Abstract
Gene knockdown by RNA interference (RNAi) in Caenorhabditis elegans is readily achieved by feeding bacteria expressing double-stranded RNA (dsRNA). Enhanced RNAi (Eri) mutants facilitate RNAi due to their hypersensitivity to dsRNA. Here, we compare eight Eri mutants for sensitivity to ingested dsRNA, targeting a variety of tissue-specific genes.
THE effectiveness of double-strand RNA (dsRNA) delivery in Caenorhabditis elegans has made high-throughput RNA interference (RNAi) screens an essential research tool (Mitani 2009). For RNAi screens, dsRNA is usually administered via feeding RNAi, whereby worms ingest bacteria expressing gene-specific dsRNA (referred to as RNAi food). This is a less potent procedure than microinjecting dsRNAs, perhaps due to the lower amounts of internalized dsRNA (Timmons and Fire 1998). The discovery of enhanced RNAi (Eri) mutants, which increases the sensitivity of worms to dsRNA, increases the discovery of RNAi phenotypes in large-scale screens. Nine Eri loci have been discovered thus far (Simmer et al. 2002; Kennedy et al. 2004; Duchaine et al. 2006; Fischer et al. 2008; Pavelec et al. 2009).
Although a variety of Eri mutants are used in RNAi screens, their selection has been ad hoc, as no systematic comparative analysis of the Eri strains has been reported. Such an analysis would provide a logical basis for selecting the most sensitive Eri mutant for general and tissue-specific screens. Here, we comprehensively characterize the tissue-specific RNAi sensitivities of eight Eri mutants. To characterize phenotypic differences among Eri mutants, we compared the relative penetrance of RNAi sensitivity at varying doses of dsRNA-expressing bacteria (Rea et al. 2007). For each bacterial strain that expresses dsRNA targeting a C. elegans gene, we scored only one defined knockdown phenotype (supporting information, Table S1, Supporting Citations). A representative dilution series is shown in Figure S1 (Table S2). We sought to use this dose-response data to compare the enhanced silencing for each Eri mutant. For all strains, the variability in penetrance is greatest at intermediate dsRNA doses, suggesting a threshold effect. This variability, best observed via coefficient of variations (Table S3), strongly interferes with determining the onset of silencing. In contrast, the trend toward reduced variability at higher dsRNA doses provides a means to discriminate among Eri mutants. On the basis of this analysis, we developed a criterion for selecting the “most effective” Eri’s: one(s) that causes near complete (upper bound of 95% confidence interval at least 100% penetrant) and robust (<10% standard deviation) silencing at the lowest dsRNA dose.
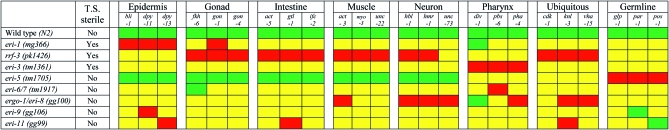
We used the methods and criterion described above (File S1) to evaluate eight Eri mutants on 24 RNAi foods in either tissues (Table S1, Supporting Citations). The results of this analysis are presented in Figure 1 (Tables S4–S27). The majority of Eri mutants enhanced RNAi for nearly all tested tissues, but all showed relative differences in RNAi hypersensitivity for some foods. Our comprehensive phenotypic analysis of the Eri mutants indicates that they are not equivalent, consistent with the reported nonoverlapping expression profiles of eri-1 and rrf-3 mutants (Lee et al. 2006).
Figure 1.—
Summary metric of tissue-specific Eri efficacy for the eight Eri strains tested on 24 RNAi foods representing eight tissues. A strain exhibiting significantly higher (t test, P < 0.05) penetrance than the N2 wild-type strain’s penetrance (green), at any tested bacterial RNAi food concentration, is marked as Eri (yellow or red). Strains exhibiting an Eri phenotype that have an upper bound of 95% confidence interval at least 100% penetrant with a <10% standard deviation are marked as the “best” Eri (red). “T.S. sterile” indicates strains that exhibit temperature-sensitive sterility at 25°.
In all experiments, we observed a sigmoidal curve for silencing penetrance vs. RNAi concentration; at intermediate concentrations, the variance was highest. Therefore, to minimize variability associated with dose, all feeding RNAi assays should be preceded by a dilution series control to ensure that the RNAi food is not used at an “inflection point” concentration. When dsRNA doses cannot be controlled, using the most appropriate Eri mutant maximizes robustness and sensitivity.
We selected RNAi targets on the basis of tissue-specific gene expression and/or phenotypes and interpreted the data based on these differences, but it is important to consider that the differences in responses might relate to unknown relationships between the genes. Consistent with our goal, most sets of tissue-specific genes show consistent phenotypes within Eri mutant classes. For rrf-3 and ergo-1/eri-8, we analyzed a second independent allele, finding similar tissue specificity (Figure S2). Therefore, the observed tissue specificity is likely a property of the eri genes rather than a consequence of unique alleles.
To further document tissue-specific Eri phenotypes, we crossed all the Eri mutants into a sur-5::gfp strain that ubiquitously expresses GFP in all cells (Gu et al. 1998). All the eri(−);sur-5::gfp doubles exhibited spontaneous transgene silencing (Figure S3), which interfered with the effect of gfp dsRNA. However, consistent with the tissue-specific effects described above, the relative differences in spontaneous gfp silencing in the intestinal nuclei among eri-1;sur-5::gfp, rrf-3;sur-5::gfp and ergo-1/eri-8;sur-5::gfp strains corresponded with their relative differences in RNAi efficacy against endogenous intestinal targets (Table S28).
A limited comparison of the Eri phenotypes of the retinoblastoma pathway mutants lin-15b(n744) and lin-35(n745) (Wang et al. 2005) with eri-1 and rrf-3 showed that their sensitivity and robustness were less than that of the Eri mutants (Figure S4).
We also found that all the Eri mutants show strong maternal rescue (Figure S5 and Figure S6 and Table S29, Table S30, Table S31, and Table S32). However, there is no maternal rescue for the temperature-sensitive (T.S.) sterility phenotype of T.S. Eri mutants (Table S33). This is not due to a perdurance problem in which the maternally loaded products are depleted before eri-related spermatogenesis begins because we utilized bli-1 RNAi—whose target is expressed only during the fourth larval stage (Page and Johnstone 1997) when spermatogenesis begins—and found penetrant maternal rescue of the Eri phenotype (Table S34). Eri maternal rescue could suggest that part of the maternal contribution to the embryo includes small RNAs or their associates.
The described tissue-specific RNAi sensitivities, T.S. sterility data, maternal rescue penetrance, brood size (Table S27), and effect on transgenes provide a practical guide to the selection of Eri mutants (File S2, Figure S7). There are other weaker enhanced RNAi mutants, including dcr-1/eri-4(mg375) (Pavelec et al. 2009), tissue-specific sid-1 overexpressers (Calixto et al. 2010), and transgene-specific silencers (Knight and Bass 2002). Although these may not be versatile genetic tools, their future phenotypic analysis is equally important because understanding the interactions among all eri genes provides insights about small RNA pathways.
Acknowledgments
We thank D. Pavelec and S. Kennedy for providing strains and mapping data for the unpublished eri-11 gene. We thank members of the Hunter lab for helpful discussions. This work was supported by National Science Foundation grant MCB 0744029 to C.P.H. and by a Graduate Research Fellowship to J.J.Z.
LITERATURE CITED
- Calixto A., Chelur D., Topalidou I., Chen X., Chalfie M., 2010. Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Methods 7: 554–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., et al. , 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Fischer S. E. J., Butler M. D., Pan Q., Ruvkun G., 2008. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature 455: 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T., Orita S., Han M., 1998. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol. 18: 4556–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S., Wang D., Ruvkun G., 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649 [DOI] [PubMed] [Google Scholar]
- Knight S. W., Bass B. L., 2002. The role of RNA editing by ADARs in RNAi. Mol. Cell 10: 809–817 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Hammell C. M., Ambros V., 2006. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani S., 2009. Nematode, an experimental animal in the national BioResource project. Exp. Anim. 58: 351–356 [DOI] [PubMed] [Google Scholar]
- Page A. P., Johnstone I. L., 1997. The cuticle (March 19, 2007), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.138.1, http://www.wormbook.org
- Pavelec D. M., Lachowiec J., Duchaine T. F., Smith H. E., Kennedy S., 2009. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 183: 1283–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S. L., Ventura N., Johnson T. E., 2007. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 5: 2312–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., et al. , 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12: 1317–1319 [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Wang D., Kennedy S., Conte D., Kim J. K., Gabel H. W., et al. , 2005. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436: 593–597 [DOI] [PubMed] [Google Scholar]