Abstract
The examination of the dermis/epidermis junction (DEJ) is clinically important for skin cancer diagnosis. Reflectance confocal microscopy (RCM) is an emerging tool for detection of skin cancers in vivo. However, visual localization of the DEJ in RCM images, with high accuracy and repeatability, is challenging, especially in fair skin, due to low contrast, heterogeneous structure and high inter- and intra-subject variability. We recently proposed a semi-automated algorithm to localize the DEJ in z-stacks of RCM images of fair skin, based on feature segmentation and classification. Here we extend the algorithm to dark skin. The extended algorithm first decides the skin type and then applies the appropriate DEJ localization method. In dark skin, strong backscatter from the pigment melanin causes the basal cells above the DEJ to appear with high contrast. To locate those high contrast regions, the algorithm operates on small tiles (regions) and finds the peaks of the smoothed average intensity depth profile of each tile. However, for some tiles, due to heterogeneity, multiple peaks in the depth profile exist and the strongest peak might not be the basal layer peak. To select the correct peak, basal cells are represented with a vector of texture features. The peak with most similar features to this feature vector is selected. The results show that the algorithm detected the skin types correctly for all 17 stacks tested (8 fair, 9 dark). The DEJ detection algorithm achieved an average distance from the ground truth DEJ surface of around 4.7μm for dark skin and around 7–14μm for fair skin.
Keywords: confocal reflectance microscopy, image analysis, skin, classification
1. INTRODUCTION
Skin cancer (melanoma or non-melanoma) is one of the most common types of cancer with a rapidly rising incidence1. Melanomas are the most deadly type of skin cancer and can be fatal if not diagnosed and excised early enough. During standard clinical screening visual examination by the naked eye aided by a dermoscope is performed. Both visual examination and dermoscopy are characterized by highly variable sensitivity and specificity that depends on clinician expertise2–3. Visual examination, when an abnormal skin lesion is suspected, is followed by biopsy and histology. Biopsies are invasive, painful, destroy the site, and leave a scar. Studies show that around 80% of biopsies return negative results. To increase the accuracy of clinical exams, reflectance confocal microscopy (RCM)4 has been under development for noninvasive imaging of skin for cancer screening and diagnosis5–9. RCM enables imaging and visualization of the epidermis and superficial dermis layers of the skin. Maximum imaging depth is limited to the papillary dermis or superficial reticular dermis, depending on the state of the overlying epidermis and the dermis/epidermis junction. Nuclear and cellular detail is imaged with nominal (instrumental) optical sectioning of 1–3 μm and lateral resolution of 0.5–1.0 μm, which is comparable to that of conventional pathology. Reflectance confocal microscopy continues to be translated and advance into clinical applications for noninvasive detection of skin cancers8,10. The reported studies in sensitivity and specificity represent a major advance for reflectance confocal microscopy (RCM) toward clinical utility, enabling noninvasive real-time screening and diagnosis, which may then serve as an adjunct to pathology, while minimizing the need for unnecessary biopsy.
RCM acquisition is performed by sequentially capturing optical sections at increasing depths in skin. Horizontal slices acquired at each depth are recorded as a stack of images. Unlike histological sections that are oriented perpendicular to the skin surface and are commonly stained purple and pink with hemotoxylin and eosin to add contrast, reflectance confocal images are oriented parallel (en face) and appear in grayscale (unstained) because they are generated with a single near-IR wavelength. The point spread function and hence optical sectioning, resolution and contrast. all degrade with depth, due to increasing aberrations and scattering. Thus, detection of certain morphologic features, based purely on visual observation, remains challenging, requiring considerable clinical training. Computer automated processing of RCM images may be able to assist visual analysis. So far there are few publications on computer automated processing of RCM images to automatically identify quantitative features11–13.
An example of a clinically important feature is the dermis/epidermis junction (DEJ), which is the 3 dimensional irregular surface separating the superficial epidermis from the underlying deeper dermis. The DEJ is clinically and pathologically important to examine, because cancers often originate and later spread from this location. Therefore, evaluation of the DEJ is important for early diagnosis.
Computer-automated image analysis may assist clinicians with the detection of the DEJ (and other morphologic features). However, in RCM images, the DEJ, like many other such features, is marked by optically subtle changes and features and is difficult to detect, with particular difficulty for lightly pigmented skin types where RCM contrast at the DEJ is poor. Additional challenges for automated-image analysis of RCM stacks from skin include heterogeneity of skin tissue, high inter- and intra-subject variability and low optical contrast. Recently, we proposed a machine learning-based image segmentation/classification algorithm for DEJ localization for lightly pigmented skin types14–16.
In this work we extend this algorithm to locate the DEJ for dark skin types. In dark skin, strong backscatter from the melanin pigment causes the basal layer right above the DEJ to appear bright and with high contrast and is thus easier to detect compared to in fair skin stacks.
In dark skin RCM stacks, the algorithm finds the appropriate peak of the smoothed average intensity depth profile of each tile. Initially we applied a method similar to the one in Gareau et al.11 to localize the basal layer as the position of the most reflective voxel along z (depth) at each lateral (x,y) position. Calculating this most reflective point at each position might in principle result in a surface of basal cells right above the DEJ. However, analyzing multiple data stacks showed that the most reflective voxel along the intensity z-profile was not necessarily the basal layer with a large amount of melanin. In some stacks, epidermis region or deep dermal collagen fibers appeared very bright and hence corresponded to the most reflective voxel. Hence, a method to select the right peak in the intensity z-profile was necessary. Based on our successful approach for light skin, we used 2-D texture features computed for each tile corresponding to a peak and automatically selected the peak corresponding to basal cells by a texture similarity analysis.
The second contribution of the paper is the implementation of a skin type detection algorithm. This algorithm first decides the skin type based on existence of basal layer and then applies the appropriate DEJ localization method.
We successfully tested the automated skin type detection algorithm on 9 RCM stacks using a leave-one-out cross validation approach. We evaluated the performance of automated DEJ detection algorithm from dark skin stacks for which manual delineation of the DEJ was available. The results indicated that the algorithm localized DEJ for those test stacks with average errors around 4.7 μm. We report similar preliminary results that were reported earlier14, 16 in which epidermis/dermis misclassification rates were around 10% and the average localization error relative to expert marked DEJ was 8.5μm.
2. METHODS
2.1 Data acquisition and preprocessing
Imaging was performed on healthy volunteers with a VivaScope 1500™ (Lucid Inc., Rochester NY, operating at 785 nm wavelength). A stack of horizontal images was captured starting at a position below the skin surface. The resolution was 0.5 μm in lateral direction and about 3 μm in the axial direction. The field-of-view was 500μm. After capturing each image, the en-face optical section was translated 1 μm deeper along the optical axis into the tissue from the starting position below the skin surface to a sufficient depth to be in the dermis. The clinician performing the imaging selected both starting and end points as arbitrary positions in epidermis and dermis respectively. After acquisition of a confocal stack, the stack of 8-bit tiff images were loaded into Matlab software for automated processing to locate the DEJ. The image stack was first converted into a volume matrix and the automated preprocessing algorithm was applied.
The images were not aligned and there was shift in lateral directions (x and y) from one image to the next due to patient movement during acquisition. To correct for this shift a standard stack registration algorithm was applied.
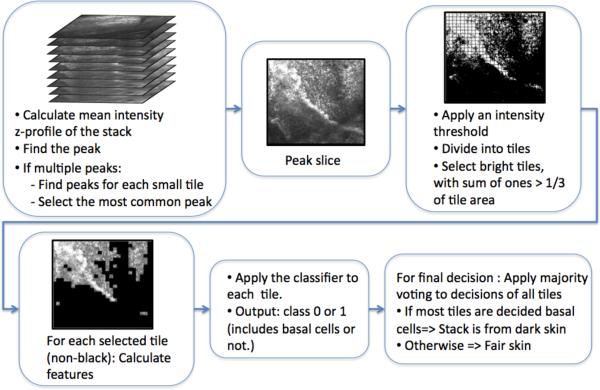
2.2 Automated Skin Type Detection
For a given RCM stack, to determine the skin type, we used an automated algorithm, which determined whether the given stack was dark skin (pigmented skin) or fair skin (very lightly pigmented skin) (See Fig. 2). To determine the skin type, one obvious useful feature is the presence of very bright basal cells in dark skin which are not present in fair skin. Therefore these basal cells need to be searched for within the given stack; if they are present, we can conclude that the stack is from dark skin. To do so, the algorithm first determined the brightest (on average) lateral slice in the stack, which potentially included the basal cells if they were present. To that end, the algorithm calculated the mean intensity for each slice in the given stack and constructed a mean intensity profile with respect to depth (z). That mean intensity z-profile was first smoothed with a Gaussian smoothing filter, then a peak detection algorithm was used to find the peak in the mean intensity z-profile. This depth, corresponding to the peak in the z-profile, determined the most overall reflective slice in the stack.
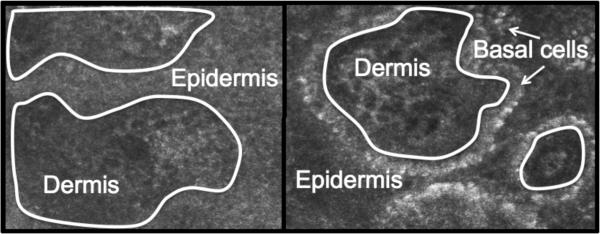
Figure 2.
The left and right panels show two slices from an RCM stack from fair skin (on the left) and dark skin (on the right) respectively. The white boundary drawn is the DEJ.
To finally determine the skin type of the stack, the algorithm further processed this slice. If the stack was from a dark skin type, this brightest slice would include basal cells. If the stack was from a fair skin type, due to the low amount of melanin in the basal cells, the basal cells would not have high reflectivity. Therefore, the brightest slice in the stack could either correspond to a superior slice including fairly bright epidermis regions or to a deeper slice including bright collagen fibers. Thus, the algorithm used a statistical classifier, built with texture features from labeled training stacks, to differentiate the brightest dark skin slice including basal cells from the brightest fair skin slice including either epidermis cells or dermis collagen fibers.
In detail, to detect bright regions in the peak slice, first the peak slice was divided into small square tiles of size 32×32 pixels and thresholded with an automatically selected initial threshold (Otsu's threshold17). Each tile was accepted as a bright tile if the number of thresholded pixels of value 1 in that tile is larger than a third of the area of the tile. After the bright tiles in the peak slice were selected using the initial threshold, the number of those tiles was calculated. If that number is too small (smaller than n=150 tiles), the intensity threshold was gradually decreased until the minimum numbers of tiles (150) were included in the set of selected tiles. Then the classifier was applied to each tile in this selected set. Majority voting, applied to decisions of all tiles, was used to make the final decision for that stack for being either a dark skin or fair skin stack.
There were certain stacks, which resulted in multiple peaks when the peak detection algorithm was applied to the mean intensity z-profiles. For those stacks, the algorithm applied a modified routine, in which the peak slice was not chosen based on global mean z-profile from each slice. Instead the mean z-profiles of all tiles were calculated. The median of the peak location distributions of all tiles was selected as the peak slice. Then, the same steps explained in the previous paragraphs were applied for skin type detection.
After skin type detection, the appropriate automated DEJ localization algorithm was run.
2.3 Automated DEJ detection for dark skin
A similar algorithm to the one explained in the skin type detection section, was used to detect the DEJ in stacks determined by the previous method to be from dark skin type, except applied at the per-tile scale instead of overall for the entire stack. The basal layer including bright basal cells that include highly reflective melanin pigment was detected first. Then DEJ was located as the lower boundary of the basal layer, separating the basal layer from the underlying dermis.
Again we divided the stack into non-overlapping tiles of size 32×32 pixels. For each tile, the location of peak of the mean intensity z-profile was detected as explained in the previous section. For some tiles multiple peaks were detected. From those peaks, some weak peaks, which have small peak height, were removed. To do so, the peak height was defined as the difference between current peak and the next closest local minima of the mean intensity z-profile. The local minimum was detected by applying a peak detection algorithm to the negative of the mean intensity profiles. After removing weak peaks, for some tiles, multiple peaks were still present. The reason for multiple peaks was that, for some tiles, either the basal cells did not appear very bright due to heterogeneity within a stack and resulted in a weaker peak and/or some collagen fibers had higher reflectivity for some regions within the stack and resulted in peaks with comparable or even higher peaks than basal layer peak. Therefore, to select the right peak, corresponding to the basal cells, a peak selection algorithm was implemented.
The peak selection algorithm selected the peak corresponding to the basal cells as follows: First, a template representing the basal layer was constructed from all tiles which had a single peak as follows: these peak tiles were selected, and a set of texture features were calculated from them. The mean feature vector calculated over all of those tiles was used as a template to represent the appearance of basal cells. For tiles with multiple peaks, the same set of features was calculated for each peak tile and the peak tile with the minimum Euclidean distance to the template was chosen as the correct peak.
The detected peak was assumed to correspond to the middle of the basal layer, so the algorithm then found the lower boundary of the basal layer, where the DEJ was located. Since the depth of the basal layer changes from site to site, instead of using a fixed height for the layer the algorithm estimated the lower boundary from the mean intensity z-profile. Inflection points were estimated for the mean intensity z-profile and the first inflection point below the peak was selected. Fig. 4 shows mean intensity profile for a sample tile for which the peak detection algorithm was applied.
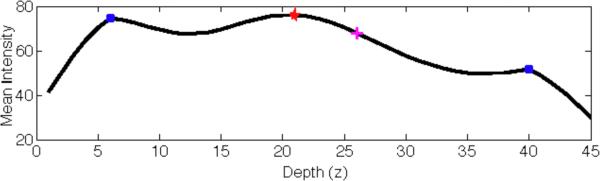
Figure 4.
The figure shows a sample mean intensity profile of a tile along depth direction (in μm). The peak detection algorithm found 3 peaks shown with blue dots and red star. The peak selection algorithm selected the right peak (red star) corresponding to the basal layer. Next, the lower boundary of the basal layer (i.e. the DEJ) was found as the closest inflection point of the mean intensity profile function to this selected peak (shown with magenta plus sign).
The DEJ surface was taken as the collection of tile DEJ depths. A cubic smoothing spline was fit to this surface18 with a surface Laplacian norm regularization term for smoothing. This smoothed spline surface was also used to compute the boundary locations for each pixel for visualization purposes.
2.4 Automated DEJ transitional zone detection for fair skin
For fair skin type, due to the small amount of melanin pigment, the DEJ detection task is harder due to the lack of contrast and strong features at the basal layer. Instead of detecting a strict DEJ, in fair skin RCM stacks, a transition zone was detected. This transition zone has an upper boundary, (i.e. a lower boundary for the epidermis layer) and a lower boundary (i.e. the upper boundary for the dermis). The DEJ is “trapped” between these two boundary surfaces.
We used the algorithm we proposed in previous work16 to detect this DEJ transitional zone in fair skin types. This algorithm is a semi-automated hybrid sequence segmentation/classification algorithm that again works on tiles. The algorithm partitioned each vertical z-stack of tiles into homogenous segments in z (depth). To do so, a model of skin layer dynamics was fitted to the features from the z-sequence of tiles. Then those tile segments were classified as epidermis, dermis, or transitional DEJ region using texture features. That classifier was trained on an RCM stack where the DEJ was manually labeled and applied to new RCM stacks to automatically locate the DEJ.
2.5 Extraction of texture features and classification
Both skin type detection and DEJ detection algorithms explained in the preceding sections use classification of tiles based on texture features. In this section, the set of texture features and the classification method used in those algorithms are explained.
The set of 170 features used in automated skin type detection and DEJ detection algorithms was the set of features we used in our previous work on DEJ detection in light skin14–16. These features, including gray level co-occurrence matrix features (contrast, energy, correlation and homogeneity), statistical metrics (mean, variance, skewness and kurtosis), features from a wavelet decomposition19, log-Gabor features and radial spectral features, were extracted from each tile.
For automated skin type detection, we needed a classifier that works on texture features extracted from each tile and differentiates bright basal cell layer in dark skin stacks from the bright collagen or epidermis regions in fair skin stacks. We used a fast version of standard support vector machine (SVM) classifier called proximal SVM for classification20.
3. RESULTS
We applied the skin type detection algorithm on 17 RCM stacks (8 fair skin stacks, 9 dark skin stacks) from our database. The skin type detection scheme was tested with a leave-one-out scheme. At each iteration, one stack was kept for testing and the remaining stacks were used for training, and we iterated through the dataset choosing different test stacks. We obtained %100 success rate in determining the skin types even for the hardest dark skin stacks where the melanin content did not result in very high reflectivity and was not easily detectable.
From these stacks we chose the five dark skin stacks for which we had ground truth (expert labeling) available and detected the DEJ. Table 1 shows the mean and standard deviation of the distances between expert labeled DEJs and the automatically located DEJs in those 5 stacks. Comparison of the DEJ found by the algorithm (dotted red) with the one marked by the expert (green) for two sample vertical cross sections (x–z) and (y–z) from the first stack in the table is shown in Fig. 5. A similar figure for the second stack is shown in Fig. 6.
Table 1.
Table shows the mean and standard deviation of the distances between expert labeled DEJs and the automatically located DEJs in 5 RCM stacks from dark skin.
| RCM Stack | Mean ± σ (μm) |
|---|---|
| 1 | 5.18 ± 4.05 |
| 2 | 3.32 ± 3.88 |
| 3 | 3.43 ± 3.07 |
| 4 | 3.28 ± 2.77 |
| 5 | 8.52 ± 7.32 |
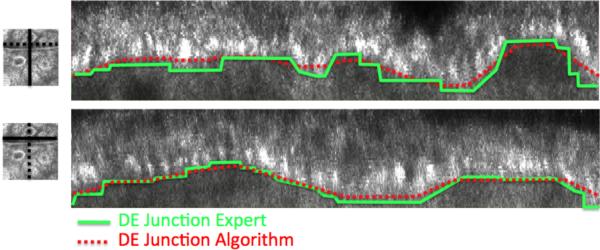
Figure 5.
The upper and lower panels on the right compare the DEJ found by the algorithm (dotted red) with the one marked by the expert (green) for two sample vertical cross sections (x–z) and (y–z) from the RCM stack 1. The solid lines in the left figures indicate the vertical slice location on a sample horizontal slice. Note that the expert marks the DEJ not on the vertical slices but on horizontal slices.
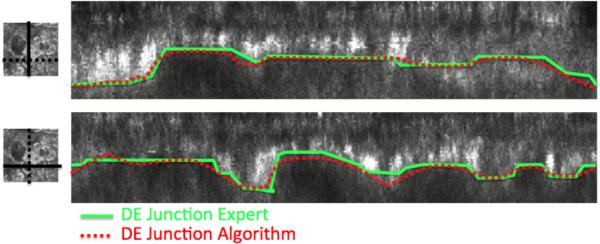
Figure 6.
The same figure as Figure 5 from a different RCM stack 2.
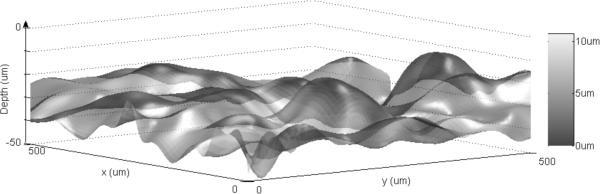
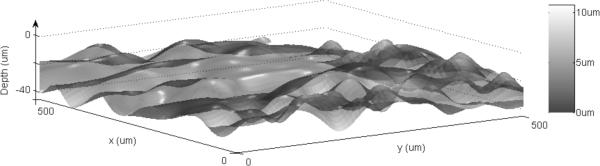
A surface plot of the DEJ automatically found by the algorithm is shown in 3D in comparison to expert labeled DEJ for the same stacks in Figs. 7 and 8 respectively. The surface itself indicates the resultant DEJ of the algorithm and the color map indicates the distance from the expert labeled DEJ (error).
Figure 7.
Surface plot of the DEJ automatically found by the algorithm is shown in 3D in comparison to expert labeled DEJ for RCM stack 1 (data1r2). The surface itself indicates the resultant DEJ of the algorithm and the color map indicates the distance from the expert labeled DEJ (error).
Figure 8.
The same figure as Fig. 6 for RCM stack 2(data3r3).
The results of the DEJ detection algorithm for fair skin were reported in our previous work. Initial results demonstrated the detectability of the DEJ with epidermis/dermis misclassification rates smaller than 10% and average distance from the expert labeled boundaries around 8.5 μm.
4. CONCLUSION AND FUTURE WORK
In this work, we developed an algorithm to detect whether the RCM stack is from light or dark skin type and to then locate the DEJ surface in RCM image stacks using the DEJ detection algorithm for either dark (this work) or fair (previous work) skin types. The proposed algorithm decided the skin type with a classifier applied to high reflectivity small tiles from the most reflective slice in the stack and classified those tiles either as basal cells or not. The stacks including reflective basal cells were classified as dark skin. The dark skin DEJ detection algorithm used a similar idea to detect the peaks in the mean intensity profiles for each tile and then select the peaks that corresponded to the basal cells. After locating the basal cells, the lower boundary of the basal cells corresponding to the DEJ was found and the DEJ surface was constructed. The results show that the algorithm detected the skin type with %100 accuracy on the 9 stacks tested, using a leave-one-out training method. The DEJ algorithm for dark skin type also resulted in reasonable performance with average distance from the ground truth DEJ surface around 4.7μm. Similar results for DEJ detection algorithm for fair skin types resulted in epidermis/dermis misclassification rates smaller than 10% and average distance from the expert labeled boundaries around 7–14 μm.
Figure 1.
The left panel shows the structure of skin in a vertical cross section diagram. The DEJ is the thin membrane, marked in red, that separates the epidermis from the dermis. The basal layer lies directly on the DEJ. The basal layer is typically at average depth of 100 μm below the surface in normal skin and 10–16 μm in thickness [1]. The right figure shows a vertical slice from an RCM volume constructed from an RCM image stack of dark skin. The layer of bright regions corresponds to the basal layer including more melanin with high reflectivity.
Figure 3.
Flow chart for the automated skin type detection algorithm
Acknowledgments
We thank Dr. Alon Scope, Dr. Juliana Casagrande and Dr. Itay Klaz for providing us the manual segmentations of DEJ in CRM images. This research was supported in part by Unilever and in part by NIH/NCRR Center for Integrative Biomedical Computing (CIBC), P41-RR12553-12.
REFERENCES
- [1].Gloster H, Brodland DG. The epidemiology of skin cancer. British Journal of Dermatology. 2008;22:217–226. doi: 10.1111/j.1524-4725.1996.tb00312.x. [DOI] [PubMed] [Google Scholar]
- [2].Vestergaard M, Macaskill P, Holt P, Menzies S. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: a meta-analysis of studies performed in a clinical setting. British Journal of Dermatology. 2008;159(3):669–676. doi: 10.1111/j.1365-2133.2008.08713.x. [DOI] [PubMed] [Google Scholar]
- [3].Rajpara S, Botello A, Townend J, Ormerod A. Systematic review of dermoscopy and digital dermoscopy/artificial intelligence for the diagnosis of melanoma. British Journal of Dermatology. 2009;161(3):591–604. doi: 10.1111/j.1365-2133.2009.09093.x. [DOI] [PubMed] [Google Scholar]
- [4].Gonzalez SG, Gill M, Halpern A. Reflectance Confocal Microscopy of Cutaneous Tumors: An Atlas with Clinical, Dermoscopic and Histological Correlations. Informa Healthcare; London: 2008. [Google Scholar]
- [5].Nori S, Rius-Díaz F, Cuevas J, Goldgeier M, Jaen P, Torres A, González S. Sensitivity and specificity of reflectance-mode confocal microscopy for in vivo diagnosis of basal cell carcinoma: A multicenter study. J. American Academy of Dermatol. 2004;51:923–930. doi: 10.1016/j.jaad.2004.06.028. [DOI] [PubMed] [Google Scholar]
- [6].Pellacani G, Guitera P, Longo C, Avramidis M, Seidenari S, Menzies S. The impact of in vivo reflectance confocal microscopy for the diagnostic accuracy of melanoma and equivocal melanocytic lesions. J. Invest. Dermatol. 2007;147(14):2759–65. doi: 10.1038/sj.jid.5700993. [DOI] [PubMed] [Google Scholar]
- [7].Guitera P, Pellacani G, Longo C, Seidenari S, Avramidis M, Menzies SW. cIn Vivo Reflectance Confocal Microscopy Enhances Secondary Evaluation of Melanocytic Lesions. J. Invest. Dermatol. 2009;149:131–138. doi: 10.1038/jid.2008.193. [DOI] [PubMed] [Google Scholar]
- [8].Guitera P, Pellacani G, Crotty KA, Scolyer RA, Li LX, Bassoli S, Vinceti M, Rabinovitz H, Longo C, Menzies SW. The Impact of In Vivo Reflectance Confocal Microscopy on the Diagnostic Accuracy of Lentigo Maligna and Equivocal Pigmented and Nonpigmented Macules of the Face. J. Invest. Dermatol. 2010 doi: 10.1038/jid.2010.84. [DOI] [PubMed] [Google Scholar]
- [9].Calzavara-Pinton P, Longo C, Venturini M, Sala R, Pellacani G. Reflectance Confocal Microscopy for In Vivo Skin Imaging. Photochemistry and photobiology. 2008;84(6):1421–1430. doi: 10.1111/j.1751-1097.2008.00443.x. [DOI] [PubMed] [Google Scholar]
- [10].Gerger A, Koller S, Weger W, Richtig E, Kerl H, Samonigg H, Krippl P, Smolle J. Sensitivity and specificity of confocal laser-scanning microscopy for in vivo diagnosis of malignant skin tumors. Cancer. 2006;107(1):193–200. doi: 10.1002/cncr.21910. [DOI] [PubMed] [Google Scholar]
- [11].Gareau DS, Hennessy R, Wan E, Pellacani G, Jacques SL. Automated detection of malignant features in confocal microscopy on superficial spreading melanoma versus nevi. J. Biomedical Optics. 2010;15(6) doi: 10.1117/1.3524301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Koller S, Wiltgen M, Ahlgrimm-Siess V, Weger W, Hofmann-Wellenhof R, Richtig E, Smolle J, Gerger A. In vivo reflectance confocal microscopy: automated diagnostic image analysis of melanocytic skin tumours. Journal of the European Academy of Dermatology and Venereology: JEADV. 2010 doi: 10.1111/j.1468-3083.2010.03834.x. [DOI] [PubMed] [Google Scholar]
- [13].Wiltgen M, Gerger A, Wagner C, Smolle J. Automatic Identification of Diagnostic Significant Regions in Confocal Laser Scanning Microscopy of Melanocytic Skin Tumors. Methods Inf. Med. 2008;47(1):14–25. doi: 10.3414/me0463. [DOI] [PubMed] [Google Scholar]
- [14].Kurugol S, Dy JG, Brooks DH, Rajadhyaksha M. Localizing the dermis/epidermis boundary in reflectance confocal microscopy images with a hybrid classification algorithm. Proc. of IEEE Int. Symposium on Biomed. Imaging: From Nano to Macro. 2009:1322–1325. [Google Scholar]
- [15].Kurugol S, Dy JG, Brooks DH, Rajadhyaksha M. Detection of the dermis/epidermis boundary in reflectance confocal images using multi-scale classifier with adaptive texture features. Proc. Of IEEE Int. Symposium on Biomed. Imaging: From Nano to Macro. 2008:492–495. [Google Scholar]
- [16].Kurugol S, Dy JG, Brooks DH, Rajadhyaksha M. A pilot study of semi-automated localization of the dermal/epidermal junction in reflectance confocal images of skin. Journal of Biomedical Optics. doi: 10.1117/1.3549740. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gonzales R, Woods R. Digital Image Processing. Prentice Hall; 2002. [Google Scholar]
- [18].Björck A. Numerical Methods for Least Squares Problems. SIAM; 1996. [Google Scholar]
- [19].Randen T, Husoy JH. Filtering for texture classification: a comparative study. IEEE Trans. Pattern Anal. Mach. Intel. 1999;21:291–310. [Google Scholar]
- [20].Fung G, Mangasarian OL. Proximal Support Vector Machine Classifiers. Proceedings KDD-2001: Knowledge Discovery and Data Mining. 2001:77–86. [Google Scholar]