Abstract
Use of the yeast two-hybrid assay to study Plasmodium falciparum protein-protein interactions is limited by poor expression of P. falciparum genes in yeast and lack of easily implemented assays to confirm the results. We report here two methods to create gene fragments – random fragmentation by partial DNAse I digestion and generation of densely overlapping fragments by PCR – that enable most portions of P. falciparum genes to be expressed and screened in the yeast two-hybrid assay. The PCR-based method is less technically challenging and facilitates fine-scale mapping of protein interaction domains. Both approaches revealed a putative interaction between PfMyb2 (PF10_0327) and PFC0365w. We developed new plasmids to express the proteins in wheat germ extracts and confirmed the interaction in both the split-luciferase assay and in co-purification experiments with glutathione-S-transferase and HA-tagged proteins. The combination of improved yeast two-hybrid screening approaches and convenient systems to validate interactions enhances the utility of yeast two-hybrid assays for P. falciparum.
Keywords: Plasmodium falciparum, protein expression, yeast, Saccharomyces cerevisiae, yeast two-hybrid assay, split luciferase assay
The yeast two-hybrid assay is a powerful tool for the discovery and characterization of protein-protein interactions. Since it was first reported more than 20 years ago [1], the assay and its many variations have been integral to the analysis of thousands of proteins. However, the yeast two-hybrid assay has not been widely used to study Plasmodium protein-protein interactions, due in large part to the poor expression of P. falciparum genes in Saccharomyces cerevisiae [2]. AT-rich P. falciparum sequences resemble S. cerevisiae motifs that specify cleavage and polyadenylation of the nascent RNA [2]. These sequences cause P. falciparum mRNAs expressed in yeast to be truncated prematurely and result in degradation by the mRNA surveillance pathway [2] [3]. To improve expression of P. falciparum genes in yeast, we previously reported the identification of yeast strains with mutations in the mRNA processing pathway [3]. Although these strains are useful for expressing P. falciparum proteins for functional studies and pair-wise yeast-two-hybrid assays, they grow more slowly and mate less efficiently than parental strains, and are not optimal for library-based yeast two-hybrid screens. Thus, alternative methods are needed to improve the yeast two-hybrid assay for P. falciparum. Here we report the development of two approaches to generate fragments of P. falciparum genes for use in the yeast two-hybrid assay. These fragmentation approaches enabled identification of interactions that could not be detected with full-length genes and are generally applicable to other systems as well.
Large-scale sequencing of clones from P. falciparum yeast two-hybrid libraries revealed gene fragments from a wide range of genes and detected no biases [4]. Since these libraries included only fragments that were expressed in yeast, most P. falciparum genes appear to contain sequences that can be expressed even if the full-length gene cannot. To more fully investigate this possibility, we tested two methods to fragment P. falciparum genes using PfMYB2 (PF10_0327, 2745 nucleotide, 74 % AT) as a test case. PfMYB2 encodes a 915-amino acid protein with two MYB DNA-binding domains at the N-terminus. Based on the presence of the MYB domains, PfMyb2 has been proposed to function as a helix-turn-helix transcription factor [5].
Random fragments of PfMYB2 were created using partial DNAse I digestion in the presence of manganese, which promotes the formation of blunt-ended fragments and fragments with one base 5’ or 3’ overhangs. After polishing the ends with T4 DNA polymerase, we ligated double-stranded DNA oligos to the fragments; the oligos were homologous to the sequences flanking the multiple cloning site in the yeast two-hybrid DNA binding domain plasmid pOBD.111 to enable cloning by recombination in yeast. The DNA was then size-fractionated on a Sephacryl S400 column to remove small fragments and unligated oligos. Fragments larger than ~300 base pairs (bp) were PCR-amplified and cloned into pOBD.111 by gap repair in the yeast strain R2HMet [4, 6]. Yeast expressing PfMYB2 fragments were selected on medium lacking tryptophan and methionine. Because the MET2 gene is fused to the 3’ end of the PfMYB2 fragment, growth of yeast in the absence of methionine indicates that the PfMYB2 fragment is expressed. Twelve fragments with diverse start and end points were identified (Fig. 1A, below bar). These fragments included most of the PfMYB2 gene except for an ~ 400 bp region near the center. Based on these data, it was not possible to determine if this region could not be expressed in yeast or if too few clones were evaluated.
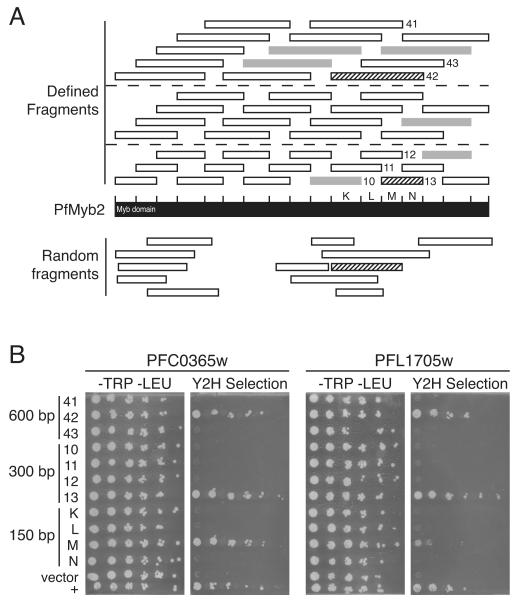
Fig. 1.
Gene fragmentation approaches to improve P. falciparum yeast two-hybrid screens. A. Gene fragments generated by partial DNAse I digestion (bottom) and PCR (top). Black bar represents PfMyb2 (PF10_0327). Bars below PfMYB2 represent fragments generated by partial DNAse I digestion of PfMYB2 that were expressed in yeast. Tick marks above the bar indicate the positions of PCR primers, which were spaced at ~150-bp intervals. Short bars above PfMyb2 represent 300-, 450-, and 600-bp fragments; dashed lines separate fragments by size. All fragments were cloned into pOBD.111 by homologous recombination in the yeast strain R2HMet and tested for expression of the fragment by growth on synthetic dropout (SD) medium lacking methionine; the MET2 gene is fused to the 3’ end of the PfMYB2 fragment and is expressed only if the PfMYB2 fragment is also expressed. White bars represent gene fragments that were expressed, whereas gray bars represent those that were not. Numbers and letters indicate fragments that were retested for interaction with PFC0365w and PFL1705w in part B; letters designate 150-bp fragments. The twelve DNAse I-generated fragments and all 600-bp defined fragments were screened against a P. falciparum yeast two-hybrid library. Striped bars indicate fragments that interacted with both PFC0365w and PFL1705w in the yeast two-hybrid assay. B. Confirmation of yeast two-hybrid interactions with PfMYB2 and mapping of the PfMYB2 protein interaction domain. PfMYB2 150-bp fragments K-N, 300-bp fragments #10-13, and 600-bp fragments #41-43 were cloned into pOBD.111 in the yeast strain R2HMet. Similarly, the PFC0365w and PFL1705w fragments that interacted with PfMYB2 fragment #42 were PCR-amplified and inserted into pOAD.102 by homologous recombination in the yeast strain BK100. Yeast harboring DNA-binding domain and activation domain plasmids were allowed to mate and then grown on SD medium lacking tryptophan and leucine to select diploid cells containing both plasmids. Yeast were grown to mid log phase, diluted to an OD600 of 1.0, serially diluted 5-fold in dH2O, and plated on SD medium lacking tryptophan and leucine or yeast two-hybrid selection medium (SD medium lacking tryptophan, leucine, methione, uracil, and histidine and containing 1 mM 3 amino-4,5-triazole (3-AT); 3-AT is a competitive inhibitor of the yeast two-hybrid reporter enzyme His3 that is added to growth medium to inhibit background yeast growth).
Because of the incomplete coverage of the random fragments and the technical challenges of the partial DNAse I digestion, we investigated an alternative approach to fragment PfMYB2. DNA oligos were designed to hybridize at ~ 150 bp intervals on PfMYB2 and used to create a mini-library of densely overlapping gene fragments (Fig. 1A, top, Supplementary Table 1). Every possible 300-, 450-, and 600-bp fragment was PCR amplified and cloned into pOBD.111 by in vivo homologous recombination. Fragments expressed in yeast were identified by growth on medium lacking methionine. Of the 45 fragments tested, 39 fragments were expressed at sufficient levels to enable yeast growth. These fragments covered the entire PfMYB2 gene, indicating that the region for which no random fragments were obtained can be expressed and that the lack of random fragments in this region was due to sampling error.
To identify P. falciparum proteins that interacted with PfMYB2, all 600-bp defined fragments and the 12 random fragments were screened against a P. falciparum activation domain library [4]. Screens were performed manually as described in supplemental methods. Both defined fragment #42 and a random fragment with nearly the same start point interacted with fragments of PFC0365w and PFL1705w; these genes were the only ones found by both a defined and a random fragment. To confirm these interactions and to further delineate the domains of PfMyb2 implicated in the interactions, we recloned the PFC0356w (nucleotides 133-615, amino acids 45-205) and PFL1705w (nucleotides 286-639, amino acids 95-213) fragments into the activation domain vector in fresh yeast and tested their ability to interact with a series of PfMYB2 fragments that overlapped with fragment #42. Both PFC0365w and PFL1705w interacted with PfMYB2 fragment 600-bp #42 and the 300-bp fragment #13 (nucleotides 1936-2235, amino acids 645-745). The 150-bp fragment M (nucleotides 1936-2085, amino acids 646-695) interacted with PFC0365w to the same extent as larger fragments #42 and #13, suggesting that the entire PFC0365w binding region was encoded by this fragment. In contrast, PFL1705w interacted poorly with fragment M and not at all with adjacent fragment N, which was encompassed by fragment #13, suggesting that the binding domain spans the junction of fragments M and N. This experiment delineated a 50 amino acid region of PfMyb2 as the binding domain for PFC0365w and a 100 amino acid region as the binding domain for PFL1705w. Surprisingly, fragment #12, which contained fragment M, and fragments #41 and #43, which included the sequences in fragments M and #13, did not interact with either PFC0365w and PFL1705w.
A second problem with yeast two-hybrid screens of P. falciparum genes is the lack of convenient systems to validate interactions. P. falciparum proteins express poorly in most heterologous systems, antibodies are not available for the vast majority of P. falciparum proteins, and generating transgenic parasites expressing epitope-tagged proteins for colocalization and co-purification experiments is still time-consuming and technically challenging. To facilitate confirmation of yeast two-hybrid interactions, we utilized wheat germ extracts, which have been reported to improve the expression of a wide range of P. falciparum proteins [7-9]. Four new plasmids were developed to express PfMyb2, PFC0356w, and PFL1705w in the wheat germ in vitro translation system for use in the split-luciferase assay and co-purifications with glutathione S transferase (GST)- and HA-epitope tagged proteins (Fig. 2A).
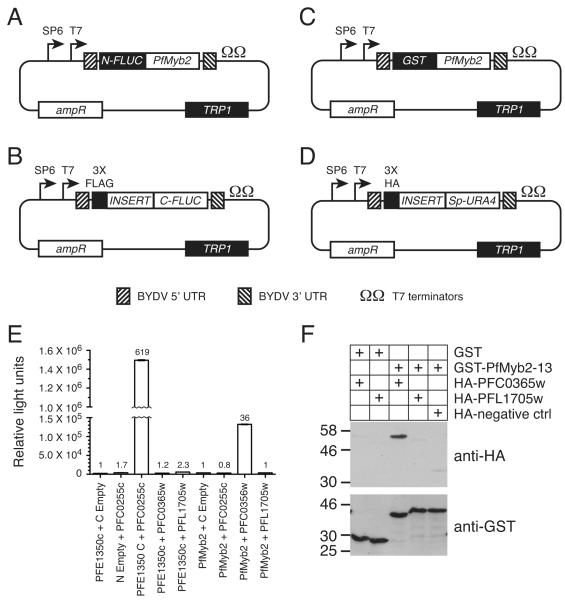
Fig. 2.
Confirmation of PfMyb2 interactions. A-D. New vectors for expressing fusion proteins in wheat germ extracts. The SP6 and T7 promoters, 5’ and 3’ UTRs from barley yellow dwarf virus (BYDV), and T7 transcription terminators from plasmid pF3A WG (BYDV) (Promega) were cloned into plasmid p424-GPD [20] in place of the GPD1 promoter and CYC1 terminator sequences to generate plasmid p424-BYDV-UTR. DNA sequences encoding the N-terminal firefly luciferase fragment (N-FLUC, amino acids 1-398) (A), C-terminal firefly luciferase fragment (C-FLUC, amino acids 394-550) (B), glutathione S transferase (GST) (C), and the URA4 gene from pOAD.102 (Sp-URA4) (D) were inserted into p424-BYDV-UTR by homologous recombination. The C-FLUC and Sp-URA4 plasmids contain the multiple cloning region and flanking sequences from pOAD.102 to facilitate cloning of fragments identified in yeast two-hybrid screens, plus sequences encoding three copies of the FLAG or HA epitope tags, respectively. Similarly, the N-FLUC and GST plasmids contain the multiple cloning region and flanking sequences from pOBD2 to facilitate transfer of genes from yeast two-hybrid DNA binding domain plasmids. All plasmids confer resistance to ampicillin in E. coli and have unique restriction sites for traditional cloning approaches. E. Confirmation of the PfMyb2-PFC0365w interaction in the split luciferase assay. PfMYB2 300-bp fragment #13 and PFE1350c were cloned into the N-FLUC plasmid, whereas the PFC0365w and PFL1705w fragments from the yeast two-hybrid screen and PFC0255c were cloned into the C-FLUC plasmid. All proteins were expressed using the TNT® SP6 High-Yield Wheat Germ Protein Expression System according to the manufacturer’s protocol (Promega). Expression was verified by western blotting with anti-FLAG (Sigma) and anti-N-terminal firefly luciferase (Santa Cruz Biotechnology) antibodies (Supplementary Fig. 1). Equal volumes of N-FLUC and C-FLUC in vitro translation reactions were added to PBS supplemented with 1% BSA and protease inhibitors (Roche), incubated at 18 h 4 °C with occasional mixing, and assayed for luciferase activity in triplicate. Graph shows the average relative light units (RLU) and standard deviation from each sample after subtracting background luminescence from the buffer only sample. The ratio of the RLU from each combination of N-FLUC and C-FLUC fusions relative to the N-FLUC fusion plus C-FLUC negative control is shown above the bar. F. Confirmation of the PfMyb2-PFC0365w interaction by co-purification. PfMYB2 300-bp fragment #13 and the PFC0365w and PFL1705w fragments from (E) were cloned into the expression vectors shown in (C) and (D), respectively. GST- and HA-tagged proteins were expressed in the TNT® SP6 High-Yield Wheat Germ Protein Expression System (Promega). A three-fold excess volume of the in vitro translation reactions of the HA-tagged proteins was added to GST or GST-PfMYB2 in PBS supplemented with 1% BSA, 0.1 % Tween 20, and protease inhibitors (Roche). After overnight incubation at 4 °C with rotation, GST tagged proteins were purified using glutathione beads and eluted by boiling in SDS sample buffer. Samples were subjected to SDS PAGE, transferred to nitrocellulose membrane, and immunoblotted with antibodies against HA (top) and GST (bottom). Proteins were imaged by chemiluminescence (GE Health Sciences). Molecular weight markers (sizes in kDa) are shown to the right of each blot.
In the split luciferase assay, proteins of interest are fused to N- and C-terminal fragments of luciferase. If the proteins interact, the N- and C-terminal luciferase fragments associate to create a functional luciferase enzyme [10]. Though a number of variants of this system have been described, for these experiments we used fragments of firefly luciferase that were optimized to yield a high signal to background ratio and sufficient enzymatic activity to be easily measured [11]. We first tested the system using the P. falciparum interacting proteins PFE1350c (nucleotides 25-405, amino acids 9-135) and PFC0255c (nucleotides 10-411, amino acids 4-137) [4, 12], which were fused to the N- and C-FLUC domains, respectively. The fusion proteins and the negative controls were expressed in wheat germ extracts, mixed, incubated overnight, and assayed for luciferase activity. The combination of N-FLUC-PFE1350c and PFC0255c-C-FLUC produced a strong signal that was more than 600-fold greater than the background signal from N-FLUC-PFE1350c and the C-FLUC negative control. Neither PFC0255c-C-FLUC plus the N-FLUC negative control, nor the combination of N-FLUC-PFE1350c with either PFC0365w-C-FLUC or PFL1705w-C-FLUC produced luciferase activity more than 2.3-fold above the signal from N-FLUC-PFE1350c plus the C-FLUC negative control. N-FLUC-PfMyb2 fragment #13 yielded a strong signal when mixed with PFC0365w-C-FLUC (36-fold greater than N-FLUC-PfMyb2 plus the C-FLUC negative control), thus confirming a physical interaction between PfMyb2 and PFC0365w. However, the combination of N-FLUC-PfMyb2 and PFL1705w-C-FLUC resulted in only background levels of luciferase activity.
We obtained similar results in co-purification experiments using GST-tagged PfMyb2 fragment #13 and HA-epitope tagged PFC0365w and PFL1705w. HA-PFC0365w copurified with GST-PfMyb2 but not GST alone, whereas HA-PFL1705w did not co-purify with either protein. Thus, we were able to validate the interaction between PfMyb2 and PFC0365w in two independent assays. In addition, the S. cerevisiae proteins with greatest homology to PfMyb2 and PFC0365w (Cef1 and Prp19) also interact with each other [13, 14]. Thus, the interaction between PfMyb2 and PFC0365w is of high confidence. In contrast, the interaction between PfMyb2 and PFL1705w was not confirmed in either secondary assay and there is no evidence for an interaction between Cef1 and the S. cerevisiae ortholog of PFL1705w, Mot2. The Saccharomyces Genome Database (www.yeastgenome.org) currently lists 16 studies that used a variety of techniques to identify 62 proteins that physically interacted with Cef1 or Mot2. None of these studies found an interaction between Cef1 and Mot2, nor is there any data to suggest a potential genetic interaction between these genes in yeast. While we cannot dismiss the possibility that the split-luciferase and the GST pulldown results are false-negatives, the interaction is not supported by other evidence and is of lower confidence.
Surprisingly, there was no detectable homology between PfMyb2 and Cef1 in the PFC0365w/PRP19 binding domains, even though the yeast orthologs of PfMyb2 and PFC0365w also interact. In fact, the only significant homology between Cef1 and PfMyb2 was in the first 233 amino acids that encode the Myb DNA-binding domain motifs (49 % identical, 70 % similar). This result was unexpected because protein interaction interfaces are generally thought to evolve more slowly, since changing residues in one protein would require compensating changes in the binding partner. The interaction between PfMyb2 and PFC0365w demonstrates that this is not always the case. A similar observation has been made for three pairs of interacting proteins from Caenorhabditis elegans [15]. Despite the lack of homology between PfMyb2 and Cef1 C-terminal to the MYB DNA-binding domain, the fact that PfMyb2 binds to the P. falciparum PRP19 homolog suggests that PfMyb2 is the functional homolog of Cef1. Since Cef1 is primarily involved in RNA splicing, we propose that PfMyb2 functions in a similar manner and is unlikely to act as a transcription factor.
In summary, we developed improved methods to identify and confirm P. falciparum protein-protein interactions in the yeast two-hybrid assay. Though these approaches were designed to address problems specific to P. falciparum, densely overlapping fragmentation and the split-luciferase assay are likely to be useful for studying protein-protein interactions in other species as well. Of the two methods investigated, the defined fragmentation approach was less technically challenging to implement, ensured that all regions of the target gene were tested for expression, and facilitated mapping of the interaction domain. The use of fragments not only enabled all regions of PfMYB2 to be expressed in yeast, but also was critical for identifying protein-protein interactions. The interaction between PfMyb2 and PFC0365w was extremely sensitive to both the start and end points of the fragments expressed in the yeast two-hybrid vectors and some fragments that completely overlapped with the protein interaction domain failed to yield positive interactions. This suggests that binding regions must be presented in the appropriate context in the yeast two-hybrid assay in order to detect a given interaction, and that small deviations can prevent detection of the interaction. It is likely that this phenomenon contributes to the large number of false-negatives in the yeast two-hybrid assay. Others have also noted that the use of fragments enables interactions to be identified that may be missed when full-length genes were used [15, 16]. Similarly, yeast two-hybrid screens of activation domain libraries containing fragments of varying sizes and sequences often yield more positives than screens of the same constructs against arrays of full-length genes [15, 17]. However, we must also add a note of caution. Though this approach successfully identified an interaction mediated by a relatively small interaction domain, some interaction domains are much larger. The densely overlapping fragmentation approach is unlikely to identify these interactions because full-length P. falciparum genes and large fragments are poorly expressed in yeast. In addition, this method will likely be limited by the same factors that affect other yeast two-hybrid approaches. Some interactions can be identified in one version of yeast two-hybrid assay but not others [18], or require the interacting partners be cloned in a particular orientation (activation domain versus DNA binding domain) or at a particular end of the activation or DNA binding domains [19].
As previously reported, cell free translation in wheat germ extracts efficiently synthesized P. falciparum fusion proteins for confirmatory assays. For example, the data shown in Fig. 2 were generated from the equivalent of 0.12 μl of in vitro translation reaction. Although both confirmation approaches yielded equivalent results, the split-luciferase assay had significant advantages over GST pulldowns. The split-luciferase assay was far easier to implement, required less hands-on time, involved no wash steps, and needed less optimization to find conditions that gave a reasonable signal to noise ratio. Because the assay can be performed in 96-well plates, many pairs can be evaluated simultaneously and multiple replicates can be performed for each pair being tested. Although it was initially envisioned solely as a tool to rapidly validate yeast two-hybrid interactions, future experiments will investigate the potential to use the split luciferase assay as a screen for novel interactions.
Supplementary Material
Supplementary Fig. 1. Expression of P. falciparum fusion proteins in wheat germ extracts. A. Immunoblot of C-FLUC fusion proteins. The indicated fusions to C-FLUC were expressed in the TNT® SP6 High-Yield Wheat Germ Protein Expression System (Promega), separated by SDS PAGE, transferred to nitrocellulose, and subjected to immunoblotting with anti-FLAG antisera (Sigma). B. Immunoblot of N-FLUC fusion proteins. The indicated fusions to N-FLUC were expressed and subjected to immunoblotting as in (A) with anti-N-terminal FLUC antisera (Santa Cruz Biotechnology). C. Immunoblot of HA epitope tagged proteins. HA epitope tagged proteins were expressed and subjected to immunoblotting as in (A) with anti-HA antisera (Rockland).
Acknowledgements
We thank L. W. Schoenfeld for technical assistance, and Drs. R. Paulmuragan and S. Gambhir for providing the split luciferase plasmids. This work was supported by grants GM092829 and P50 GM64655 from the NIH. S.F. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–6. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- [2].Sibley CH, Brophy VH, Cheesman S, Hamilton KL, Hankins EG, Wooden JM, et al. Yeast as a model system to study drugs effective against apicomplexan proteins. Methods. 1997;13:190–207. doi: 10.1006/meth.1997.0511. [DOI] [PubMed] [Google Scholar]
- [3].LaCount DJ, Schoenfeld LW, Fields S. Selection of yeast strains with enhanced expression of Plasmodium falciparum proteins. Mol Biochem Parasitol. 2009;163:119–22. doi: 10.1016/j.molbiopara.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–7. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- [5].Bischoff E, Vaquero C. In silico and biological survey of transcription-associated proteins implicated in the transcriptional machinery during the erythrocytic development of Plasmodium falciparum. BMC Genomics. 2010;11:34. doi: 10.1186/1471-2164-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–16. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- [7].Mudeppa DG, Pang CK, Tsuboi T, Endo Y, Buckner FS, Varani G, et al. Cell-free production of functional Plasmodium falciparum dihydrofolate reductase-thymidylate synthase. Mol Biochem Parasitol. 2007;151:216–9. doi: 10.1016/j.molbiopara.2006.10.016. [DOI] [PubMed] [Google Scholar]
- [8].Tsuboi T, Takeo S, Iriko H, Jin L, Tsuchimochi M, Matsuda S, et al. Wheat germ cell-free system-based production of malaria proteins for discovery of novel vaccine candidates. Infect Immun. 2008;76:1702–8. doi: 10.1128/IAI.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tsuboi T, Takeo S, Sawasaki T, Torii M, Endo Y. An efficient approach to the production of vaccines against the malaria parasite. Methods Mol Biol. 2010;607:73–83. doi: 10.1007/978-1-60327-331-2_8. [DOI] [PubMed] [Google Scholar]
- [10].Ozawa T, Kaihara A, Sato M, Tachihara K, Umezawa Y. Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Anal Chem. 2001;73:2516–21. doi: 10.1021/ac0013296. [DOI] [PubMed] [Google Scholar]
- [11].Paulmurugan R, Gambhir SS. Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal Chem. 2007;79:2346–53. doi: 10.1021/ac062053q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Alexandrov A, Vignali M, LaCount DJ, Quartley E, de Vries C, De Rosa D, et al. A facile method for high-throughput co-expression of protein pairs. Mol Cell Proteomics. 2004;3:934–8. doi: 10.1074/mcp.T400008-MCP200. [DOI] [PubMed] [Google Scholar]
- [13].Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. Rna. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tsai WY, Chow YT, Chen HR, Huang KT, Hong RI, Jan SP, et al. Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J Biol Chem. 1999;274:9455–62. doi: 10.1074/jbc.274.14.9455. [DOI] [PubMed] [Google Scholar]
- [15].Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–45. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Flajolet M, Rotondo G, Daviet L, Bergametti F, Inchauspe G, Tiollais P, et al. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene. 2000;242:369–79. doi: 10.1016/s0378-1119(99)00511-9. [DOI] [PubMed] [Google Scholar]
- [17].Pilot-Storck F, Chopin E, Rual JF, Baudot A, Dobrokhotov P, Robinson-Rechavi M, et al. Interactome mapping of the phosphatidylinositol 3-kinase-mammalian target of rapamycin pathway identifies deformed epidermal autoregulatory factor-1 as a new glycogen synthase kinase-3 interactor. Mol Cell Proteomics. 2010;9:1578–93. doi: 10.1074/mcp.M900568-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stanyon CA, Liu G, Mangiola BA, Patel N, Giot L, Kuang B, et al. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen YC, Rajagopala SV, Stellberger T, Uetz P. Exhaustive benchmarking of the yeast two-hybrid system. Nat Methods. 2010;7:667–8. doi: 10.1038/nmeth0910-667. author reply 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–22. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Expression of P. falciparum fusion proteins in wheat germ extracts. A. Immunoblot of C-FLUC fusion proteins. The indicated fusions to C-FLUC were expressed in the TNT® SP6 High-Yield Wheat Germ Protein Expression System (Promega), separated by SDS PAGE, transferred to nitrocellulose, and subjected to immunoblotting with anti-FLAG antisera (Sigma). B. Immunoblot of N-FLUC fusion proteins. The indicated fusions to N-FLUC were expressed and subjected to immunoblotting as in (A) with anti-N-terminal FLUC antisera (Santa Cruz Biotechnology). C. Immunoblot of HA epitope tagged proteins. HA epitope tagged proteins were expressed and subjected to immunoblotting as in (A) with anti-HA antisera (Rockland).