Abstract
[URE3] is a prion (infectious protein) of the Saccharomyces cerevisiae Ure2p, a regulator of nitrogen catabolism. We show that wild S. paradoxus can be infected with a [URE3] prion, supporting the use of S. cerevisiae as a prion test bed. We find that the Ure2p of Candida albicans and C. glabrata also regulate nitrogen catabolism. Conservation of amino acid sequence within the prion domain of Ure2p has been proposed as evidence that the [URE3] prion helps its host. We show that the C. albicans Ure2p, which does not conserve this sequence, can nonetheless form a [URE3] prion in S. cerevisiae, but the C. glabrata Ure2p, which does have the conserved sequence, cannot form [URE3] as judged by its performance in S. cerevisiae. These results suggest that the sequence is not conserved to preserve prion forming ability.
THE Saccharomyces cerevisiae Ure2 protein is central in nitrogen catabolite repression—the ability to repress the uptake and utilization systems for poor nitrogen sources when a good nitrogen source is present (Cooper 2002). The C-terminal domain of Ure2p (amino acids 94–354) is sufficient for its regulatory function when overproduced (Masison and Wickner 1995), although the first 93 amino acids are important to stabilize the protein against degradation (Shewmaker et al. 2007). However, these proximal 93 amino acids allow Ure2p to form a prion, an infectious inactive form of the protein (Masison and Wickner 1995; Wickner 1994). The first 65 amino acids have been shown sufficient to propagate the prion form of Ure2p (Masison et al. 1997), and form infectious amyloid with a parallel in-register β-sheet structure (Brachmann et al. 2005; Baxa et al. 2007; reviewed in Wickner et al. 2008).
The C-terminal domains of the Ure2 proteins of ascomycete yeasts, starting at amino acid 100 of S. cerevisiae, show strong conservation (Edskes and Wickner 2002; Baudin-Baillieu et al. 2003; Harrison et al. 2007). For instance the C-terminal domains of the Ure2 proteins of the human pathogenic yeasts Candida albicans and C. glabrata share, respectively, 80 and 92% sequence identity with the S. cerevisiae protein, and each can functionally substitute for S. cerevisiae Ure2p (Edskes and Wickner 2002).
In contrast to the conservation in the C-terminal domains the N-terminal domains of these three proteins are poorly conserved, although they all contain a substantial amount of asparagine residues (Edskes and Wickner 2002; Baudin-Baillieu et al. 2003; Harrison et al. 2007). In addition, the S. cerevisiae and C. glabrata proteins share a 30-amino-acid domain found in some but not all Ure2 proteins (residues 10–39 of the S. cerevisiae Ure2p) (Edskes and Wickner 2002). This domain, in isolation, has a strong propensity to form amyloid (Baxa et al. 2005; Chan et al. 2005), but that amyloid is not infectious (Brachmann et al. 2005).
Ure2p is thought to transmit information about the nitrogen state of the environment through an interaction with the transcription factor Gln3p (reviewed in Cooper 2002; Magasanik and Kaiser 2002). Curiously, in contrast to Ure2 proteins of different ascomycete yeasts, the Gln3 proteins show limited sequence conservation. Identity is limited to ∼52 centrally located amino acids comprising a zinc finger domain. This zinc finger domain is characteristic of fungal transcription factors involved in nitrogen catabolite repression. In addition, the three Gln3 proteins share a 7- or 8-residue C-terminal sequence. Overall identity between S. cerevisiae and C. glabrata Gln3 proteins is 34%, between S. cerevisiae and C. albicans Gln3 proteins is 24%, and between C. glabrata and C. albicans Gln3 proteins is also 24%. While the Gln3p of C. albicans is known to regulate nitrogen catabolism (Liao et al. 2008), it is not clear that the albicans Ure2p does so, or does so through Gln3p.
C. albicans and C. glabrata are predominantly found as commensals of warm-blooded animals. Both species can proliferate in healthy people and, when the immune system is weakened, can cause mucosal and bloodstream infections (Kaur et al. 2005; Bialkova and Subik 2006; Noble and Johnson 2007; Brisse et al. 2009; Lewis 2009). C. albicans causes 45–60% of invasive candidiasis while C. glabrata contributes 20–22% of the cases. Bloodstream infections of either organism have a death rate of 40–50% (Lewis 2009). These organisms are distantly related, with C. glabrata closer to S. cerevisiae. C. glabrata is a haploid yeast, whereas C. albicans, like S. cerevisiae, is found in the wild as a diploid. Although an incomplete sexual cycle has been described for C. albicans, no mating has been observed in C. glabrata (Nielsen and Heitman 2007). Nonetheless, loci with sequence similarity to the S. cerevisiae MATa and MATα are found in C. glabrata populations.
Prions (infectious proteins) of yeast and fungi include [URE3], [PSI+], [PIN+], [SWI+], [MCA], [OCT+], [MOT+], and [ISP+] of S. cerevisiae and [Het-s] of Podospora anserina, which are amyloids of Ure2p, Sup35p, Rnq1p, Swi1p, Mca1p, Cyc8p, Mot3p, Sfp1p, and HETs, respectively (Wickner 1994; Coustou et al. 1997; Derkatch et al. 2001; Du et al. 2008; Alberti et al. 2009; Nemecek et al. 2009; Patel et al. 2009; Rogoza et al. 2010). Sup35p is a subunit of the translation termination factor; Rnq1p has no known function; Swi1p, Cyc8p, Mot3p, and Sfp1 are transcription factors; and Mca1p is a metacaspase (putative protease). HETs, in its prion form, is involved in heterokaryon incompatibility in P. anserina. In yeast and fungi, prions are both infectious proteins (the definition of a prion), spreading horizontally by cytoplasmic mixing (cytoduction), and heritable as genes, passing vertically to mitotic offspring.
The first suggestion that prions might be beneficial was the discovery of a prion underlying a heterokaryon incompatibility phenomenon in P. anserina (Coustou et al. 1997; comment by Wickner 1997), but this prion also induces a meiotic drive system, calling that interpretation into question (Dalstra et al. 2003). It was then proposed that [PSI+] had a beneficial effect on yeast, enabling it to resist stress (Eaglestone et al. 1999) and thereby promoting evolvability (True and Lindquist 2000). It has likewise been suggested that [URE3] is beneficial (Shorter and Lindquist 2005), and the conservation, in some yeasts, of a 30-residue region in the Ure2p prion domain (see above) was cited as evidence for the usefulness of prion formation (Harrison et al. 2007). Since the Ure2p of C. glabrata has this conserved region, but it is missing from that of C. albicans, we undertook a study of their prion forming abilities in part as a test of these hypotheses.
MATERIALS AND METHODS
Strains and media:
Media are as described by Sherman (1991), except YES medium, which contains 5 g/liter yeast extract, 30 g/liter dextrose, and 30 mg/liter tryptophan. Strains are listed in Table 1. All C. albicans strains were derived from SN148 (Noble and Johnson 2005). Disruption of genes and marker exchange were performed according to Dennison et al. (2005). Transformation of C. albicans was performed using the Li acetate method at 44° (Ramon and Fonzi 2009). The URA2 gene was disrupted with LAL and LHL (Dennison et al. 2005). Subsequently, IRO1 and URA3 were restored through transformation with a PCR product originating 418 bp upstream of IRO1 and terminating 408 bp downstream of URA3. As template, genomic DNA from the Darlington strain of C. albicans was used (Kakeya et al. 2000). After URA3 was restored, CRE-mediated recombination was initiated [to remove the arginine (LAL) and histidine (LHL) markers] using a LEU2-marked CRE gene controlled by the MET3 promoter and directed to the ARG4 locus. This produced strain HCA17. Sequences containing 5′ ARG4 and the MET3 promoter as well as ARG4 3′ sequences were amplified from pCAD (Dennison et al. 2005). LEU2 was amplified from SN100 genomic DNA (Noble and Johnson 2005). The whole CRE cassette was cloned into the EcoRV site of pBC KS+ (Stratagene) resulting in pH948. The URE2 gene was then disrupted with LAL and LHL and the markers again removed through CRE-mediated recombination giving strains HCA37 and HCA40, which both grew slightly slower than the parental strain HCA17.
TABLE 1.
Strains of S. cerevisiae and S. paradoxus
| Strain | Genotype | Reference |
| Strains used for S. paradoxus [URE3] study | ||
| MA544 | S.p. MATα ade1 leu1 trp5 lys1 ho::kanMX | Talarek et al. (2005) |
| MA578 | S.p. MATaade2 his4 ura3 leu2 ho::kanMX PDAL5:ADE2 | Talarek et al. (2005) |
| LM156 | S.c. MATα ura2 leu2 his3 PDAL5:ADE2 URE2paradoxus [URE3para156] | Edskes et al. (2009) |
| 4884 | S.c. MATatrp1 ura2 his3 leu2 kar1∆15hygr PDAL5:ADE2 URE2paradoxus [ure-o] | This work |
| 4891 | S.c. MATatrp1 ura2 his3 leu2 kar1∆15hygr PDAL5:ADE2 URE2paradoxus [URE3para156] | This work |
| 4888 | S.c. MATα ura2 his3leu2 kar1-1 PDAL5:ADE2 URE2paradoxus [URE3para156] | This work |
| 4899 | S.p. MATα ura3 leu1 trp5 ade2-1 PDAL5:ADE2 ho::kanMX [ure-o] | Meiotic segregant of MA544 and MA578 |
| YJM498 | Wild-type S.p. isolated from a patient | J. McCusker; Nakayashiki et al. (2005) |
| Y-1548 | Wild-type S.p. isolated from oak exudate in The Netherlands | C. Kurtzman; Nakayashiki et al. (2005) |
| Strains of Candida albicans | ||
| HCA17 (wt) | MATa/MATα arg4∆/arg4∆ leu2∆/leu2∆ his1∆/his1∆ ura2::loxP/ura2::loxP ura3::imm434/URA3 iro1::imm434/IRO1 | This work |
| HCA37, HCA40 (HCA17 ure2Δ) | MATa/MATα arg4∆/arg4∆ leu2∆/leu2∆ his1∆/his1∆ ura2::loxP/ura2::loxP ura3::imm434/URA3 iro1∆::imm434/IRO1 ure2::loxP/ure2::loxP | |
| Strains of C. glabrata | ||
| BG14 (wt) | ura3∆::TN903NeoR | Cormack and Falkow (1999) |
| BG88b = BG14his3Δ | ura3∆::TN903NeoR his3∆ | Cormack and Falkow (1999) |
| HCg1 = BG88b ure2Δ | ura3∆::TN903NeoR his3∆ ure2::HIS3 | |
| HCg7 = BG88b ure2Δ | ura3∆::TN903NeoR his3∆ ure2::HIS3 | |
| Strains of S. cerevisiae | ||
| YHE711 (wt) | MATα ura2 leu2::hisG | Edskes et al. (1999b) |
| TIFY3 | MATα ura2 leu2::hisG ure2::G418 | Tiffany Weinkopff |
| BY256 | MATahis3 leu2 trp1 ure2::kanMX PDAL5:ADE2 PDAL5:CAN1 kar1 | Brachmann et al. (2005) |
| BY302 | MATahis3 leu2 trp1 URE2albicans PDAL5:ADE2 PDAL5:CAN1 kar1 | |
| BY304 | MATahis3 leu2 trp1 URE2glabrata PDAL5:ADE2 PDAL5:CAN1 kar1 | |
| YHE1178 | MATα ura2 leu2 URE2albicans PDAL5:ADE2 | BY302 × YHE887 |
| YHE1174 | MATα ura2 leu2 kar1 URE2glabrata PDAL5:ADE2 | BY304 × YHE887 |
| YHE887 | MATα ura2 leu2 ure2::kanMX (sigma background) | |
| YHE1265 | BY304 with URE2glabrata replaced with URE2cerevisiae | |
| YHE1207 | BY302 [URE3alb] | |
| YHE1271 | YHE1265 [URE3cer] | |
HCA37 and HCA40 share the first disrupted URE2 allele but were independently disrupted in the second URE2 allele. The “156” in [URE3para156] identifies a particular isolate.
URE2 was disrupted in C. glabrata strain BG88b (Cormack and Falkow 1999) using a PCR product obtained from genomic DNA of C. glabrata strain 37A (Miyazaki et al. 1998) that contained the HIS3 gene (bordered by loxP sites), starting 443 bp upstream of the start codon and terminating at the HIS3 stop codon. C. glabrata strains BG14 and BG88b are both derived from clinical isolate BG2 (Fidel et al. 1996). C. glabrata was transformed by the Li acetate method (Geitz and Woods 2002). Both HCg1 and HCg7 grew slightly slower than the parental strain BG14.
URE2 of YHE711 was replaced with kanMX4 by transformation with a PCR- amplified ure2::kanMX4 from the corresponding S. cerevisiae Genome Deletion Project strain. S. cerevisiae was transformed by the Li acetate method (Geitz and Woods 2002). TIFY3 grew slightly slower than parental strain YHE711.
We integrated URE2albicans and URE2glabrata into the URE2 locus of strain BY256 replacing the kanMX gene present there (ure2::kanMX). The ORF of URE2albicans or URE2glabrata flanked by 179 bp 5′-UTR and 463 bp 3′-UTR from the S. cerevisiae URE2 was transformed into strain BY256. Transformants containing URE2albicans (BY302) or URE2glabrata (BY304) at the S. cerevisiae URE2 locus were selected using their resistance to canavanine, sensitivity to G418, and inability to grow on medium lacking adenine. There are no CTG codons in URE2albicans.
Assay of [URE3]:
DAL5, encoding alantoate permease, is strongly repressed by Ure2p. Dal5p also recognizes and takes up ureidosuccinate (USA), an intermediate in uracil biosynthesis. As a result, inactivity of Ure2p may be detected by ability of ura2 cells to grow on USA (33 μg/ml) in the presence of ammonia as nitrogen source (e.g., Difco yeast nitrogen base without amino acids). Alternatively, the DAL5 promoter may be fused to ADE2 and adenine prototrophy, and a change from red to white colony color on adenine-limiting media indicates Ure2p inactivity.
Cytoduction:
Cytoplasm is transferred from one strain to another using the kar1 mutation defective in nuclear fusion (Conde and Fink 1976). The donor of cytoplasm is ρ+ and the recipient, of opposite mating type, is made ρo by growth on ethidium bromide. Cells are mixed in water and incubated on rich medium for ∼7 hr. The mixture is then streaked for single colonies on plates selecting against the donor strain. Donor or recipient must have a kar1 mutation to largely prevent nuclear fusion. Mating occurs and the unfused nuclei separate at the next cell division. Clones with the nuclear markers of the recipient (but not diploid) and ρ+ are cytoductants and are scored for the prion phenotype.
Microarray protocols:
Total RNA was isolated from log phase cultures of S. cerevisiae (strains YHE711 and TIFY3), C. albicans (strains HCA17, HCA37, and HCA40) and C. glabrata (strains BG88b, HCg1, and HCg7) grown in YPAD at 30° using Trizol (Invitrogen) and purified using the RNeasy MiniElute cleanup kit (Qiagen). S. cerevisiae microarrays were purchased from Agilent. For C. glabrata, Agilent custom microarrays were used as described (Tsai et al. 2010) C. albicans microarrys (Brown et al. 2006) were purchased from the Genome Sequencing Center at Washington University, St. Louis, MO. Microarray hybridization was performed by the Genomic Technologies Section, Research Technologies Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Bethesda, MD (Tim Myers) as described by Tsai et al. (2010). Statistical calculations were performed on the processed signal data by using the mAdb analysis system provided by the BIMAS group at the Center for Information Technology, NIH. Data were filtered with the parameters that included genes present in three or more arrays per group and each array with 80% or more genes present. Genes not present in both URE2 deletion strains of C. albicans and C. glabrata were discarded. Signals of genes scoring consistently <100 pixels on all arrays were discarded as background. Genes for which reciprocal labeling showed an inconsistent trend were discarded as well as genes positive in three or more arrays per group but not positive in the majority of arrays.
RESULTS
Nomenclature:
As previously described (Edskes et al. 2009), we indicate a [URE3] prion originating in cells expressing the C. albicans Ure2p and propagating in cells expressing the S. cerevisiae Ure2p by the symbol [URE3alb]cer (if there were such a strain). In this work, [URE3alb]alb will usually be abbreviated to [URE3alb]. When we refer to [URE3] of the S. paradoxus Ure2p in S. paradoxus cells, we will be explicit.
Wild S. paradoxus strains can propagate [URE3]:
Because of the difficulties of Candida genetics, we chose to examine the prion-forming abilities of C. albicans and C. glabrata Ure2p in S. cerevisiae, an approach often used by others as well (Chernoff et al. 2000; Kushnirov et al. 2000; Santoso et al. 2000; Nakayashiki et al. 2001). However, while the Ure2p of S. paradoxus forms a prion in S. cerevisiae (Edskes and Wickner 2002; Edskes et al. 2009), it was found to not form [URE3] in S. paradoxus itself (Talarek et al. 2005), casting doubt on this approach.
As reported by Talarek et al. (2005), we found that we could not select [URE3para]para in S. paradoxus strain MA578, even on overproduction of Ure2ppara. We tested whether the [URE3para]para, generated in S. cerevisiae carrying the paradoxus URE2 gene in place of that of cerevisiae, could be transmitted to S. paradoxus by cytoduction (cytoplasmic mixing) to strain MA578, and found that none of several [URE3para] variants were transmitted (Table 2 and data not shown). However, using genetic crosses, we constructed several other S. paradoxus strains with the PDAL5:ADE2 construct used for scoring [URE3] as Ade+, and found that, for example, S. paradoxus strain 4899 could be made [URE3para] by cytoduction from S. cerevisiae [URE3para] donors (Table 2). Each of these Ade+ cytoductants was cured by growth on 3 mm guanidine, an inhibitor of the disaggregating chaperone Hsp104, whose activity is necessary for propagating the S. cerevisiae [URE3]. Further, these cytoductants could transmit their Ade+ trait back to S. cerevisiae strain 4884 by cytoduction. Thus, S. paradoxus strain 4899 can propagate [URE3para].
TABLE 2.
S. paradoxus can propagate [URE3paradoxus]
| Donors | Recipients | Cytoductants |
| S.c.4888[URE3para] | S.p.MA544 | 25 all Ade− |
| S.c.4888[URE3para] | S.p.4899 | 7 Ade+, 5 Ade− |
| S.c.4891[URE3para156], | 20 segregants of S.p. YJM498 spores | >20 Ade+ in each cytoduction, rare Ade− |
| S.c.4888[URE3para156] | X S.p. 4899 | |
| S.c.4891[URE3para156], | 8 segregants of S.p. Y-1548 spores | >20 Ade+ in each cytoduction, rare Ade− |
| S.c.4888[URE3para156] | X S.p. 4899 |
S.c., Saccharomyces cerevisiae; S.p., Saccharomyces paradoxus. An interspecies cytoduction of [URE3para] from S.c. URE2paradoxus [URE3para] strains to S.p. strains was carried out as for S. cerevisiae. The PDAL5:ADE2, with the DAL5 promoter controlling the ADE2 gene, was used as a reporter of Ure2p activity. [ure-o] cells are Ade−, while [URE3] cells are Ade+ because active Ure2p prevents transcription from the DAL5 promoter (Rai et al. 1987).
To determine whether wild S. paradoxus can propagate [URE3para], we carried out meiotic crosses between S.p.4899 and germinating meiotic spores from the wild S.p. strains YJM498 and Y-1548. From the former cross we chose 20 and from the latter 8 suitably marked (ade2-1 PDAL5:ADE2ho::kanMX) segregants, and used each as a cytoduction recipient from a S.c. [URE3para] donor. We found that each of the 28 segregants from the two crosses could stably maintain [URE3para], indicating that there is at least no single gene defect in either wild strain preventing [URE3para] propagation (Table 2). These results indicate that wild S. paradoxus can be [URE3] and suggest that the use of S. cerevisiae as a test bed for potential prion proteins appears to be valid.
C. albicans and C. glabrata Ure2p regulate N-catabolism genes:
GLN3 is known to have an important role in nitrogen regulation in C. albicans (Dabas and Morschhauser 2007; Dabas and Morschhauser 2008; Liao et al. 2008). As the Ure2 proteins from C. glabrata and C. albicans fully restore nitrogen regulation in a S. cerevisiae ure2Δ strain (Edskes and Wickner 2002), it seems likely that nitrogen regulation in these two Candida species is also directed by Ure2p. However, the limited sequence identity found among the Gln3 proteins from S. cerevisiae, C. albicans, and C. glabrata (supporting information, Figure S1) brings into question whether the response of these organisms to Ure2p inactivation parallel each other. To record the transcriptional response of these three organisms to Ure2p inactivation, we disrupted URE2 in S. cerevisiae, C. albicans, and C. glabrata.
We find that among the 182 genes of S. cerevisiae whose transcription is repressed twofold or more by Ure2p in rich medium, 73 are involved in nitrogen utilization and amino acid metabolism (File S1). Of these 182 genes, 47 were reported by Godard et al. (2007) to be nitrogen catabolite repression (NCR) target genes. None of the 22 genes that we found twofold or more upregulated by Ure2p were NCR targets. These results are similar to previous work in S. cerevisiae (Cox et al. 1999; Shamji et al. 2000; Ross and Wickner 2004).
Of the 46 C. albicans genes derepressed by ure2Δ, 16 are homologous to Godard et al.'s (2007) NCR target genes. Of the 209 genes upregulated in ure2Δ C. glabrata, 19 are NCR targets, but 5 genes listed by Godard et al. (2007) as NCR targets are downregulated in C. glabrata (File S1).
Overall, the roles of Ure2p in the three yeasts seem to be quite similar, but must differ in some details. All three organisms respond to deletion of URE2 by changing uptake and metabolism pathways of amino acids. They also enhance uptake and metabolism of nucleotides. In addition, all three organisms promote the uptake and utilization of urea as a nitrogen source. These shared responses are highlighted by the eight genes whose expression we find changes in the same direction in S. cerevisiae, C. albicans, and C. glabrata: CAR1 (arginase), DIP5 (dicarboxylic amino acid permease), DUR1 (urea amidolyase), GAP1 (general amino acid permease), GLN1 (glutamine synthase), UGA1 (γ-aminobutyrate transaminase), FCY2 (purine-cytosine permease), and YPS1 (aspartyl protease) (File S1, Figure S2).
C. albicans Ure2p can form a prion in S. cerevisiae:
Overproduction of a prion protein increases the frequency of de novo formation of the prion form (Wickner 1994), thus providing both a means of obtaining cells with the prion and evidence that it is indeed a prion. Neither C. albicans nor C. glabrata Ure2p overproduction induces the appearance of S. cerevisiae [URE3] (Edskes and Wickner 2002). Further, when C. albicans or C. glabrata Ure2p expression was directed from a plasmid in a S. cerevisiae strain carrying a chromosomal ure2 deletion neither protein could propagate [URE3] derived from S. cerervisiae Ure2p introduced by cytoduction. These results indicate that there is at least a substantial species barrier between S. cerevisiae Ure2p and the two Candida Ure2ps.
Using pH563 (pPGAL1URE2albicans) in BY302 (URE2albicans), we transiently overproduced Ure2palbicans by growing cells in galactose-containing medium. [URE3]-containing cells were detected using a PDAL5:ADE2 fusion. DAL5, encoding allantoate permease, is repressed by Ure2p when a rich nitrogen source, such as ammonia, is present. Dal5p can also transport USA, the product of Ura2p (aspartate transcarbamylase). Thus, deficiency of Ure2p (as when [URE3] is present) derepresses DAL5 transcription, allowing a ura2 mutant to grow on USA in place of uracil. Alternatively, placing the ADE2 gene under the control of the DAL5 promoter results in normal cells being Ade− and [URE3] cells being Ade+. After growth on galactose, cells were plated on −Ade plates with glucose as the carbon source to shut off the overproduction of Ure2p. The frequency of Ade+ cells was increased >100-fold by overproduction of Ure2palbicans, indicating that [URE3alb] had formed (Table 3).
TABLE 3.
Prion induction by transient overexpression of full-length Ure2palbicans or Ure2pglabrata
| Host | Plasmid | ADE+ per 106 cells plated |
| BY302 albicans URE2 | pH317 (vector) | 11 ± 8 |
| pH563 (albicans) | 1680 ± 490 | |
| BY304 glabrata URE2 | pH317 (vector) | 20 ± 15 |
| pH659 (glabrata) | 16 ± 16 |
BY302 (or BY304) with a chromosomal copy of URE2albicans (or URE2glabrata) with the S. cerevisiae URE2 promoter was transformed with a copy of the same URE2 controlled by the GAL1 promoter or the vector control. Transformants were grown in leucine dropout media containing 2% galactose and 1% rafinose as carbon source. Cells were washed, diluted, and plated onto adenine dropout plates containing glucose as the carbon source. Colonies were counted after 5 days at 30°. pH317 2μ, LEU2 PGAL1 (Edskes and Wickner 2002); pH563 pH317 with URE2albicans (Edskes and Wickner 2002); pH659 pH317 with URE2albicans (Edskes and Wickner 2002).
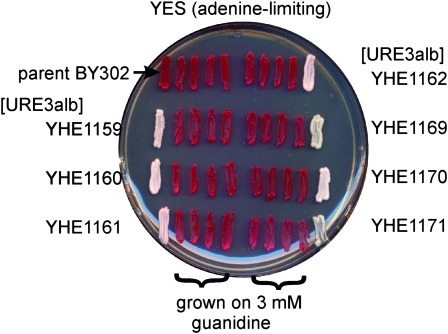
Many of the Ade+ clones induced by Ure2albicans overexpression were stable on growth under nonselective conditions (YPAD), but were then cured by growth on YPAD containing 3 mm guanidine HCl (Figure 1), a known feature of S. cerevisiae [URE3] (Moriyama et al. 2000).
Figure 1.—
Curing of [URE3alb] by growth on 3 mM guanidine. The parent strain BY302 and seven Ade+ derivatives were streaked for single colonies on YPAD with 3 mM guanidine HCl. Four single colonies from each were streaked on adenine-limiting medium (YES). Red clones are Ade−. The white clones on the left and right are the Ade+ derivatives of BY302 not exposed to guanidine.
Prions are, in most cases, readily transmitted by cytoduction. Seven stably Ade+ guanidine-curable putative [URE3alb] clones were used as cytoduction donors to a rhoo [ure-o] recipient. In four cases, all (20) or nearly all (19 or 20) of the cytoductants had become USA+, confirming that these isolates were [URE3alb].
Two variants of [URE3alb]:
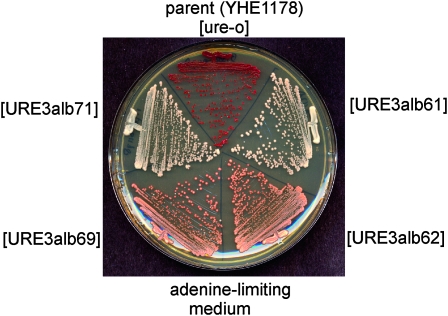
A single protein can adopt different amyloid structures and thus show distinct heritable and infectious biological properties (reviewed by Collinge and Clarke 2007). Cytoductants of [URE3alb] isolates YHE1161 and YHE1171 into the common recipient strain YHE1178 were slow growing and only weakly Ade+ (red on adenine-limiting media), while cytoductants of isolates YHE1162 and YHE1169 into the same recipient were fast-growing and strongly Ade+ (Figure 2). Thus, this prion, as other yeast prions, can display variants.
Figure 2.—
Two [URE3alb] prion variants. [URE3alb] in strains YHE1171, YHE1169, YHE1161, and YHE1162 was cytoduced into YHE1178. Strain YHE1178 and cytoductants from each were streaked on adenine-limiting medium (YES). Two strains may be distinguished by colony color.
C. glabrata Ure2p cannot form a [URE3] prion:
While transient overexpression of Ure2palbicans induced the formation of [URE3alb], the same was not true of overexpression of Ure2pglabrata in cells expressing only Ure2pglabrata (Table 3), although Ure2pglabrata was indeed strongly overexpressed (Figure S2). The frequency of Ade+ clones did not increase, and none of the stably Ade+ clones were cured by growth on guanidine, so they were not prions. Six Ade+ clones were used as cytoduction donors with strain YHE1174 [MATα ura2leu2kar1 URE2glabrata PDAL5:ADE2] as recipient. Not a single case of transfer of the ADE+ phenotype from donor cells to recipient cells was observed (total of 79 cytoductants with 8–17 cytoductants per BY304 ADE+ isolate).
It has been shown that the prion forming domain of S. cerevisiae Ure2p is far more efficient in inducing [URE3] when transiently overexpressed than the full-length protein (Masison and Wickner 1995). We thus transiently overexpressed the N-terminal domains from the S. cerevisiae, C. albicans, and C. glabrata Ure2 proteins and measured their ability to induce [URE3] (Table 4). While the cerevisiae and albicans N-terminal domains' overproduction dramatically increased Ade+ colony formation, only a small increase was seen for glabrata. Guanidine curing experiments confirmed that all of the albicans Ade+ colonies contained prions (12 of 12 tested cured by guanidine). Tests of 40 of the glabrata Ade+ clones showed that none were guanidine curable, although some were completely converted to Ade− on growth in rich medium.
TABLE 4.
[URE3] induction assays using N-terminal domains
| Host | Plasmid | ADE+/per million cells plated ± SD |
| BY302 albicans URE2 | Vector (pH317) | 3 ± 3 |
| cerevisiae 1–65 (pH382) | 3 ± 3 | |
| cerevisiae 1–99 (pH565) | 1 ± 1 | |
| albicans1–89 (pH564) | 224,000 ± 39,000 | |
| glabrata1–100 (pH1026) | 2 ± 4 | |
| BY304 glabrata URE2 | Vector | 13 ± 5 |
| cerevisiae 1–65 | 18 ± 4 | |
| cerevisiae 1–99 | 11 ± 3 | |
| albicans1–89 | 15 ± 14 | |
| glabrata1–100 | 85 ± 25 | |
| YHE1265 cerevisiae URE2 | Vector | 36 ± 15 |
| cerevisiae 1–65 | 29,900 ± 6000 | |
| cerevisiae 1–99 | 23,700 ± 14,000 | |
| albicans1–89 | 1070 ± 500 | |
| glabrata1–100 | 32 ± 9 |
Cells were grown to saturation in leucine dropout medium containing 2% galactose and 1% rafinose as carbon sources. Tenfold dilutions were plated onto dextrose medium lacking adenine and incubated for 5 days at 30°. Viable cells were measured by plating of dilutions on YPAD. YHE1265 was derived from BY304 by replacing URE2glabrata with URE2cerevisiae.
In a further attempt to induce [URE3gla] formation, we introduced cytoplasm with [URE3cerevisiae] or [URE3alb] by cytoplasmic mixing. Neither prion was successfully transferred to cells expressing Ure2pglabrata, although each was efficiently transmitted to a recipient expressing a Ure2p identical to the donor (Table 5). [URE3alb] was not transmitted to cells expressing Ure2p from several Saccharomyces species.
TABLE 5.
Species barriers among [URE3] prions
| Cytoductants |
|||
| Donor [URE3]/URE2 | Recipient/URE2 | Total | USA+ |
| LM69 S. cerevisiae | YHE1202 C. glabrata | 26 | 0 |
| LM69 S. cerevisiae | YHE1177 C. albicans | 26 | 0 |
| LM69 S. cerevisiae | LM60 S. cerevisiae | 19 | 19 |
| YHE1211 C. albicans | LM11 S. bayanus | 8 | 0 |
| YHE1211 C. albicans | LM14 S. kudriavzevii | 10 | 0 |
| YHE1211 C. albicans | LM16 S. mikatae | 7 | 0 |
| YHE1211 C. albicans | LM18 S. paradoxus | 8 | 0 |
| YHE1211 C. albicans | LM27 S. castellii | 7 | 0 |
| YHE1211 C. albicans | LM41 S. cariocanus | 13 | 0 |
| YHE1211 C. albicans | LM45 S. cerevisiae | 16 | 0 |
| YHE1211 C. albicans | YHE1203 S. glabrata | 45 | 0 |
| YHE1211 C. albicans | YHE1181 C. albicans | 85 | 85 |
All strains are S. cerevisiae with URE2 from the indicated species. Donor strains have the corresponding [URE3cer] or [URE3alb].
We considered the possibility that in the process of constructing BY304 some other change had been inadvertently introduced that made the cells unable to propagate a prion. We therefore, replaced URE2glabrata with URE2cerevisiae forming YHE1265. Overexpression of Ure2cerevisiae1-65 or 1-99 each stimulated prion formation (Table 4), indicating that no such mutation was present.
Aggregate formation in vivo:
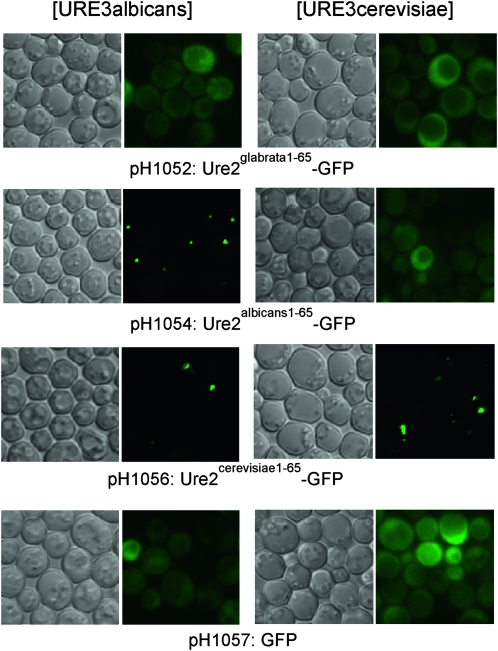
Ure2-GFP fusion proteins that include the prion domain can be incorporated into the prion filaments in vivo and allow visualization of the prion aggregates (Edskes et al. 1999a) (Figure 3). The fusion protein expression also may cure the prion (Edskes et al. 1999a). We found that when Ure2albicans1-65-GFP (Figure 3) was expressed from the S. cerevisiae URE2 promoter in a [URE3alb] cell, and cells were grown selecting for the prion, dot-like aggregates were observed in a large fraction of cells. Unexpectedly, Ure2cerevisiae1-65-GFP or Ure2cerevisiae1-99-GFP (also expressed from the S. cerevisiae URE2 promoter) also showed the dot appearance in [URE3alb] strains (Figure 3). No foci were observed in isogenic [ure-o] cells.
Figure 3.—
Ure2palbicans-GFP fusion protein shows aggregation in [URE3alb] strains. YHE1207 (URE2albicans [URE3alb]) or YHE1272 (URE2cerevisiae [URE3cer]) were transformed with the indicated plasmids expressing Ure2-GFP fusion proteins and observed after for 3 days on medium without adenine (selective for retention of the [URE3alb] prion).
Overexpression of many proteins, including prion proteins, often produce an aggregated appearance of protein-GFP fusions even without prion formation. Overexpressing Ure2cerevisiae1-99 from a GAL1 promoter produces aggregates that are revealed by Ure2cerevisiae1-65-GFP expressed at a low level from the URE2 promoter. However, Ure2glabrata1-100-GFP or Ure2glabrata1-65-GFP did not detect any aggregates, when Ure2glabrata1-100 was overexpressed from a GAL1 promoter, indicating that Ure2glabrata had little tendency to aggregate.
URE2-GFP curing:
We found that expression of URE2albicans1-65 from the URE2 promoter cured [URE3alb] (in strain YHE1207), but not [URE3cer] (in strain YHE1271) (Table S1).
DISCUSSION
S. cerevisiae and S. paradoxus are separated by ∼5–20 million years of evolution (Kellis et al. 2003). We find that the S. paradoxus Ure2p can form a prion in S. paradoxus itself, thus resolving an apparent disparity with results in S. cerevisiae. While it may be reassuring that S. cerevisiae turns out to be a good test bed for prion formation in this case, there are numerous chromosomal genes affecting prion generation and propagation, so that this assumption may not always prove to be valid in the many studies which have used this approach. The Ure2p of S. castellii is unable to form a prion in S. cerevisiae (Edskes et al. 2009), but it has not yet been tested in S. castellii itself. Engineering other organisms to detect prions is a daunting task. We found that the Ure2p of C. albicans and C. glabrata have nitrogen regulation functions, like that of S. cerevisiae. However, when we constructed ura2Δ/ura2Δ ure2Δ/ure2Δ homozygous knockout mutants of C. albicans, we found that they did not utilize ureidosuccinate in place of uridine, so we could not use this approach to test for [URE3] in C. albicans. C. albicans has a DAL5 homolog, but C. glabrata does not, so we did not attempt this approach.
A protein domain that can substitute for the Sup35 prion domain in prion formation may not be able to do so in its native context. Since prion formation by prion domains of Ure2p or Sup35p are hundreds or thousands of times more likely to form prions than the full-length proteins expressed at similar levels (Masison and Wickner 1995; Kochneva-Pervukhova et al. 1998), it is possible that some domains that can be prions in isolation or a foreign environment may be completely stabilized in their native context.
The Ure2p of C. albicans forms a [URE3alb] prion with properties similar to those of the [URE3] of S. cerevisiae. We observed elevation of prion generation frequency by overproducing the full-length Ure2albicans and even greater induction by overexpressing just the prion domain. The prion is cured by guanidine, transmitted by cytoduction, and shows at least two prion variants.
In contrast, the Ure2p of C. glabrata does not detectably form a prion under the conditions tested. Even with overproduction of Ure2glabrata or its prion domain, we could not detect [URE3gla] formation. Attempts to observe aggregated Ure2glabrata using fusions with GFP were likewise unsuccessful. Because of the substantial dependence of prion propagation on chaperones and other cellular components, it remains possible that Ure2glabrata can form a prion in C. glabrata itself.
It has been suggested that the conservation of sequence in the prion domains of Ure2p and Sup35p are indications that the prion must have some function beneficial to the cell that has been selected, thus resulting in the conserved prion domain sequence (Shorter and Lindquist 2005; Harrison et al. 2007). However, we find that the C. glabrata Ure2p, which has the conserved prion domain sequence (residues 10–39 in S. cerevisiae), cannot form a prion. Similarly, the Kluyveromyces lactis Ure2p, which also has the conserved prion domain sequence, cannot form a [URE3] prion as tested in K. lactis itself (Safadi et al. 2011). In contrast, the Ure2p of C. albicans, which lacks the conserved sequence, forms a prion much like that of S. cerevisiae. Thus the sequence conservation is not a sign of conservation of prion-forming ability. In fact, prion-forming ability is not even conserved within Saccharomyces. The Ure2p of S. castellii cannot form [URE3] in S. cerevisiae (Edskes et al. 2009), and one-fourth of S. cerevisiae strains tested in one study had a large deletion in the prion domain of Sup35p, making them unable to propagate [PSI+] (Resende et al. 2003).
Beyond these facts, it should be noted that this argument that prions must be adaptive assumes that the conservation of sequence in the prion domain would be necessary for, or at least favor, prion formation. In fact, the amino acid sequences of the Ure2p or Sup35p prion domains may be randomly shuffled and each of the five shuffled domains of each domain can form prions (Ross et al. 2004, 2005). Thus, the conservation of sequence is not an argument for conservation of prion forming ability.
Why are there conserved sequences in the prion domains? The Sup35p prion domain is important for the normal process of mRNA turnover in yeast and humans (Hoshino et al. 1999; Hosoda et al. 2003; Funakoshi et al. 2007), while the Ure2p prion domain is important for the stability against degradation of the full-length protein (Shewmaker et al. 2007). It is likely that these important functions are the basis for the conservation of sequence of these prion domains.
Does prion function benefit the yeast cells or are prions diseases? Since [PSI+] and [URE3] can be quite stable, if they were beneficial, they would spread rapidly in wild populations and, like mitochondrial DNA, be found in most wild strains. In fact, the prion state of Ure2p or Sup35p is rare in nature, as none of the many wild strains examined carry the [URE3] or [PSI+] prions (Nakayashiki et al. 2005). The [PIN+] prion is found at a frequency comparable to that of the mildly detrimental RNA viruses and 2-μm DNA plasmid (Nakayashiki et al. 2005). Since deletion of RNQ1 has no phenotype (Sondheimer and Lindquist 2000), it is possible that there is not enough of a detriment of carrying [PIN+] to eliminate it from the population.
Another test of whether or not cells are happy to be [PSI+] or [URE3] is the response of their stress reaction system, the heat shock proteins. Transcription of HSP104 and SSA1 are stimulated by a variety of stresses, including heat, high ethanol, high salt, high osmolarity, heavy metals, dessication, or starvation (among other conditions). Masison and coworkers have found that Hsp104 and Ssa1 protein levels are consistently elevated by the presence of the [PSI+] or [URE3] prions (Jung et al. 2000; Schwimmer and Masison 2002), suggesting that these prions are recognized by the cell as stress conditions. The occasional broken limb trait is widely conserved among vertebrates, but one would not argue that the limb structures are conserved for this purpose.
On the basis of the very sparse evidence so far available, [URE3] and [PSI+] prion-forming ability appears to be sporadically distributed among species, as its occurence in the wild is, at most, sporadic. Further work will be necessary to explore the properties of prions in other organisms.
Acknowledgments
We thank Brendan Cormack for C. glabrata strains and plasmids, Alexander D. Johnson for C.albicans strains, Alistair J. P. Brown for the C. albicans cre-loxP plasmid system, and Janyce A. Sugui for help with microscopy. We are grateful to Michel Aigle for marked S. paradoxus strains and to Clete Kurtzman for wild S. paradoxus isolates. This work was supported by the Intramural Program of the National Institute of Diabetes, Digestive, and Kidney Diseases of the National Institutes of Health.
LITERATURE CITED
- Alberti S., Halfmann R., King O., Kapila A., Lindquist S., 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A., Fernandez-Bellot E., Reine F., Coissac E., Cullin C., 2003. Conservation of the prion properties of Ure2p through evolution. Mol. Biol. Cell 14: 3449–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U., Cheng N., Winkler D. C., Chiu T. K., Davies D. R., et al. , 2005. Filaments of the Ure2p prion protein have a cross-beta core structure. J. Struct. Biol. 150: 170–179 [DOI] [PubMed] [Google Scholar]
- Baxa U., Wickner R. B., Steven A. C., Anderson D., Marekov L., et al. , 2007. Characterization of β-sheet structure in Ure2p1–89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry 46: 13149–13162 [DOI] [PubMed] [Google Scholar]
- Bialkova A., Subik J., 2006. Biology of the pathogenic yeast Candida glabrata. Folia Microbiol. 51: 3–20 [DOI] [PubMed] [Google Scholar]
- Brachmann A., Baxa U., Wickner R. B., 2005. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24: 3082–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisse S., Pannier C., Angoulvant A., De Meeus T., Diancourt L., et al. , 2009. Uneven distribution of mating types among genotypes of Candida glabrata isolates from clinical samples. Eukaryot. Cell 8: 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V., Sexton J. A., Johnston M., 2006. A glucose sensor in Candida albicans. Eukaryot. Cell 5: 1726–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. C. C., Oyler N. A., Yau W.-M., Tycko R., 2005. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry 44: 10669–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y. O., Galkin A. P., Lewitin E., Chernova T. A., Newnam G. P., et al. , 2000. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol. Microbiol. 35: 865–876 [DOI] [PubMed] [Google Scholar]
- Collinge J., Clarke A. R., 2007. A general model of prion strains and their pathogenicity. Science 318: 930–936 [DOI] [PubMed] [Google Scholar]
- Conde J., Fink G. R., 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. USA 73: 3651–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to th GATA factors: connecting the dots. FEMS Microbiol. Rev. 26: 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B. P., Falkow S., 1999. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151: 979–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V., Deleu C., Saupe S., Begueret J., 1997. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA 94: 9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox K. H., Pinchak A. B., Cooper T. G., 1999. Genome-wide transcriptional analysis in S. cerevisiae by mini-array membrane hybridization. Yeast 15: 703–713 [DOI] [PubMed] [Google Scholar]
- Dabas N., Morschhauser J., 2007. Control of ammonium permease expression and filamentous growth by the GATA transcription factors GLN3 and GAT1 in Candida albicans. Eukaryot. Cell 6: 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabas N., Morschhauser J., 2008. A transcription factor regulatory cascade controls secreted aspartic protease expression in Candida albicans. Mol. Microbiol. 69: 586–602 [DOI] [PubMed] [Google Scholar]
- Dalstra H. J. P., Swart K., Debets A. J. M., Saupe S. J., Hoekstra R. F., 2003. Sexual transmission of the [Het-s] prion leads to meiotic drive in Podospora anserina. Proc. Natl. Acad. Sci. USA 100: 6616–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison P. M., Ramsdale M., Manson C. L., Brown A. J., 2005. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet. Biol. 42: 737–748 [DOI] [PubMed] [Google Scholar]
- Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W., 2001. Prions affect the appearance of other prions: the story of [PIN]. Cell 106: 171–182 [DOI] [PubMed] [Google Scholar]
- Du Z., Park K.-W., Yu H., Fan Q., Li L., 2008. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 40: 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone S. S., Cox B. S., Tuite M. F., 1999. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 18: 1974–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H. K., Gray V. T., Wickner R. B., 1999a. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. USA 96: 1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H. K., Hanover J. A., Wickner R. B., 1999b. Mks1p is a regulator of nitrogen catabolism upstream of Ure2p in Saccharomyces cerevisiae. Genetics 153: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H. K., Wickner R. B., 2002. Conservation of a portion of the S. cerevisiae Ure2p prion domain that interacts with the full - length protein. Proc. Natl. Acad. Sci. USA 99(Suppl. 4): 16384–16391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H. K., McCann L. M., Hebert A. M., Wickner R. B., 2009. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics 181: 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidel P. L., Cutright J. L., Tait L., Sobel J. D., 1996. A murine model of Candida glabrata vaginitis. J. Infect. Dis. 173: 425–431 [DOI] [PubMed] [Google Scholar]
- Funakoshi Y., Doi Y., Hosoda N., Uchida N., Osawa M., et al. , 2007. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 21: 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitz R. D., Woods R. A., 2002. Transformation of yeast by the Liac/SS carrier DNA/PEG method. Methods Enzymol. 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Godard P., Urrestarazu A., Vissers S., Kontos K., Bontempi G., et al. , 2007. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 3065–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison L. B., Yu Z., Stajich J. E., Dietrich F. S., Harrison P. M., 2007. Evolution of budding yeast prion-determinant sequences across diverse fungi. J. Mol. Biol. 368: 273–282 [DOI] [PubMed] [Google Scholar]
- Hoshino S., Imai M., Kobayashi T., Uchida N., Katada T., 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. J. Biol. Chem. 274: 16677–16680 [DOI] [PubMed] [Google Scholar]
- Hosoda N., Kobayashii T., Uchida N., Funakoshi Y., Kikuchi Y., et al. , 2003. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278: 38287–38291 [DOI] [PubMed] [Google Scholar]
- Jung G., Jones G., Wegrzyn R. D., Masison D. C., 2000. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakeya H., Miyazaki Y., Miyazaki H., Nyswaner K., Grimberg B., et al. , 2000. Genetic analysis of azole resistance in the Darlington strain of Candida albicans. Antimicrob. Agents Chemother. 44: 2985–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R., Domergue R., Zupancic M. L., Cormack B. P., 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 8: 378–384 [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S., 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N. V., Poznyakovski A. I., Smirnov V. N., Ter-Avanesyan M. D., 1998. C-terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+] determinant in Saccharmyces cerevisiae. Curr. Genet. 34: 146–151 [DOI] [PubMed] [Google Scholar]
- Kushnirov V. V., Kochneva-Pervukhova N. V., Cechenova M. B., Frolova N. S., Ter-Avanesyan M. D., 2000. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 19: 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R. E., 2009. Overview of the changing epidemiology of candidemia. Curr. Med. Res. Opin. 25: 1732–1740 [DOI] [PubMed] [Google Scholar]
- Liao W. L., Ramon A. M., Fonzi W. A., 2008. GLN3 encodes a global regulator of nitrogen metabolism and virulence of C. albicans. Fungal Genet. Biol. 45: 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B., Kaiser C. A., 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290: 1–18 [DOI] [PubMed] [Google Scholar]
- Masison D. C., Wickner R. B., 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270: 93–95 [DOI] [PubMed] [Google Scholar]
- Masison D. C., Maddelein M.-L., Wickner R. B., 1997. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc. Natl. Acad. Sci. USA 94: 12503–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki H., Miyazaki Y., Gerber A., Parkinson T., Hitchcock C., et al. , 1998. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PHD1, in Candida glabrata. Antimicrob. Agents Chemother. 42: 1695–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H., Edskes H. K., Wickner R. B., 2000. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20: 8916–8922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T., Ebihara K., Bannai H., Nakamura Y., 2001. Yeast [PSI+] “prions” that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol. Cell 7: 1121–1130 [DOI] [PubMed] [Google Scholar]
- Nakayashiki T., Kurtzman C. P., Edskes H. K., Wickner R. B., 2005. Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. USA 102: 10575–10580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek J., Nakayashiki T., Wickner R. B., 2009. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc. Natl. Acad. Sci. USA 106: 1892–1896 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nielsen K., Heitman J., 2007. Sex and virulence of human pathogenic fungi. Adv. Genet. 57: 143–173 [DOI] [PubMed] [Google Scholar]
- Noble S. M., Johnson A. D., 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4: 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S. M., Johnson A. D., 2007. Genetics of Candida albicans, a diploid human fungal pathogen. Annu. Rev. Genet. 41: 193–211 [DOI] [PubMed] [Google Scholar]
- Patel B. K., Gavin-Smyth J., Liebman S. W., 2009. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 11: 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F., Lea H. Z., Cooper T. G., 1987. Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae. J. Bacteriol. 169: 3521–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon A. M., Fonzi W. A., 2009. Genetic transformation of Candida albicans. Methods Mol. Biol. 499: 169–174 [DOI] [PubMed] [Google Scholar]
- Resende C. G., Outeiro T. F., Sands L., Lindquist S., Tuite M. F., 2003. Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol. Microbiol. 49: 1005–1017 [DOI] [PubMed] [Google Scholar]
- Rogoza T., Goginashvili A., Rodionova S., Ivanov M., Viktorovskaya O., et al. , 2010. Non-mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc. Natl. Acad. Sci. USA 107: 10573–10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E. D., Wickner R. B., 2004. Prions of yeast fail to elicit a transcriptional response. Yeast 21: 963–972 [DOI] [PubMed] [Google Scholar]
- Ross E. D., Baxa U., Wickner R. B., 2004. Scrambled prion domains form prions and amyloid. Mol. Cell. Biol. 24: 7206–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E. D., Edskes H. K., Terry M. J., Wickner R. B., 2005. Primary sequence independence for prion formation. Proc. Natl. Acad. Sci. USA 102: 12825–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safadi R. A., Talarek N., Jacques N., Aigle M., 2011. Yeast prions: Could they be exaptations? The URE2/[URE3] system in Kluyveromyces lactis. FEMS Yeast Res. 11: 151–153 [DOI] [PubMed] [Google Scholar]
- Santoso A., Chien P., Osherovich L. Z., Weissman J. S., 2000. Molecular basis of a yeast prion species barrier. Cell 100: 277–288 [DOI] [PubMed] [Google Scholar]
- Schwimmer C., Masison D. C., 2002. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22: 3590–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamji A. F., Kuruvilla F. G., Schreiber S. L., 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the TOR proteins. Curr. Biol. 10: 1574–1581 [DOI] [PubMed] [Google Scholar]
- Sherman F., 1991. Getting started with yeast, pp. 3–21 in Guide to Yeast Genetics and Molecular Biology, edited by C. Guthrie and G. R. Fink Academic Press, San Diego; [DOI] [PubMed] [Google Scholar]
- Shewmaker F., Mull L., Nakayashiki T., Masison D. C., Wickner R. B., 2007. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics 176: 1557–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Lindquist S., 2005. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6: 435–450 [DOI] [PubMed] [Google Scholar]
- Sondheimer N., Lindquist S., 2000. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5: 163–172 [DOI] [PubMed] [Google Scholar]
- Talarek N., Maillet L., Cullin C., Aigle M., 2005. The [URE3] prion is not conserved among Saccharomyces species. Genetics 171: 23–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- True H. L., Lindquist S. L., 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483 [DOI] [PubMed] [Google Scholar]
- Tsai H.-F., Sammons L. R., Zhang X., Suffis S. D., Su Q., et al. , 2010. Microarray and molecular analyses of the azole resistance mechanism in Candida glabrata oropharyngeal isolates. Antimicrob. Agents Chemother. 54: 3308–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science 264: 566–569 [DOI] [PubMed] [Google Scholar]
- Wickner R. B., 1997. A new prion controls fungal cell fusion incompatibility. Proc. Natl. Acad. Sci. USA 94: 10012–10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Shewmaker F., Kryndushkin D., Edskes H. K., 2008. Protein inheritance (prions) based on parallel in-register β-sheet amyloid structures. Bioessays 30: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]