination of whether the three tumor types reflect a progression from adenosquamous to adenocarcinoma to anaplastic tumors, though in many cases two of these histological patterns were observed in the same lesion. Remarkably, Mad2 heterozygosity resulted in a shift to a more differentiated tumor spectrum, with 48% adenosquamous tumors, 51% adenocarcinomas and only 1.4% (1/65) anaplastic (Figure 5D). This represents a 12-fold reduction in anaplastic lesions that results from the loss of one copy of the Mad2 gene. Furthermore, regardless of genotype, adenosquamous tumors and adenocarcinomas showed baseline moderate levels of aneuploidy as detected by chromosome FISH for three separate chromosomes while the anaplastic tumors seen in the Mad2 wild-type background were more markedly aneuploid (Figure 6A and 6B). This increase in total aneuploidy reflected both an increase in near-tetraploid as well as of near-diploid aneuploid cells within anaplastic tumors (Figure S5A). All Mad2+/+ tumors had average Mad2 expression levels roughly two-fold that of Mad2+/− tumors (Figure S5B, left), while levels of BubR1, another component of the mitotic checkpoint, remained equivalent between the two genotypes (Figure S5B, right). Thus, while there is an equivalent Mad2-induced drive towards aneuploidy in all the tumor subtypes in the Mad2+/+ background, it is likely that only the anaplastic lesions have lost the proposed aneuploidy surveillance mechanism (Torres et al., 2010; Torres et al., 2007; Williams et al., 2008) which would facilitate their survival and propagation. Alternatively, Mad2 levels may be higher only transiently in the anaplastic tumors during the early stages of tumor progression which could enhance the acquisition of CIN. This is consistent with our previous observation that transient pulses of Mad2 overexpression can confer significant instability at later stages of the disease (Sotillo et al., 2010).
Figure 6. Anaplastic Mammary Tumors in WAPT121;Mad2+/+ Mice Are Aneuploid, Invasive and More Metastatic.
(A) Percent of aneuploid cells as determined by chromosome FISH in Mad2+/+ and Mad2+/− WAPT121 animals distributed by tumor type. p values calculated using unpaired t-test.
(B) Representative examples of chFISH in adenosquamous WAPT121;Mad2+/− and anaplastic WAPT121;Mad2+/+ tumors. Scale bar indicates 10 μm.
(C) Representative example of epithelial to mesenchymal transition (EMT) as determined by loss of E-cadherin and gain of Vimentin immunofluorescence staining in an anaplastic tumor. Normal appearance (E-cadherin positive and Vimentin negative) of an adenocarcinoma is seen in lower panels.
(D) Representative examples of H&E and CyclinD1, Keratin8 and Ki67 immunohistochemistry examples from adenosquamous WAPT121;Mad2+/− and anaplastic WAPT121;Mad2+/+ tumors.
(E) Frequency of spontaneous metastasis by tumor subtype in WAPT121;Mad2+/+ mice. p value for adenocarcinomas and anaplastic tumors = 0.0342 (unpaired t-test). Error bars indicate SEM.
(F) Representative H&E examples of metastatic lesions in WAPT121;Mad2+/+ animals harboring anaplastic tumors.
See also Figure S5.
The spindle shaped cells observed in the anaplastic tumors suggested the presence of an epithelial to mesenchymal transition (EMT), an event associated with the ability of tumors to locally invade and metastasize. In fact, induction of EMT in mammary epithelial cells has been shown to drive morphological transformation, enhanced cell migration and tumorigenic cooperation with known oncogenes (Yu et al., 2009). Anaplastic tumors in the WAP-T121 setting stained positive for markers of EMT; loss of the epithelial marker E-cadherin and gain of the mesenchymal marker Vimentin, as detected by co-immunofluorescence (Figure 6C). Conversely, adenocarcinomas remained E-cadherin positive and Vimentin negative regardless of genotype. Anaplastic tumors retained the mammary specific marker Keratin 8 (TROMA) as seen by immunohistochemistry, excluding the possibility that they were stromal fibroblast reactions to an epithelial lesion (Figure 6D). We did not observe notable differences in proliferation rate nor T121 transgene activation between tumors of different histology or genotype, as determined by immunohistochemistry for Ki67 and CyclinD1, respectively (Figure 6D). When we examined serial lung sections of all animals in order to determine the metastatic frequency per total lung area, anaplastic tumors in the Mad2+/+ background were significantly more likely to metastasize compared to adenocarcinomas and adenosquamous tumors (p = 0.0342) (Figures 6E and 6F).
Inhibition of p53 function by mutation or loss often correlates with tumor progression and could explain the appearance of anaplastic lesions. Three out of 10 WAP-T121;Mad2+/+ tumors showed p53 hotspot mutations (one adenocarcinoma and two anaplastic tumors) but 7/10 showed no evidence of p53 deletion or hotspot missense mutations (Figure S5C and S5D) that could account for the distribution of the different tumor subtypes. Overall then, these results show that in the context of Rb pathway inhibition in the mammary gland, Mad2-mediated CIN accelerates the progression to anaplastic adenocarcinomas, a clinical entity that is associated with triple negative state, poor prognosis and metastatic progression.
Normalization of Elevated Mad2 Levels Rescues CIN in a p53 Mutant Mouse Model
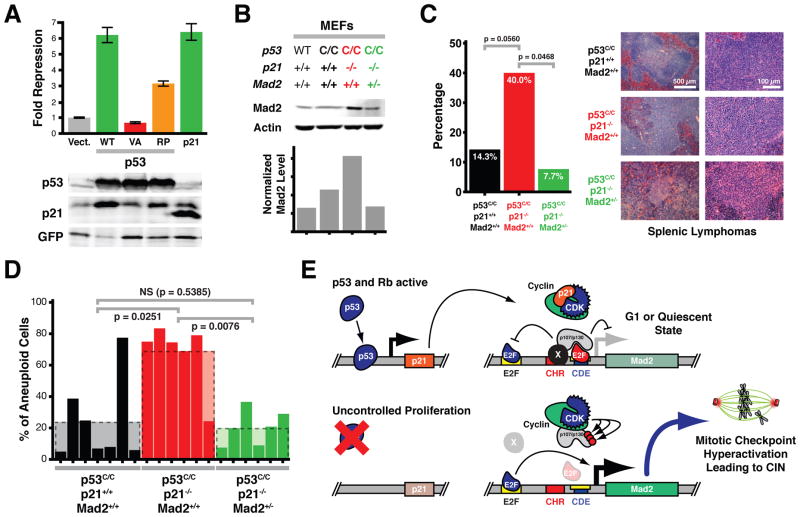
Given that p53 represses Mad2 expression largely, if not exclusively, through p21 and crosstalk to the Rb pathway, we wondered whether the CIN phenotype in a p53 mutant mouse model could be rescued or alleviated by normalization of Mad2 levels. We used a previously developed p53 knock-in model in which both p53 copies are replaced by alleles expressing the p53R172P mutant (p53C/C) (Liu et al., 2004). This mutant is defective in inducing apoptosis, yet retains the ability to induce cell cycle arrest through induction of p21 expression (Liu et al., 2004). Consistent with this, we found that p53R175P, the human ortholog of murine p53R172P, repressed Mad2 promoter activity in human cells (Figures 1F, 7A and data not shown). P53C/C mice develop lymphomas and sarcomas with stable, diploid genomes (Liu et al., 2004). Interestingly, when these mice are crossed into a p21−/− background, lymphomas and sarcomas not only develop significantly earlier, but they are also aneuploid (Barboza et al., 2006). We hypothesized that the latter is a consequence of upregulated Mad2 levels, as we had previously observed in p21−/− MEFs (Figure 1C). Western blot analysis of MEF lysates derived from p53C/C and p53C/C;p21−/− mice confirmed that Mad2 levels were elevated in the absence of p21 (Figure 7B). In order to reduce these elevated levels of Mad2, we crossed p53C/C;p21−/− animals with Mad2+/− mice. In a Mad2+/− background, p53C/C;p21−/− MEFs showed reduced Mad2 levels, comparable to those in wild-type controls, while phospho-histone H3 levels were similar (Figure 7B, S6A).
Figure 7. Normalization of Elevated Mad2 Levels Rescues CIN in a p53 Mutant Mouse Model.
(A) Reporter assays as in Figure 1B comparing p53R175P repressive activity to wild-type p53 and p21. Means ± SD are shown. Western blots below show relative levels of exogenous p53 and p21 and relative induction of endogenous p21 expression.
(B) Western blot analysis and quantification of wild-type, p53C/C, p53C/C p21−/− and p53C/C p21−/− Mad2+/− MEF lysates.
(C) Lymphoma incidence in p53C/C, p53C/C p21−/− and p53C/C p21−/− Mad2+/− animals. p values calculated using one-sided Fisher’s exact tests. Right, representative H&E from splenic lymphomas from mice with indicated genotypes.
(D) Percent of aneuploid cells in tumors from p53C/C, p53C/C p21−/− and p53C/C p21−/− Mad2+/− mice as determined by chromosome FISH. p values calculated using one-sided Fisher’s exact tests.
(E) Model of Mad2 expression regulation showing crosstalk between p53 and Rb pathways via p21 action on the Mad2 promoter. An unknown protein(s) (X) stabilizes the interaction of the repressor E2F (red)/p107 or p130 (p107/p130) complex through the CHR element. Activator E2Fs (blue) serve as transcriptional activators at the upstream E2F binding site.
See also Figure S6.
To assess whether this normalization of Mad2 levels was sufficient to rescue the CIN phenotype observed in p53C/C;p21−/− tumors, we monitored tumor development in p53C/C, p53C/C;p21−/− and p53C/C;p21−/−;Mad2+/− mice. Animals predominantly developed histiocytic sarcomas or lymphomas in the spleen, thymus, and liver with no significant differences in overall tumor latency (Figure S6B). We note that this may be due to the fact that in our mixed genetic background a much smaller difference between latencies in p53C/C and p53C/C p21−/− animals was observed relative to the reported study in which all animals were in a pure C57BL/6 background (Barboza et al., 2006). However, 14% of p53C/C mice (2 out of 14) developed lymphomas, whereas this fraction was increased to 40% in p53C/C;p21−/− mice (8 out of 20). In the p53C/C;p21−/−;Mad2+/− background significantly fewer animals were prone to lymphomagenesis (8%; 1 out of 13 mice; p=0.0468, one-sided Fisher’s exact test) (Figure 7C, S6C), suggesting that elevated Mad2 levels stimulated the development of lymphomas.
To determine whether elevated Mad2 levels were mediating the CIN in p53C/C;p21−/− tumors, we analyzed the ploidy status of the tumors using locus-specific interphase FISH on tumor sections. In line with previous observations (Barboza et al., 2006), tumors from p53C/C mice typically had stable, diploid genomes, whereas in a p21−/− background tumors were highly aneuploid (Figure 7D). Normalization of Mad2 levels in p53C/C;p21−/−;Mad2+/− mice led to a significant reduction in the number of aneuploid tumors (p = 0.0076; one-sided Fisher’s exact test). When we subdivided the total number of aneuploid cells into near-diploid cells and near-tetraploid cells, we noticed that in p53C/C;p21−/− tumors the numbers of near-diploid aneuploid cells and near-tetraploid aneuploid cells were both higher than in tumors that had developed in p53C/C or p53C/C;p21−/−;Mad2+/− animals (Figure S6D). These results indicate that elevated Mad2 levels are largely responsible for the CIN caused by the absence of p21 and suggest that Mad2 overexpression is a crucial mediator of CIN in tumors with defective p53 pathway signaling (Barboza et al., 2006; Liu et al., 2004).
Discussion
While CIN has been observed in a large percentage of human solid tumors, how it arises is now first being explored mechanistically. It has been suggested that the mitotic checkpoint, whose function is to maintain a normal chromosome complement, is lost or weakened by mutation of its key players. Nevertheless, it is difficult to reconcile this with the finding that mutations or deletions of mitotic checkpoint genes are exceedingly rare in human tumors (Perez de Castro et al., 2007; Schvartzman et al., 2010). In fact, cancer cell lines often have a robust checkpoint (Tighe et al., 2001). We have argued that overactivation of the mitotic checkpoint is a much more frequent cause of CIN than loss or partial loss of function. Mitotic checkpoint genes are frequently overexpressed in human tumors (Rhodes et al., 2007), an event that has been shown in vitro and in murine models to lead to CIN (Hernando et al., 2004; Sotillo et al., 2007; Thompson and Compton, 2008).
Here we provide direct evidence that inhibition of the p53 or Rb pathways, events that are widespread in human malignancy, lead to upregulation of Mad2 and that this upregulation is required for generating CIN. This is directly demonstrated by the fact that Mad2 normalization rescues the instability observed in these model systems.
Three recent studies show that pRb loss leads to defects in chromosome condensation and cohesion, abnormal centromere structure and accumulation of DNA damage in vitro (Coschi et al., 2010; Manning et al., 2010; van Harn et al., 2010) and suggest that these chromosomal abnormalities lead to the observed CIN. These seemingly contradictory results can be reconciled if in fact mitotic checkpoint overactivation, as seen with Mad2 upregulation, leads to these abnormalities, a hypothesis that can now be tested directly. Manning et al. (2010) reported that overexpression of E2F1 did not result in centromeric defects but it remains possible that this did not lead to sufficiently high Mad2 levels to induce the effect. Alternatively, the chromosome dynamic changes observed in vitro may not actually be causative of the observed instability. It is also important to note that inactivation of pRb alone leads to only a modest increase in Mad2 (Figure 3A) (Hernando et al., 2004) and in vivo this might not be sufficient to drive instability. Indeed, pituitary tumors that develop in Rb+/− mice (and have lost the wild-type Rb allele) have largely normal karyotypes (Purdie et al., 1994). Centromeric decondensation may well lead to mitotic abnormalities independent of Mad2 upregulation and be responsible for the residual instability that was not rescued by Mad2 normalization in TKO cells (Figure 4B).
We have previously reported that overexpression of Mad2 also leads to chromosome breaks and deletions (Sotillo et al., 2007). However, it is still unclear how these are formed. We speculate that stabilization of Securin leads to impaired Separase-dependent cleavage of Cohesin. Amphitelic attachments would then result in stress at kinetochore-microtubule attachments and at pericentromeric chromatid regions harboring the cohesins that were not removed by dephosphorylation during prophase. Excess force at these points would presumably result in DNA breaks. Pericentromeric DNA breaks of this sort have in fact been seen in cells with spindle defects (Guerrero et al., 2010).
Homozygous loss of all three Rb family members (p107, p130 and pRb) leads to the highest Mad2 levels (Figure 3A) and extensive CIN but p130−/− cells also show strong Mad2 activation. Whether p130−/− tumors are genomically unstable has not been studied extensively but there may be a threshold level of Mad2 required for instability achieved only upon complete pathway inhibition. Complete pathway inhibition can be achieved in a variety of ways, including loss of p16 or amplification of Cyclin D1, and this may account for the high percentage of human tumors that overexpress Mad2 and display CIN (Wiedemeyer et al., 2010).
CIN has long been associated with tumor progression, aggressiveness and invasion but the causative nature of the effect is first beginning to be established by a wide range of mouse modeling experiments (Schvartzman et al., 2010). Further support is provided by our observation here that the growth of allograft fibrosarcomas derived from HrasV12-transformed TKO MEFs is delayed by Mad2 normalization. In addition, that Mad2 heterozygosity in the context of Rb pathway inhibition in vivo leads to the disappearance of anaplastic lesions and a reduction in metastatic capacity points to a role for CIN as the driver of later tumor events, among them invasion and metastasis. We have recently shown that Mad2 overexpression and CIN in a KrasG12D-driven model of lung tumorigenesis promotes tumor recurrence after oncogene withdrawal (Sotillo et al., 2010). Interestingly, recent studies in yeast also suggest that aneuploid cells show growth advantages in non-ideal growth conditions or conditions of stress (Pavelka et al., 2010).
We also show here that p53 represses expression of the Mad2 gene via p21 induction and canonical Rb pathway signaling. The strongest evidence for this is provided by p21 overexpression in TKO MEFs, demonstrating a requirement of Rb pocket proteins for Mad2 repression. That p21 mutants that are impaired in cyclin/Cdk binding also fail to repress Mad2 promoter activity further supports this conclusion. As shown in Figure S2E, p21 overexpression can only repress the Mad2 promoter when the CHR and CDE/E2F sites are intact, suggesting that cell cycle dependent and/or independent effects must be working through these promoter elements. Our identification of repressor E2Fs, pocket proteins, cyclin B1 and cyclin-dependent kinase-1 (CDK-1) as proteins whose binding is dependent on the CHR site in the Mad2 promoter indicates that an as yet unidentified protein directly stabilizes pocket protein-repressor E2F binding to DNA via the CHR site. It is therefore tempting to speculate that these proteins are all part of the CDF1 complex (Figure 7E). This multiprotein repressor complex would also act to inhibit the transcriptional enhancer located upstream, as suggested by the observation that loss of the CHR and E2F/CDE sites leads to constitutive promoter activation only in the presence of upstream E2F sites (Figure S2D). Two observations further suggest that this mechanism of transcriptional regulation is not unique to Mad2, but likely also applies to other mitotic checkpoint genes. First, the promoters of at least three mitotic checkpoint genes, Mad2, BubR1 and CENPE contain E2F binding sites as well as CHR and CDE sites which are highly conserved among various vertebrate species (Figure S2F, G) (Hernando et al., 2004; Polager et al., 2002; Ren et al., 2002). Second, loss of p53 or p21 in primary cells results in an upregulation of both Mad2 and BubR1, independent of the cell cycle (Figures 1A, and 1C). Given the widespread prevalence of CIN in human solid tumors and its role in contributing to escape from oncogene addiction (Sotillo et al., 2010), our findings provide a mechanistic link between necessary tumor driving events (inhibition of the p53 or Rb pathway), tumor progression and escape from therapeutic intervention. Moreover, our results underscore the direct interconnection of tumor suppressor loss and genomic instability. Strategies for targeting aneuploid cells that are likely under stress (Tang et al., 2011) may yield drugs which will have a significant impact on cancer progression and relapse in patients.
Experimental Procedures
Primary Cells, Cell Lines and Cell Culture
TKO (Rb−/−, p107−/−, p130−/−) MEFs were derived from TKO ES cells as described (Sage et al., 2000) (Supplemental Experimental Procedures). All other MEFs were derived from day 12.5 embryos. Primary cells were grown at early passages unless indicated. Mad2 knockdown was achieved with lentiviral vectors from Open Biosystems. Mad2 knock-down, 3T3, seeding efficiency, anchorage independence and focus formation assays were carried out as described in Supplemental Experimental Procedures. Transformation of TKO MEFs for allograft injection was carried out with a pBabe-HrasV12 retroviral vector. For CIN assays, TKO MEFs were plated at low density on glass chamber slides, grown until colonies of 100–500 cells were visible and fixed with 3:1 Methanol:Acetate added dropwise. FISH analysis was performed with two pericentromeric probes from chromosomes 12 and 17.
Reporter Assays
HCT116 or Saos-2 cells were plated in 24-well plates and transfected using Lipofectamine 2000 (Invitrogen) and calcium-phosphate precipitation (Sambrook and Russel, 2001), respectively. Luciferase constructs contained 235 or 204 base pairs of the human Mad2 promoter upstream of the firefly luciferase reporter in the pGL3-Basic vector (Promega). P53 and p21 were expressed from the pcDNA3 vector (Invitrogen). The pRL construct (Promega), from which Renilla luciferase was constitutively expressed, was co-transfected to correct for transfection efficiency. Firefly and Renilla luciferase activities were measured using the Dual Luciferase Assay Reporter System (Promega) and a Bio-Tek Clarity microplate luminometer (Bio-Tek Instruments).
Protein Analysis
Mad2 levels were determined by Western blot of RIPA lysates. Antibodies used are described in Supplemental Information.
Animal Husbandry
All mice were in a 129/C57BL6 mixed genetic background and kept in pathogen-free housing under guidelines approved by the MSKCC Institutional Animal Care and Use Committee and Research Animal Resource Center.
Histopathology
Tissues were formalin-fixed, paraffin-embedded and sectioned at 5 μm. The primary antibodies used for immunohistochemistry were Ki67 (Vector, 1:10000), CyclinD1 (Thermo Scientific, 1:500) and Keratin 8 (TROMA). Immunofluorescence was carried out using anti E-cadherin (BD Transduction 2.5 μg/ml) and anti-Vimentin (Progen 0.1 μg/ml) antibodies. Hematoxylin and Eosin staining was used for standard histology analysis and all tumors assessed without prior knowledge of genotype.
Chromosome Fluorescent In Situ Hybridization (FISH)
Probes were synthesized from three pericentromeric BAC clones of chromosome 12 (Chr 12-RP23-54G4, RP23-41E22 and RP23 16809 5a), chromosome 16 (Chr 16-RP23-290E4, RP23-356A24 and RP24 258J4 4a) and chromosome 17 (Chr 17-RP23-354J18-6c, RP23-73N16 and RP23-202G20) and labelled with SpectrumGreen-dUTP, SpectrumRed-dUTP and SpectrumOrange-dUTP (Vysis), respectively. The BAC DNAs were labelled by nick translation according to standard protocols. The number of hybridization signals for these probes was assessed in a minimum of 150 interphase nuclei with well-delineated contours.
Supplementary Material
Highlights.
p53 represses Mad2 expression via induction of p21 and crosstalk to the Rb pathway
Mad2 upregulation mediates the genomic instability seen upon Rb pathway inhibition
Elevated Mad2 enhances transformation and mediates aggressive mammary tumor formation
Normalization of elevated Mad2 rescues genomic instability in a p53 tumor model
Acknowledgments
We thank J. Sage and A. Koff for TKO ESCs/MEFs and mutant p21 constructs, T. van Dyke for WAPT121 mice, G. Lozano for p53C/C mice, J. van Deursen for BubR1 antibody, J. McGuire and M. Leversha for FISH, B. Vogelstein and K.W. Kinzler for p53+/+, p53−/−, p21+/+ and p21−/− HCT116 cells, E. Hernando for the pGL3-Mad2 promoter(1287)-luciferase construct, M. Higgins for Mad2 promoter analysis, H. Erdjument-Bromage for mass spectrometry analysis, S. Couto and V. Gillespie for histopathological analysis and Y. Chin for technical assistance. We are also grateful to P.J. Cook, C. Mayr and A.M. Unni for critical reading of the manuscript. J.M.S. is supported by a Breast Cancer Research Program (BCRP) Predoctoral Traineeship Award from the US Department of Defense (Congressionally Directed Medical Research Programs), P.H.G.D. is supported by a postdoctoral fellowship from Susan G. Komen for the Cure and a Young Investigator Award from Alex’s Lemonade Stand Foundation, R.S. is funded by the Thomas G. Labrecque Foundation and R.B. is supported by the National Institutes of Health.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, Supplemental Figures S1–S6 and Supplemental Table S1 (related to Figure 4).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G. p21 delays tumor onset by preservation of chromosomal stability. Proc Natl Acad Sci U S A. 2006;103:19842–19847. doi: 10.1073/pnas.0606343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc Natl Acad Sci U S A. 2005;102:11296–11301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Saha P, Kornbluth S, Dynlacht BD, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschi CH, Martens AL, Ritchie K, Francis SM, Chakrabarti S, Berube NG, Dick FA. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2010;24:1351–1363. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Godley LA, Aldaz CM, Pyle R, Shi YP, Pinkel D, Gray J, Bradley A, Medina D, Varmus HE. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev. 1995;9:882–895. doi: 10.1101/gad.9.7.882. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer. 2009:831–841. doi: 10.1038/nrc2731. [DOI] [PubMed] [Google Scholar]
- Flint SJ. Principles of virology: molecular biology, pathogenesis, and control. Washington, D.C: ASM Press; 2000. [Google Scholar]
- Fung TK, Poon RY. A roller coaster ride with the mitotic cyclins. Semin Cell Dev Biol. 2005;16:335–342. doi: 10.1016/j.semcdb.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, Blasco MA. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- Guerrero AA, Gamero MC, Trachana V, Futterer A, Pacios-Bras C, Diaz-Concha NP, Cigudosa JC, Martinez AC, van Wely KH. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Proc Natl Acad Sci U S A. 2010;107:4159–4164. doi: 10.1073/pnas.0912143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- Ikeda MA, Jakoi L, Nevins JR. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci U S A. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila MR, Evan GI. p53--a Jack of all trades but master of none. Nat Rev Cancer. 2009;9:821–829. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- Liu G, Parant JM, Lang G, Chau P, Chavez-Reyes A, El-Naggar AK, Multani A, Chang S, Lozano G. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet. 2004;36:63–68. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
- Liu N, Lucibello FC, Korner K, Wolfraim LA, Zwicker J, Muller R. CDF-1, a novel E2F-unrelated factor, interacts with cell cycle-regulated repressor elements in multiple promoters. Nucleic Acids Res. 1997;25:4915–4920. doi: 10.1093/nar/25.24.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010;24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel L, Diaz-Rodriguez E, Narayan G, Hernando E, Murty VV, Benezra R. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc Natl Acad Sci U S A. 2004;101:4459–4464. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Moberg K, Starz MA, Lees JA. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez de Castro I, de Carcer G, Malumbres M. A census of mitotic cancer genes: new insights into tumor cell biology and cancer therapy. Carcinogenesis. 2007;28:899–912. doi: 10.1093/carcin/bgm019. [DOI] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- Polager S, Kalma Y, Berkovich E, Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21:437–446. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- Purdie CA, Harrison DJ, Peter A, Dobbie L, White S, Howie SE, Salter DM, Bird CC, Wyllie AH, Hooper ML, et al. Tumour incidence, spectrum and ploidy in mice with a large deletion in the p53 gene. Oncogene. 1994;9:603–609. [PubMed] [Google Scholar]
- Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel D. Molecular Cloning: A Laboratory Manual. 3. CSHL Press; 2001. [Google Scholar]
- Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simin K, Wu H, Lu L, Pinkel D, Albertson D, Cardiff RD, Van Dyke T. pRb inactivation in mammary cells reveals common mechanisms for tumor initiation and progression in divergent epithelia. PLoS Biol. 2004;2:E22. doi: 10.1371/journal.pbio.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Williams BR, Siegel JJ, Amon A. Identification of Aneuploidy-Selective Antiproliferation Compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A, Johnson VL, Albertella M, Taylor SS. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2001;2:609–614. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Mak TW, Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol. 2008;18:244–252. doi: 10.1016/j.tcb.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, te Riele H. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010;24:1377–1388. doi: 10.1101/gad.580710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron A, Cherier J, Barre B, Gamelin E, Coqueret O. The cell cycle inhibitor p21waf1 binds to the myc and cdc25A promoters upon DNA damage and induces transcriptional repression. J Biol Chem. 2006;281:34742–34750. doi: 10.1074/jbc.M602492200. [DOI] [PubMed] [Google Scholar]
- Wiedemeyer WR, Dunn IF, Quayle SN, Zhang J, Chheda MG, Dunn GP, Zhuang L, Rosenbluh J, Chen S, Xiao Y, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci U S A. 2010;107:11501–11506. doi: 10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Smolen GA, Zhang J, Wittner B, Schott BJ, Brachtel E, Ramaswamy S, Maheswaran S, Haber DA. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev. 2009;23:1737–1742. doi: 10.1101/gad.1809309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambetti GP, Levine AJ. A comparison of the biological activities of wild-type and mutant p53. FASEB J. 1993;7:855–865. doi: 10.1096/fasebj.7.10.8344485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.