Abstract
In maize, mutations in the pr1 locus lead to the accumulation of pelargonidin (red) rather than cyanidin (purple) pigments in aleurone cells where the anthocyanin biosynthetic pathway is active. We characterized pr1 mutation and isolated a putative F3′H encoding gene (Zmf3′h1) and showed by segregation analysis that the red kernel phenotype is linked to this gene. Genetic mapping using SNP markers confirms its position on chromosome 5L. Furthermore, genetic complementation experiments using a CaMV 35S::ZmF3′H1 promoter–gene construct established that the encoded protein product was sufficient to perform a 3′-hydroxylation reaction. The Zmf3′h1-specific transcripts were detected in floral and vegetative tissues of Pr1 plants and were absent in pr1. Four pr1 alleles were characterized: two carry a 24 TA dinucleotide repeat insertion in the 5′-upstream promoter region, a third has a 17-bp deletion near the TATA box, and a fourth contains a Ds insertion in exon1. Genetic and transcription assays demonstrated that the pr1 gene is under the regulatory control of anthocyanin transcription factors red1 and colorless1. The cloning and characterization of pr1 completes the molecular identification of all genes encoding structural enzymes of the anthocyanin pathway of maize.
THE flavonoid biosynthetic pathway is ubiquitous in higher plants and leads to the synthesis of a variety of pigmented and nonpigmented compounds. The pigmented flavonoid metabolites have been used as phenotypic markers in many model plant species (Holton and Cornish 1995; Chopra et al. 2006) and have proven to be an excellent tool to study the genetic, molecular, and biochemical processes underlying the regulation of tissue-specific gene expression patterns (Koes et al. 2005). Flavonoids have important biological functions during the growth and development of a plant (Stafford 1990; Shirley 1996; Dixon and Steele 1999; Taylor and Grotewold 2005) and have many pharmacological and dietary benefits for humans and animals (Miyagi et al. 2000). Flavonoids are produced through the phenylpropanoid pathway (Figure 1A), and depending upon the genetic constitution of the plant system, naringenin has several different fates including formation of anthocyanins, flavans, flavones, condensed tannins, and phlobaphenes (Winkel-Shirley 2001). In maize, purple and red anthocyanins are derived from 3-hydroxyflavonoids (Styles and Ceska 1989) and their tissue-specific accumulation is regulated by pairs of duplicated transcription factors red1/booster1 (r1/b1) and colorless1/purple plant1 (c1/pl1). The R1 and B1 genes encode bHLH transcription factors (Ludwig et al. 1990; Goff et al. 1992) and C1 and PL1 encode MYB-homologous DNA binding domain proteins (Cone et al. 1993). Anthocyanin accumulation in aleurone requires the joint action of R1 and C1, while B1 and PL1 together are needed for anthocyanin biosynthesis in vegetative plant parts (Chandler et al. 1989).
Figure 1.—
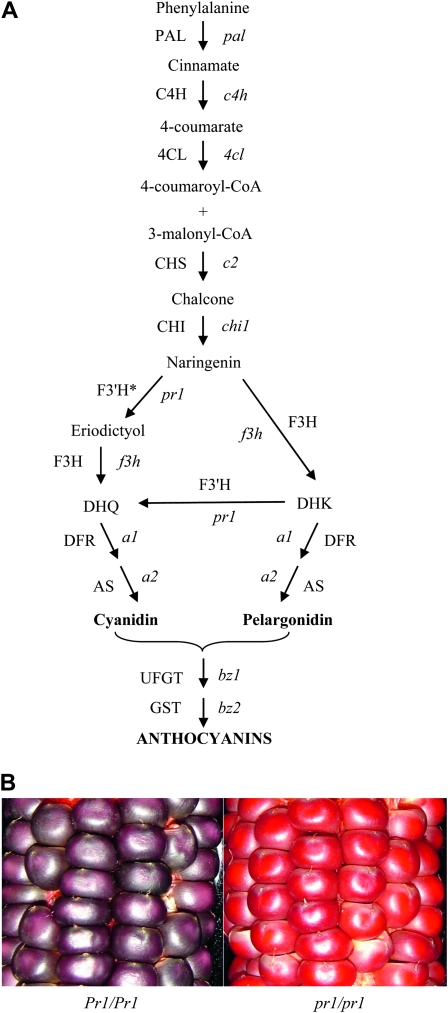
Biosynthesis and accumulation of anthocyanins in maize. (A) Phenylpropanoid biosynthetic pathway leading to the production of anthocyanins. Genes (enzymes) in the pathway are: pal (PAL), phenylalanine-ammonia lyase; c4h (C4H), cinnamic acid hydroxylase; 4cl (4CL), 4-coumaryl:CoA ligase; c2 (CHS), chalcone synthase; chi1 (CHI), chalcone isomerase; f3h (F3H), flavanone 3-hydroxylase; pr1 (F3′H), flavonoid 3′-hydroxylase (* = based on enzyme activity assay); a1 (DFR), dihydroflavanone reductase; a2 (AS), anthocyanidin synthase; bz1 (UFGT), UDP-glucose flavonoid 3-O-glucosyltransferase; and bz2 (GST), glutathione S-transferase. (B) Close-up photos show red and purple aleurones in kernels of pr1 and Pr1 ears, respectively.
We are interested in sorghum and maize 3-deoxyanthocyanidins, 3-hydroxyanthocyanidins, and C-glycosyl flavones that have a role in tolerance to fungal pathogens (Snyder and Nicholson 1990) and insect pests (Snook et al. 1994). It was shown that several flavonoid branches leading to these compounds require the activity of a flavonoid 3′-hydroxylase in sorghum and maize (Cortes-Cruz et al. 2003; Boddu et al. 2004). Similarly, formation of anthocyanidins in the kernel aleurone requires the activity of a flavonoid 3′-hydroxylase (F3′H), and this activity has been attributed to the functional red aleurone1 locus, also known as purple aleurone1 and designated as pr1 in maize (Larson et al. 1986). Since the early 20th century, the pr1 locus has been used as a marker in maize genetics for the identification, characterization, and mapping of several loci. During the mid 20th century, genetic and biochemical studies established that the red and purple aleurone color difference is due to the presence (Pr1) or absence (pr1) of a hydroxyl group in the pigment and that a single gene is responsible for this phenotype (McClary 1942; Zarudnaya 1950; Coe 1955). Maize F3′H is a NADPH-dependent cytochrome P450 enzyme, which can act on a wide range of substrates: the flavonols, flavones, and flavanones, all of which are intermediates in the flavonoid biosynthetic pathway (Larson and Bussard 1986). Although the availability of mutants at different catalytic steps has led to the isolation of most of the structural genes required for maize anthocyanin biosynthesis (Dooner et al. 1991), the sequence of the pr1 gene has remained elusive (Cone 2007). The pr1 mutants produce a red pigment, pelargonidin, due to the failure to hydroxylate the B ring of dihydrokaempferol (DHK) (Forkmann 1991) to yield the purple cyanidin pigment produced in maize lines carrying a functional Pr1 allele (Figure 1B). Using mutants of pr1, Larson et al. (1986) demonstrated that the pr1 locus may encode or regulate an F3′H-mediated conversion of DHK to dihydroquercitin (DHQ) in vitro. To define the role of the pr1 gene in maize anthocyanin biosynthesis, we isolated and characterized a putative maize f3′h1 (Zmf3′h1) sequence. Our results provide the historic missing link between red and purple anthocyanin accumulation in maize. Analysis of Zmf3′h1 and its transcriptional regulation in distinct tissue types was performed to understand the intermediate steps leading to the synthesis of anthocyanins in maize. Gene expression results and genetic data presented here demonstrate that the pr1 gene is regulated by transcription factors that control the synthesis of anthocyanins in silk, husk, and aleurone tissues.
MATERIALS AND METHODS
Maize genetic stocks:
Seeds of the following maize inbred lines and genetic stocks were kindly provided by the Maize Genetics Cooperation Stock Center (US Department of Agriculture-Agricultural Research Service, University of Illinois, Urbana, IL): W23 (genotype P1-wr c1 r-g), W22 (Pr1 A1 C1 R1), Mp708 (pr1 c1 r1 p1), MGS14273 (pr1 A1 A2 C1 R1), MGS130543 (pr1 A1 A2 C1 R1), MGS14284 (pr1 A1 A2 C1 R1), MGS131036 (c1 R1-g b1 pl1), MGS14633 (c1 R1-r B1 Pl1), MGS167054 (r1-g C1 b1 pl1), and MGS14638 (r1-r C1 B1 Pl1). To develop F2 populations, plants of pr1-MGS14273, pr1-MGS130543, and pr1-MGS14284 alleles were crossed by W22 and progenies were grown from the selfed F1 plants. F2 populations segregated 3:1 for purple to red aleurone, indicating the recessive nature of all pr1 alleles studied. Segregating plants were used for RNA expression and cosegregation analysis using PCR-based polymorphism. To study the regulation of pr1 by anthocyanin regulatory genes, crosses were made between pr1-MGS14273 and c1-MGS131036, c1-MGS14633, r1-MGS167054, and r1-MGS14638 alleles. The F2 progenies were grown from the selfed F1 plants. The mutant stocks of the regulatory genes c1 and r1 carried functional alleles of the biosynthetic genes required for anthocyanin synthesis.
To create the Ds insertion allele described in this study, test-cross progeny were generated by crossing females carrying a recessive pr1 allele (pr1 A1 C1 R1 y1; kindly provided by Dr. Erik Vollbrecht) by closely linked hemizygous Ds pollen donors (Ds/+). The inbred W22 Ds donor was estimated to be ∼3.8 Mb, or 1.8 cM distal to pr1, and carried a single copy of Ac-im (Conrad and Brutnell 2005) (Pr1 Ds/+; A1; C1; r1; Y1; Ac-im/+). Putative pr1 loss-of-function alleles were identified as kernels with exceptional red aleurone among the darkly purple-pigmented aleurones of sibling progeny. One insertion allele was confirmed and named as pr1-Ds1:W22.
Genomic, cDNA cloning, and sequence characterization:
During the onset of this project a full-length maize f3′h sequence was not available in the GenBank. Putative f3′h sequences (http://www.ncbi.nlm.nih.gov) were used to design PCR primers to isolate the full-length Zmf3′h1 sequence. Oligonucleotides ZF3F2 and ZF3R2 (see supporting information, Table S2 for primer sequences) were based on the alignment of the maize partial EST sequence (accession no. BG873885) with sorghum f3′h sequence (Boddu et al. 2004) and used to PCR amplify a 387-bp fragment (F387). Fragment FR is a partial genomic sequence from the 5′ half of the gene, which was PCR amplified using forward primer OSF1 (designed from the rice f3′h gene) and reverse primer ZMR4. All DNA fragments used as probes were labeled with α-32P-dCTP using the Prime-a-Gene Labeling System (Promega, Madison, WI). A λ-FIX II (Stratagene, La Jolla, CA) library prepared from seedling leaf DNA of maize inbred line W23 was screened to isolate the full-length Zmf3′h1 gene (accession no. HQ699781). The full-length maize f3′h1 cDNA was obtained by performing reverse transcription (RT)–PCR using gene-specific primers SBF12 and SBR22 on total RNA isolated from maize silks of W23. Standard PCR buffer and reaction conditions were followed with the modified annealing temperature of 60° for 2 min, followed by polymerization at 72° for 2 min. All DNA sequencing reactions were performed at the Pennsylvania State University's Nucleic Acid Facility, using a method of dye primer cycle sequencing, and reactions were analyzed on the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Sequence assembly and analysis was performed using tools available from the NCBI at www.ncbi.nih.gov (Altschul et al. 1990). Conserved domain searches were done using the NCBI CD Search tool at http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
Multiple sequence alignment:
Amino acid sequences were aligned using the ClustalX program (version 1.81; multiple alignment parameters: gap opening 10, gap extension 0.20; DNA weight matrix: IUB; Protein weight matrix: Gonnet series) (Thompson et al. 1994). Additional F3′H amino acid sequences included: Arabidopsis thaliana (AF271651), Callistephus chinensis (AF313488), Glycine max (AF499731), Matthiola incana (AF313491), Oryza sativa (AC021892), Pelargonium × hortorum (AF315465), Perilla frutescens (AB045593), Petunia hybrida (AF155332), Torenia hybrida (AB057673), and Sorghum bicolor (AY675075).
PCR amplification:
Positions of PCR primers used for the successful identification of polymorphisms in Pr1 and pr1 alleles are shown in Figure 3 (see Table S2 for primer sequences). The standard reaction (50 μl) contained 0.1 μg of genomic DNA, 25 μl of GoTaq green master mix (Promega), and 200 nM each of the two oligonucleotide primers. A typical reaction consisted of 35 cycles of denaturation (1 min, 94°), annealing (1 min, 57°), and extension (2 min 30 sec, 72°) in a thermal cycler (MJ Research). PCR products from both mutant and wild-type pr1 alleles were cloned in pCR2.1-TOPO cloning vector (Invitrogen, Carlsbad, CA) and sequenced. Nucleotide sequences were aligned using ClustalW program (Larkin et al. 2007).
Figure 3.—
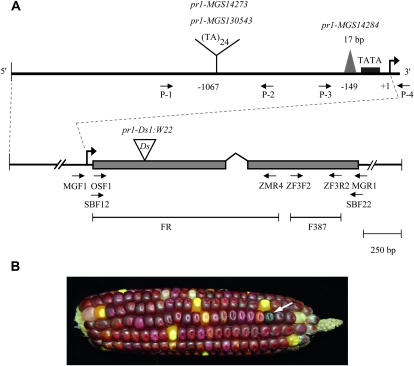
Characterization of Zmf3′h1, isolation of pr1 insertion, and deletion alleles. (A) A segment of λ-clone shows Zmf3′h1 and its 5′- and 3′-flanking sequences. FR and F387 are the probes used to screen the genomic library. Shaded boxes represent two exons that are joined by a bent line, which corresponds to the single intron of the Zmf3′h1 gene. Enlarged 5′-flanking region shows the location of 24 dinucleotide repeats and a deletion near the TATA box in mutant pr1 alleles. Positions of TA repeats and a 17-bp deletion are marked with respect to transcription start site (shown as bent arrow and marked +1). The position of the Ds insertion in exon 1 is shown and allele's name is indicated. Small arrows below the map illustrate the orientation and position of the PCR primers. (B) Self-pollinated ear of a plant with the genotype pr1::Ds/pr1-ref. Note the purple revertant kernel (arrow), indicating a potential excision event of Ds, which would result in the restoration of Pr1 gene expression.
Mapping of Zmf3′h1 gene:
A single nucleotide polymorphism (SNP)-based assay was developed to map the Zmf3′h1 sequence. The following primers, 5′-AGGTGGACGGGTT CCGCATC-3′ and 5′-GTATGCCTCCTCCATGTCTAGC-3′ were used to amplify and sequence a segment of the Zmf3′h1 gene from DNA of the inbred lines Tx501, NC7A, and Mp708. Examination of the sequence alignments revealed a C-to-G polymorphism in Tx501 relative to NC7A and Mp708. The primer 5′-GATGAGCTCGAAGTCGCT-3′ was designed to interrogate the polymorphic site by primer extension. SNP markers were genotyped with the SNaPshot system (Applied Biosystems, Foster City, CA) and assayed on an ABI 3700 as previously described (Vroh Bi et al. 2006). The genotypes for SNP were derived from 346 (Tx501 × NC7A) F2 individuals and 246 (Tx501 × Mp708) F2 individuals obtained from previously characterized quantitative trait locus mapping populations (Cortes-Cruz et al. 2003). The linkage maps were constructed with Mapmaker/Exp, version 3.0 (Whitehead Institute, Cambridge MA).
RNA gel blot analysis:
For RNA isolation, silk and husk tissues were collected at the silk emergence. To isolate total RNA, tissues were ground in liquid nitrogen and then extracted using Tri-Reagent (Molecular Research Center). The RNA was separated on denaturing gels and RNA gel blot hybridizations were performed following standard methods described previously (Boddu et al. 2006).
RT–PCR expression assay:
Kernel aleurones were peeled from endosperm tissues at 24–28 days after pollination (DAP) from pr1 segregating plants, c1 and r1 mutants, and from wild-type W22 inbred line. RNA was isolated from three biological replicates of each line and three samples were analyzed from each replicate. To isolate total RNA, tissues were ground in liquid nitrogen and then extracted using Tri-Reagent (Molecular Research Center, Cincinnati, OH). First strand cDNA was synthesized using SuperScript II cDNA synthesis system (Invitrogen) and oligo-dT primer using conditions as described by the manufacturer. One microgram of total RNA from aleurones was used for each reverse transcription reaction. First strand cDNA was diluted to a final volume of 100 μl with sterile ddH2O. Five microliters of first strand cDNA was PCR amplified using gene-specific primers. Plasmid DNA that contained the complete Zmf3′h1 genomic sequence was used as an internal control. This amplified a PCR product of 996 bp vs. the observed 771-bp product amplified from Zmf3′h1 cDNA. Primers used for detection of anthocyanin gene transcripts are shown in Table S1 (Piazza et al. 2002). PCR reactions were performed in a total volume of 25 μl with GoTaq green master mix (Promega) using the manufacturer's instructions. Conditions were as follows: an initial denaturation of 94° for 4 min, denaturation 94° for 1 min, annealing 55°–60° for 1 min, and extension 72° for 1 min 30 sec, repeated for 30 cycles, and a final extension step of 72° for 10 min. PCR products were analyzed on 1.0% agarose gel.
Extraction, TLC, and HPLC analysis of flavonoid compounds:
Thin layer chromatography (TLC) was carried out using silica 60 plates (Sigma Aldrich, St. Louis, MO) and a mobile solvent containing ethyl acetate:formic acid:acetic acid:water (100:11:11:27, v/v) (Holton 1995; Dong et al. 2001). Approximately 2 g of chopped aleurones and endosperms were allowed to soak in 1 ml of methanol with 1% HCl for 48 hr before transferring the extracts to a fresh tube. A total of 30 μl extract was spotted per lane. For high performance liquid chromatography (HPLC) analysis of dihydroflavonols, maize silk samples (300 mg) taken at the time of silk emergence were imbibed in 1-ml mixture of methanol:HCl (99:1,v/v) and compounds were allowed to leach for 48 hr at 4°. The anthocyanin pigments were extracted by grinding 300 mg of aleurone tissue in 1 ml of 80% methanol. Extracts were acid hydrolyzed with an equal volume of 4 N HCl to prepare aglycones (Burbulis et al. 1996; Nyman and Kumpulainen 2001). The extracts were then filtered through 0.45 μm Acrodisc LC 13-mm syringe filters (Gelman Laboratory, Ann Arbor, MI). Reverse phase HPLC analysis was performed on a Shimadzu high-performance liquid chromatograph (Shimadzu, Columbia, MD) using an Ascentis C18 column (25 cm × 4.6 mm, 5 μm; Supelco, Bellefonte, PA). Pigment separation was performed at 35° by gradient elution using 0.2% formic acid (solution A) and 100% methanol (solution B) at a flow rate of 1 ml/min. The injection volume was 50 μl and spectral measurements were taken over a wavelength range of 230–550 nm, which is known to detect flavonoid compounds (Grotewold et al. 1998). Flavonoid standards of cyanidin and pelargonidin were purified from maize husk and pelargonium flowers, respectively.
Arabidopsis seed stocks, growth conditions, and stable transformation:
For genetic complementation experiments, seeds of Landsberg erecta and transparent testa7 (tt7) mutants were obtained from the Arabidopsis Biological Resource Center at Ohio State University, Columbus, OH. The mutant tt7 used in this study was in a Landsberg erecta background. Seeds were surface sterilized with 10% bleach and germinated in pots filled with steam-sterilized Metro Mix 300. Soil was misted with water after seeding, then covered with plastic wrap, and kept in a cold room at 4° for 3–5 days to break seed dormancy and to promote uniform germination. Plastic wrap was removed and pots were transferred to growth chamber and maintained at 22° and 70% relative humidity (RH). Plants were kept under short day conditions (10 hr light and 14 hr dark) for the first 3 weeks and then transferred to a greenhouse under a long day cycle (16 hr light and 8 hr dark) to induce flowering. An overnight-grown Agrobacterium tumefaciens strain GV3101 carrying a CaMV 35S::ZmF3′H1 construct cloned into a pSR3000 vector was harvested by centrifugation and suspended in 200-ml solution containing 5% sucrose and 0.002% Silwet l-77 (Lehle Seeds, Round Rock, TX). Plants were transformed using the floral dip method (Clough and Bent 1998), in which each plant inflorescence was immersed for at least 10 sec. Transformants were screened on Murashige and Skoog (MS) medium (Murashige and Skoog 1962) that contained 50 μg/ml kanamycin and 100 μg/ml ampicillin. Green seedlings were transferred to soil mix and grown in a growth chamber at 22° with a 10 hr light and 14 hr dark cycle.
Spectrophotometric analysis:
Seeds of Landsberg erecta, tt7 and T2 seeds from two independent transformation events were germinated on minimal medium containing 3% sucrose and 0.5% (w/v) agar. These were kept at 25° in continuous light; seedlings were collected after 10 days, and pigments were extracted as described in Dong et al. (2001). Briefly, seedlings were homogenized in 1.5 ml of 1% (v/v) HCl in methanol, then 1 ml of double-distilled water was added, and chlorophyll was separated from the extracts with chloroform. Pigment analysis was done on UV spectrophotometer, UV-mini 1240 (Shimadzu) and measurements were taken over wavelength range of 400–600 nm. HPLC analysis of extracts was performed with the same column and solvent program used for aleurone anthocyanin analysis.
Dissection of pr1 promoter:
The analysis of the pr1 promoter for the location and distribution of cis-regulatory sequence elements was performed using the Plant Care database (http://bioinformatics.psb.ugent.be/webtools/plantcare.html/) and MetInspector (Genomatrix, Munich, Germany) (Quandt et al. 1995). The cis-binding sites were identified on the basis of their similarity with sites present in promoters of other anthocyanin genes as well as similarity with MYB and MYC protein binding sequences (Sainz et al. 1997).
RESULTS
Isolation of a putative flavonoid 3′-hydroxylase from maize:
To identify the locus encoding flavonoid 3′-hydroxylase from maize, λ-phage libraries were screened with probes designed from putative partial Zmf3′h1 sequences from the maize genome (see materials and methods). The isolated Zmf3′h1 sequence corresponds to GRMZM2G025832, an annotated gene in B73 RefGen_v2 (chromosome 5: 180038179–180040249, www.maizegdb.org). Comparison between Zmf3′h1 [W23] and GRMZM2G025832 shows 92% identity for gene sequence and 95% similarity for peptide sequence.
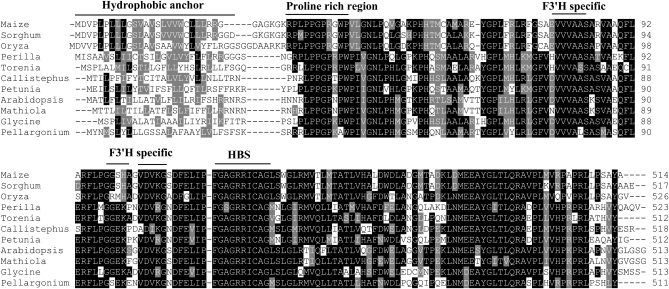
Sequence alignments with other plant F3′H's showed that the isolated sequence encodes a predicted F3′H peptide of 517 amino acids (Figure 2). Overall, the maize F3′H shares ∼55% amino acid identity with dicot F3′H's and high sequence similarity with monocot F3′H's: 78% with rice, 90% with sugarcane, 65% with barley, and 91% with sorghum. The phylogenetic tree (data not shown) shows that maize F3′H resides in the same clade as other monocots such as rice and the closely related species sorghum and is clearly divergent from the dicot clade that includes Arabidopsis and Matthiola. Conserved cytochrome P450-dependent monoxygenase domains, hallmark of F3′H's, were observed in the putative maize F3′H sequence. These domains include a heme-binding site (HBS), oxygen-binding site (OBS), and the characteristic hydrophobic membrane anchor present at the amino terminus. A dicot F3′H-specific motif (GGEK) with unknown function had previously been reported (Brugliera et al. 1999; Toda et al. 2002), and this sequence was found to be modified to GGSH in maize and sorghum F3′H (Boddu et al. 2004).
Figure 2.—
Multiple sequence alignments of deduced amino acid sequences of F3′H from maize and other plant species using the ClustalX program. Lightly shaded areas are F3′H-specific sequences and darkly shaded regions are conserved domains found in the CYP450 proteins. Other regions shown are: hydrophobic anchor, proline-rich region, and heme-binding site (HBS).
Characterization of the lesion in pr1 mutants:
To characterize the lesion present in three different mutant alleles of pr1 (pr1-MGS14273, pr1-MGS130543, and pr1-MGS14284), we used a PCR-based approach. Using Pr1 gene-specific primers, we detected polymorphisms between wild-type and mutant pr1 alleles (Figure 3A). Sequencing results indicated the presence of a 24 TA dinucleotide repeat insertion in the upstream promoter region of pr1-MGS14273 and pr1-MGS130543, and a 17-bp deletion near the TATA box of pr1-MGS14284. Inbred MP708, which carries a recessive allele of pr1, also showed the presence of 24 TA dinucleotide repeats. PCR screening of the genic region did not show any sequence polymorphisms between Pr1 and pr1 alleles. This suggests that the mutant pr1 alleles may be defective due to these insertion and deletion events in the 5′ region. To determine whether the isolated Zmf3′h1 sequence is linked to the pr1 locus, we developed a population segregating for Pr1 and pr1 by crossing pr1 plants with W22 inbred. A total of 40 segregating plants from each population were characterized for PCR-based polymorphisms. A 1:2:1 ratio for Pr1/Pr1:Pr1/pr1:pr1/pr1 showed a molecular cosegregation of the polymorphisms defined for each of the tested alleles (pr1-MGS14273, pr1-MGS130543, and pr1-MGS14284) with the pr1 mutant phenotype.
To unambiguously confirm the identity of the pr1 locus, an independent pr1 allele was characterized, which resulted from a Ds insertional mutagenesis. Following a directed tagging experiment (see materials and methods) five candidate pr1 kernels were identified from 37 test-cross ears (∼7400 kernels). Genomic DNA was extracted from four of these individual seedlings (one did not germinate) and used as templates in a series of PCR assays containing multiple pr1 gene-specific and Ds-specific primer pairs (see Table S2 for primer sequences), in a screen similar to that described previously for the Ds tagging of bz2 (Ahern et al. 2009). The primer set MGR1/JSR01 amplified a product that indicated a single Ds insertion at pr1. Complementary PCR products were amplified using the additional primer sets MGR2/JSR01 and MGF1/JSR05. The latter two products, representing the regions flanking either side of the Ds insertion site, were purified and sequenced. Both sequences independently confirmed the insertion of a Ds element in the first exon of the Zmf3′h1 gene and thus confirmed the identity of the pr1 locus. This seedling was transplanted, grown to maturity, and self-pollinated to create an ear segregating for the Ds insertion at pr1 (Figure 3B). The identification of a Ds insertion in the Zmf3′h1 gene that cosegregated with the pr1 mutant phenotype thus confirmed the molecular identity of the pr1 locus. The position of the Ds insertion in the pr1 sequence is shown in Figure 3A.
The Zmf3′h1 gene sequence maps to the pr1 locus on chromosome 5L:
A genetic linkage map was constructed for chromosome 5 containing an Zmf3′h1 SNP based on the scoring of 346 and 246 individuals for the (Tx501 × NC7A) F2 and (Tx501 × Mp708) F2 populations, respectively (Figure S1). The position of the Zmf3′h1 SNP corresponds to the genetic RFLP-placed position of the pr1 locus (MaizeGDB, http://www.maizegdb.org) and interestingly, it was also detected as a major QTL for the synthesis of apimaysin vs. maysin in these same populations (Cortes-Cruz et al. 2003). Therefore, these genetic mapping results conclusively show that the Zmf3′h1 gene sequence isolated in this study corresponds to the pr1 locus on chromosome 5L.
The Zmf3′h1 transcripts are absent in tissues accumulating red anthocyanins:
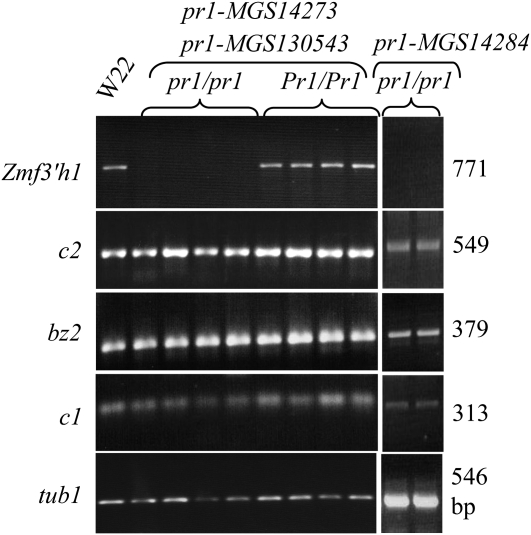
RT–PCR and RNA gel blot analysis of steady state transcription of Zmf3′h1 were performed for kernel aleurone, young maize silk, and husk tissues (Figure 4, Figure S2) collected from plants segregating for Pr1 or pr1 alleles. Pr1 plants produced ears with all purple kernels and accumulated Zmf3′h1 transcripts. Zmf3′h1-specific transcripts could not be detected in RNA from homozygous pr1 plants that produced ears with red kernels. RT–PCR using gene-specific primers for other flavonoid pathway genes as well as for c1 and r1, the anthocyanin regulatory genes, showed steady state transcripts of these genes in both Pr1 and pr1 plants. Similarly, RNA gel blots were stripped and rehybridized with probes of other genes required for anthocyanin biosynthesis. C1, a transcription factor required for the expression of genes encoding structural enzymes of the anthocyanin pathway, as well as the enzyme-encoding genes c2 and bz2, were expressed in both functional and mutated pr1 plants. In summary, these results show that Zmf3′h1-specific transcripts were absent in mutant pr1 plants while expression of other genes of the pathway were detected.
Figure 4.—
Mutant pr1 alleles do not accumulate Zmf3′h1 transcripts in aleurone tissues. Expression of Zmf3′h1, c2, bz2, and c1 in aleurones was detected by RT–PCR from three pr1/pr1 mutant alleles and Pr1/Pr1 wild-type plants. Purple aleurones from W22 were used as positive control and α-tubulin1 was used as a loading control.
Zmf3′h1 is required for the formation of cyanidin:
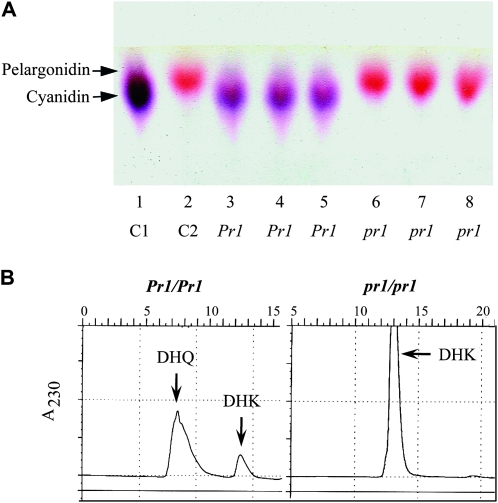
Methanolic extracts of pr1 and Pr1 endosperms and aleurones were separated using TLC (Figure 5A). pr1 aleurones accumulated red pelargonidin glycosides, while Pr1 showed the presence of purple cyanidin glycosides. None to very little cyanidin pigment was detected in the pr1 plants. While the yellow tetrahydroxychalcone can sometimes be detected in the extracts, numerous other compounds in the pathway are colorless and were not detected in our TLCs (Holton and Cornish 1995; Brugliera et al. 1999). HPLC analysis of silk methanolic extracts from lines containing the mutant pr1 allele revealed the expected DHK peak at 12 min, while lines containing Pr1 showed a peak at 8 min, the peak expected for DHQ as measured at 230 nm (Figure 5B). HPLC analysis of acid hydrolyzed extracts from the aleurone tissues at 530 nm revealed that the pr1 plants accumulated mostly the pelargonidin (not shown). Pr1 plants accumulated high levels of cyanidin (data not shown). Overall, chromatography results show that Pr1 is required for the formation of DHQ from DHK and subsequently downstream cyanidin synthesis, which causes the purple pigmentation in the aleurone tissue in Pr1 plants. The accumulation of the respective anthocyanidins and dihydroflavonols in the mutant and wild-type plants confirms that the pr1 gene is responsible for the formation of 3′, 4′-hydroxylated flavonoids.
Figure 5.—
pr1 aleurones produce pelargonidin, while Pr1 aleurones produce cyanidin. (A) TLC of extracts from Pr1 or pr1 kernels. Samples shown in each lane are: 1, control C1 for cyanidin; 2, control C2 for pelargonidin; 3–5, Pr1; and 6–8, pr1. (B) HPLC analysis of silk methanolic extracts from sibling plants carrying pr1 or Pr1. Extracts were analyzed at wavelength 230 nm for DHK and DHQ. pr1 shows single peak representing DHK, while Pr1 has two peaks representing both compounds; thus DHK is converted into DHQ through the action of F3′H (Figure 1).
Zmf3′h1 complements Arabidopsis tt7 mutant phenotype:
The A. thaliana mutant tt7 is defective in F3′H activity (Shirley et al. 1995; Peer et al. 2001). Mutant seeds have a yellow coat color when compared to wild-type seeds with a brown seed coat and tt7 seeds fail to accumulate brown tannins in the testa (Koornneef 1990; Shirley et al. 1995). In addition, tt7 seedlings do not produce anthocyanin pigments in the cotyledon or hypocotyl when grown on a nitrogen-deficient minimal medium (Hsieh et al. 1998). The development of anthocyanin pigments in wild-type Arabidopsis seedlings in response to stress has been used to study the anthocyanin biosynthetic pathway (Grotewold et al. 1998). We have used this assay to ascertain the role of isolated Zmf3′h1 in the development of anthocyanins. tt7 plants were transformed with the 35S::ZmF3′H1 transgene (see materials and methods). The T1 seeds were screened on kanamycin-containing medium, and resistant plants were grown to maturity in the growth chamber. Seeds collected from kanamycin-resistant T2 plants showed complete restoration of brown seed coat color. In the seedling assay, T2 seedlings produced purple cotyledons when grown on a minimal medium (Figure 6). Spectrophotometric analysis of the extracts from 10-day-old Landsberg erecta- and Zmf3′h1-complemented tt7 seedlings grown on minimal medium showed the characteristic absorbance of anthocyanidin pigments at a wavelength of 530 nm, while tt7 mutant seedlings did not show this peak (Figure S3). Anthocyanin compounds present in wild-type, mutant, and Zmf3′h1-expressing seedlings were further characterized by HPLC. The peaks for different cyanidin glycosides were detected in both the wild-type and the Zmf3′h1-complemented tt7 mutant and were absent in tt7 mutant seedlings (Figure S3). In summary, these results demonstrate that the putative Zmf3′h1 sequence encodes a functional F3′H enzyme capable of rescuing the tt7 mutant phenotype in Arabidopsis.
Figure 6.—
Complementation of tt7 mutant with Zmf3′h1 resulted in accumulation of pigments in seedling and seed coat. Seedling and seed phenotypes are shown for wild-type Landsberg erecta, tt7 mutant, and 35S::ZmF3′H1-complemented tt7.
Anthocyanin regulatory genes c1 and r1 are required for pr1 expression in kernel aleurone pigmentation:
To genetically place pr1 in the C1/R1 regulatory network, crosses were made between pr1 (pr1/pr1; C1/C1; R1/R1) and c1 (c1/c1; R1/R1) and pr1 and r1 (r1/r1; C1/C1) mutants (see materials and methods). Two independent mutant alleles of each regulatory gene were used (c1-MGS131036, c1-MGS14633, r1-MGS167054, and r1-MGS14638). The F1 ear phenotype indicated that both the c1 and r1 mutant stocks carry a functional Pr1 allele. A representative F2 ear from each cross is shown in (Figure S4). Purple and red kernels were due to Pr1/−; C1/−; R1/− and pr1/pr1; C1/−; R1/−, respectively. Colorless kernels indicated the presence of mutant alleles of c1/c1 or r1/r1. If the regulation of pr1 is downstream of both the c1 and r1 genes, we expect to see kernel phenotypes segregating as nine purple: three red: and four colorless. Chi-square analysis of F2 ear phenotypes showed that purple, red, and colorless kernels segregated in a 9:3:4 ratio (Table 1). To confirm these results, we analyzed the segregation ratio of kernel aleurone phenotype on test-cross ears. A segregation ratio of 1:1 for purple and colorless kernels was observed on progeny ears from F1 plants test-crossed to c1 or r1 mutant plants (see Table 2). Results from these genetic assays showed that the functional c1 and r1 alleles were required for pr1 expression in the formation of purple aleurone pigment.
TABLE 1.
Regulation of pr1 by c1 and r1 during purple anthocyanin synthesis in the kernel aleurone: number of kernels on F2 progeny ears with specific aleurone color
| c1 allele | Purple (9) | Red (3) | Colorless (4) | P-value χ2 | r1 allele | Purple (9) | Red (3) | Colorless (4) | P-value χ2 |
| c1-MGS131036 (c1 R1-g b1 pl1) | 465 | 185 | 216 | 0.13 | r1-MGS167054 (r1-g C1 b1 pl1) | 688 | 265 | 336 | 0.10 |
| c1-MGS14633 | 270 | 86 | 141 | 0.20 | r1-MGS14638 | 614 | 232 | 298 | 0.20 |
χ2 analysis was performed to detect significant deviation (P < 0.01) of observed phenotypic classes from expected.
TABLE 2.
Regulation of pr1 by c1 and r1 during purple anthocyanin synthesis in the kernel aleurone: number of kernels on test-cross ears with specific aleurone color
| c1 allele | Purple (1) | Colorless (1) | P-value χ2 | r1 allele | Purple (1) | Colorless (1) | P-value χ2 |
| c1-MGS131036 (c1 R1-g b1 pl1) | 446 | 475 | 0.34 | r1-MGS167054 (r1-g C1 b1 pl1) | 466 | 447 | 0.53 |
| c1-MGS14633 (c1 R1-r B1 Pl1) | 430 | 450 | 0.50 | r1-MGS14638 (r1-r C1 B1 Pl1) | 290 | 325 | 0.16 |
χ2 analysis was performed to detect significant deviation (P < 0.01) of observed phenotypic classes from expected.
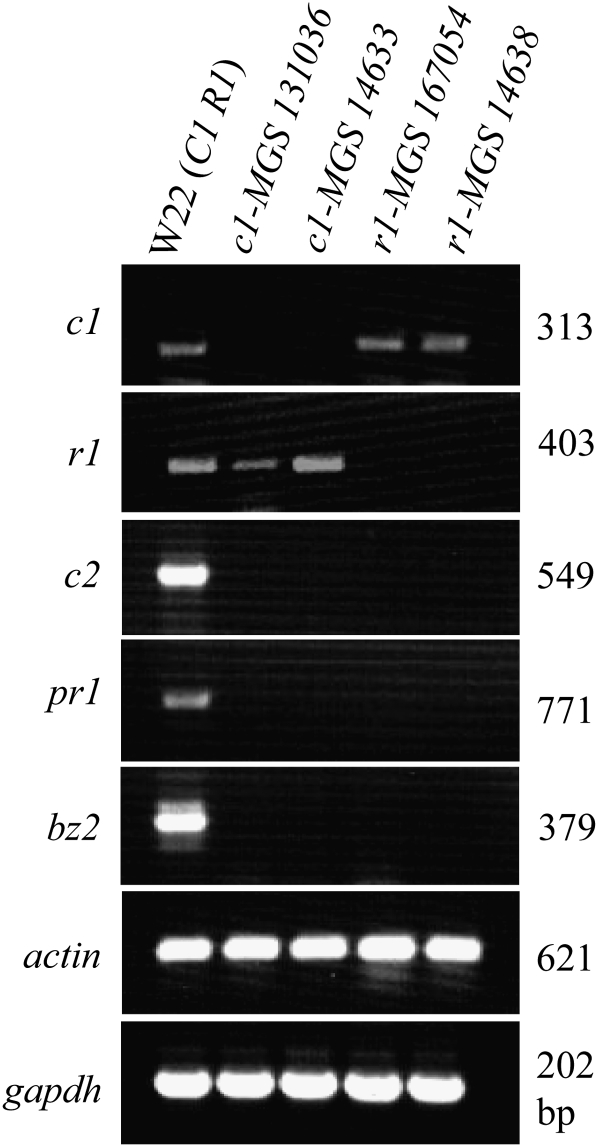
These genetic results were verified at the molecular level by studying the expression of c1, r1, and pr1 genes in mutant as well as wild-type plants. Steady-state transcripts of c1, r1, and pr1 were measured in the aleurones of a wild-type W22 line and two each of c1 and r1 mutant alleles using RT–PCR (Figure 7). W22 kernel aleurones showed steady-state transcripts of all three genes tested, while c1 and r1 mutants did not accumulate c1 and r1 transcripts, respectively. Interestingly, the pr1 gene-specific transcripts could not be detected in colorless kernels of mutant c1 and r1 plants. Similarly, RT–PCR of two other anthocyanin pathway genes known to be regulated by c1 and r1, c2, and bz2, showed steady-state transcripts in W22 kernel aleurones, but these transcripts were absent in mutant c1 or r1 aleurones. These results demonstrated the requirement of functional c1 and r1 alleles for pr1 transcript accumulation in the kernel aleurone. Furthermore, in silico analysis of the promoter sequence of the pr1 gene identified a putative cis-regulatory sequence required for the binding of c1 and r1 encoded proteins. The sequence “AGGTGGTAGCTGGGA” lies 126 bp upstream of the putative transcription start site (TSS), which was previously shown to be required for binding of a C1 MYB protein and was designated as the C1-binding site (CBS) (MetInspector) (Sainz et al. 1997). In addition, a cis-sequence similar to an anthocyanin regulatory element (ARE), is present at −106 position (Tuerck and Fromm 1994). The ARE has been demonstrated to play an important role in the activation of anthocyanin genes (Tuerck and Fromm 1994; Lesnick and Chandler 1998). In conclusion, our results from genetic, molecular, and promoter sequence analyses show that c1 and r1 are required for pr1 expression in the anthocyanin biosynthetic pathway and suggest a direct binding of C1 to the Pr1 promoter region.
Figure 7.—
Mutant c1 and r1 plants carrying Pr1 do not accumulate pr1 transcripts. Expression of pr1 and other anthocyanin genes in aleurones were analyzed by RT–PCR in c1-MGS131036, c1-MGS14633, r1-MGS167054, and r1-MGS14638 alleles. Purple aleurones from W22 were used as positive control for anthocyanin genes’ expression, while actin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as loading controls.
DISCUSSION
The pr1 locus has been widely used as a phenotypic marker in maize; however, the molecular difference between Pr1 and pr1 plants has never been investigated. The results presented in this study show that transcripts for Zmf3′h1 accumulate in aleurones of Pr1, but not in pr1 aleurone tissue. Expression of Zmf3′h1 in the silk and husk shows similar trend. The gene expression pattern of Zmf3′h1 correlates with the accumulation of anthocyanidins in aleurone and dihydroflavonols in silk tissue. However, in their study on F3′H enzyme in maize, Larson et al. (1986) found some F3′H activity in seedlings of homozygous mutant pr1 plants. They suggested that either the mutant pr1 allele is hypomorphic i.e., it has a reduced level of pr1 gene expression or there is a duplicate f3′h gene present in maize. Despite these previous findings, we did not observe evidence of reduced levels of pr1 gene expression or a second f3′h gene in any of the tissues studied. This discrepancy may be the result of sequence differences between the Zmf3′h1 and the proposed second hydroxylase gene or it could be that they had detected the activity of a P450 hydroxylase other than F3′H in mutant pr1 plants. Our results from the expression analysis confirm that the aleurone color difference between Pr1 and pr1 is due to the lack of Zmf3′h1 transcripts in pr1 and not due to any absence or change in the expression of other anthocyanin biosynthetic or regulatory genes.
We showed using TLC and HPLC analysis of kernel aleurone extracts that purple cyanidin glycosides and red pelargonidin glycosides accumulated in Pr1 and pr1 lines, respectively. This indicates a probable block in the formation of DHQ in mutant lines. DHQ can be formed from DHK by F3′H or from naringenin to eriodictyol via F3′H followed by F3H activity. Chromatographic analyses of the silk extracts from Pr1 plants show the presence of DHQ and DHK as well, while the pr1 plants have DHK only. The correlation of Pr1 and pr1 alleles to the types of flavonoid compounds produced is consistent with the role of pr1 as F3′H encoding gene and is able to perform the 3′-hydroxylation reaction in the 3-hydroxyanthocyanin branch pathway. This was further confirmed by the presence of dominant peaks for cyanidin and pelargonidin (which differ by one hydroxyl group attached at the 3′ position in cyanidin) in the methanolic extracts from purple and red aleurones, respectively.
Anthocyanin biosynthesis in maize is transcriptionally regulated by an interaction between two sets of transcription factors encoded by c1/pl1 and r1/b1; c1 and r1 are required in the kernel aleurone, while pl1 and b1 function to activate these genes in the plant body (Chandler et al. 1989). Our genetic analysis revealed that the pr1 gene plays a role in the c1- and r1-regulated anthocyanin biosynthetic pathway. RNA expression of pr1 and other biosynthetic genes was not detected in the absence of functional alleles of c1 and r1. R1 and C1 proteins physically interact; additionally, C1 binds directly to specific cis-regulating elements present in the promoters of biosynthetic genes (Sainz et al. 1997). In vitro protein–DNA binding assays using point mutations, or in vivo transposon insertions in these cis-binding sites disrupt gene expression (Grotewold et al. 1994; Pooma et al. 2002). Similar elements were found in the pr1 promoter, indicating that pr1 may be regulated by C1 and R1 in a fashion similar to other anthocyanin biosynthetic genes (Sainz et al. 1997; Lesnick and Chandler 1998).
Our SNP-based genetic linkage analysis mapped the Zmf3′h1 coding region to the pr1 locus on chromosome 5L and this is in agreement with the B73 RefGen_v2 genome sequence. The proposed function of Zmf3′h1 was verified by both the in vitro and in vivo complementation of the pr1 phenotype. In transient assays, cotransformation of 35S::ZmF3′H1 and 35S::C1+R1 plasmid constructs causes the accumulation of purple pigments in Zea mays Black Mexican Sweet (BMS) cells as well as in kernel aleurones of MP708 (data not shown), both of which carry nonfunctional pr1 alleles in their genetic background (Grotewold et al. 1998). Our results from complementation of Arabidopsis tt7 mutation by the Zmf3′h1 explicitly confirm that the isolated Zmf3′h1 gene encodes for a functional F3′H enzyme. Finally, the identification of a Ds-induced insertion reveals independently the role of Zmf3′h1 in producing purple cyanidin glycosides from red pelargonidin glycosides.
The flavonoid biosynthetic pathway is present in both monocots and dicots, but the structural enzymes in this pathway are divergent in these two groups (Dong et al. 2001). One theory proposes that because of less stringent selective pressure on the enzymes of secondary metabolic pathways, these enzymes have diverged rapidly during evolution (Pichersky and Gang 2000). In contrast, mutations in Arabidopsis and petunia (both are dicots) reveal that flavonoid biosynthetic genes can be complemented by orthologs from Z. mays, a monocot (Dong et al. 2001). Similarly, we found that Zmf3′h1-transformed tt7 seedlings produced cyanidin and its glycosylated derivatives. These results are surprising given that many maize genes, including Zmf3′h1 (which shares 57% identity to Arabidopsis F3′H) have only moderate sequence identity to homologous genes in Arabidopsis. Additionally, the regulatory genes c1 and r1 from maize when expressed in transgenic Arabidopsis and tobacco plants resulted in increased anthocyanin pigmentation in both dicot species (Lloyd et al. 1992). These prior results along with the results of the current study demonstrate that many enzymes of the flavonoid biosynthetic pathway are conserved between monocots and dicots and that they can be transferred between these two divergent groups of flowering plants.
In this study, three spontaneous “loss-of-function” pr1 alleles were genetically characterized to further confirm the function of the pr1 gene. Two of these pr1 alleles show an insertion of 24 TA dinucleotide repeats in the promoter region, while a 17-bp deletion was detected in the putative 5′-UTR of the third allele. Further, these deletion and insertion polymorphisms cosegregated with the red aleurone phenotype in a F2 population segregating for Pr1 and pr1. The variation of dinucleotide repeats (also known as tandem repeats) in the promoter region has been shown to interfere with expression levels of certain genes (Martin et al. 2005). In yeast, differences in promoter tandem repeat length affect transcription levels, which elevated with the increase in the length of tandem repeats to a certain number and then diminished (Vinces et al. 2009). Tandem repeats can also reduce the promoter activity by changing the spacing between two binding sites or by inserting into a cis-element and making it unavailable for binding of an important trans-factor (Raijmakers et al. 2000). However, we were unable to demonstrate the role of the tandem repeats in limiting expression from the pr1 locus.
In addition to elucidating the molecular identity of the pr1 locus, this study provides a foundation to examine the role of DHK and DHQ as antifungal and insecticidal compounds (Hammerschmidt and Nicholson 1977; Hedin et al. 1983; Hipskind 1996; Karageorgou and Manetas 2006). F3′H enzyme activity is required for hydroxylation at the 3′ position on the B ring of flavonoid compounds and was observed to act on a wide range of substrates including flavonols, flavones, and flavanones (Larson and Bussard 1986). Interestingly, the hydroxylation pattern of the B ring is also the key determinant in the antioxidant property of anthocyanins; cyanidin, because of an additional hydroxyl group at its 3′ B-ring position, has a higher oxygen radical absorbing capacity as compared to pelargonidin and, therefore, has higher anticarcinogenic activity (Wang et al. 1997). It has been demonstrated that the f3′h gene in sorghum is induced in response to fungal infection, and its expression is correlated with the biosynthesis of flavonoid phytoalexins (Boddu et al. 2004). Moreover, pr1 was detected as a major QTL for the synthesis of C-glycosyl flavones that have insecticidal activity against corn earworm (Lee et al. 1998; Cortes-Cruz et al. 2003). We are thus further interested in investigating the role of pr1 in plant disease and insect resistance through the biosynthesis of different flavonoids that act as defense compounds.
Acknowledgments
We thank Catherine Svabek for her excellent technical assistance, Scott Harkcom, Jim Breining, John Shaffer, and the Penn State Agronomy Farm staff for their help with land preparation and tending of maize genetic nurseries. This work was supported in part by research support to S.C. under Hatch projects 4144 and 4154 of the College of Agricultural Sciences, Pennsylvania State University, and a United States Department of Agriculture–National Research Initiative-2007-35318-17795 award. This work was also supported by funding from the National Science Foundation to T.P.B. (D.B.I. 0501713). M.S. was supported by a doctoral assistantship from the Department of Crop and Soil Sciences, Pennsylvania State University.
LITERATURE CITED
- Ahern K. R., Deewatthanawong P., Schares J., Muszynski M., Weeks R., et al. , 2009. Regional mutagenesis using dissociation in maize. Methods 49: 248–254 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Boddu J., Jiang C., Sangar V., Olson T., Peterson T., et al. , 2006. Comparative structural and functional characterization of sorghum and maize duplications containing orthologous myb transcription regulators of 3-deoxyflavonoid biosynthesis. Plant Mol. Biol. 60: 185–199 [DOI] [PubMed] [Google Scholar]
- Boddu J., Svabek C., Sekhon R., Gevens A., Nicholson R., et al. , 2004. Expression of a putative flavonoid 3′-hydroxylase in sorghum mesocotyls synthesizing 3-deoxyanthocyanidin phytoalexins. Physiol. Mol. Plant Pathol. 65: 101–113 [Google Scholar]
- Brugliera F., Barri-Rewell G., Holton T. A., Mason J. G., 1999. Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J. 19: 441–451 [DOI] [PubMed] [Google Scholar]
- Burbulis I. E., Iacobucci M., Shirley B. W., 1996. A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell 8: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler V. L., Radicella J. P., Robbins T. P., Chen J., Turks D., 1989. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell 1: 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra S., Hoshino A., Boddu J., Iida S., 2006. Flavonoid pigments as tools in molecular genetics, pp. 147–173 in The Science of Flavonoids, edited by Grotewold E. The Ohio State University, Columbus, OH. [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coe E. H., Jr., 1955. Anthocyanin synthesis in maize, the interaction of A2 and Pr in leucoanthocyanin accumulation. Genetics 40: 568 [Google Scholar]
- Cone K. C., 2007. Anthocyanin synthesis in maize aleurone tissue. Plant Cell Monogr. 8: 121–139 [Google Scholar]
- Cone K. C., Cocciolone S. M., Burr F. A., Burr B., 1993. Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad L. J., Brutnell T. P., 2005. Ac-immobilized, a stable source of Activator transposase that mediates sporophytic and gametophytic excision of dissociation elements in maize. Genetics 171: 1999–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Cruz M., Snook M., McMullen M. D., 2003. The genetic basis of C-glycosyl flavone B-ring modification in maize (Zea mays L.) silks. Genome 46: 182–194 [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Steele C. L., 1999. Flavonoids and isoflavonoids: a gold mine for metabolic engineering. Trends Plant Sci. 4: 394–400 [DOI] [PubMed] [Google Scholar]
- Dong X., Braun E. L., Grotewold E., 2001. Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol. 127: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H. K., Robbins T. P., Jorgensen R. A., 1991. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25: 173–199 [DOI] [PubMed] [Google Scholar]
- Forkmann G., 1991. Flavonoids as flower pigments: the formation of the natural spectrum and its extension by genetic engineering. Plant Breed. 106: 1–26 [Google Scholar]
- Goff S. A., Cone K. C., Chandler V. L., 1992. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6: 864–875 [DOI] [PubMed] [Google Scholar]
- Grotewold E., Drummond B. J., Bowen B., Peterson T., 1994. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553 [DOI] [PubMed] [Google Scholar]
- Grotewold E., Chamberlin M., Snook M., Siame B., Butler L., et al. , 1998. Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10: 721–740 [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt R., Nicholson R. L., 1977. Resistance of maize to anthracnose: changes in host phenols and pigments. Phytopathol. 67: 251–258 [Google Scholar]
- Hedin P. A., Jenkins J. N., Collum D. H., White W. H., Parrott W. L., et al. , 1983. Cyanidin-3-b-glucoside, a newly recognized basis for resistance in cotton to the tobacco budworm Heliothis virescens (Fab.)(Lepidoptera: Noctuidae). Experientia 39: 799–801 [Google Scholar]
- Hipskind J., Wood M., Nicholson R. L., 1996. Localized stimulation of anthocyanin accumulation and delineation of pathogen ingress in maize genetically resistant to Bipolaris maydis race O. Physiol. Mol. Plant Pathol. 49: 247–256 [Google Scholar]
- Holton T. A., 1995. Modification of flower colour via manipulation of P450 gene expression in transgenic plants. Drug Metabol. Drug Interact. 12: 359–368 [DOI] [PubMed] [Google Scholar]
- Holton T. A., Cornish E. C., 1995. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M. H., Lam H. M., van de Loo F. J., Coruzzi G., 1998. A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc. Natl. Acad. Sci. USA 95: 13965–13970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgou P., Manetas Y., 2006. The importance of being red when young: anthocyanins and the protection of young leaves of Quercus coccifera from insect herbivory and excess light. Tree Physiol. 26: 613–621 [DOI] [PubMed] [Google Scholar]
- Koes R., Verweij W., Quattrocchio F., 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Koornneef M., 1990. Mutations affecting the testa colour in Arabidopsis. Arabidopsis Info. Serv. 27: 1–5 [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., et al. , 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2949 [DOI] [PubMed] [Google Scholar]
- Larson R., Bussard J., 1986. Microsomal flavonoid 3′-monooxygenase from maize seedlings. Plant Physiol. 80: 483–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson R., Bussard J. B., Coe E. H., Jr., 1986. Gene-dependent flavonoid 3′-hydroxylation in maize. Biochem. Genet. 24: 615–624 [DOI] [PubMed] [Google Scholar]
- Lee E. A., Byrne P. F., McMullen M. D., Snook M. E., Wiseman B. R., et al. , 1998. Genetic mechanisms underlying apimaysin and maysin synthesis and corn earworm antibiosis in maize (Zea mays L.). Genetics 149: 1997–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnick M. L., Chandler V. L., 1998. Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters. Plant Physiol. 117: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A. M., Walbot V., Davis R. W., 1992. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258: 1773–1775 [DOI] [PubMed] [Google Scholar]
- Ludwig S. R., Bowen B., Beach L., Wessler S. R., 1990. A regulatory gene as a novel visible marker for maize transformation. Science 247: 449–450 [DOI] [PubMed] [Google Scholar]
- Martin P., Makepeace K., Hill S. A., Hood D. W., Moxon E. R., 2005. Microsatellite instability regulates transcription factor binding and gene expression. Proc. Natl. Acad. Sci. USA 102: 3800–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClary J. E., 1942. The anthocyanin pigments of corn. Maize Genet. Coop. News Lett. 16: 30 [Google Scholar]
- Miyagi Y., Om A. S., Chee K. M., Bennink M. R., 2000. Inhibition of azoxymethane-induced colon cancer by orange juice. Nutr. Cancer 36: 224–229 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F., 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15: 473–497 [Google Scholar]
- Nyman N. A., Kumpulainen J. T., 2001. Determination of anthocyanins in berries and red wine by high-performance liquid chromatography. J. Agricult. Food Chem. 49: 4183–4187 [DOI] [PubMed] [Google Scholar]
- Peer W. A., Brown D. E., Tague B. W., Muday G. K., Taiz L., et al. , 2001. Flavonoid accumulation patterns of transparent testa mutants of arabidopsis. Plant Physiol. 126: 536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza P., Procissi A., Jenkins G. I., Tonelli C., 2002. Members of the c1/pl1 regulatory gene family mediate the response of maize aleurone and mesocotyl to different light qualities and cytokinins. Plant Physiol. 128: 1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E., Gang D. R., 2000. Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 5: 439. [DOI] [PubMed] [Google Scholar]
- Pooma W., Gersos C., Grotewold E., 2002. Transposon insertions in the promoter of the Zea mays a1 gene differentially affect transcription by the Myb factors P and C1. Genetics 161: 793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K., Frech K., Karas H., Wingender E., Werner T., 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23: 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raijmakers M. T., Jansen P. L., Steegers E. A., Peters W. H., 2000. Association of human liver bilirubin UDP-glucuronyltransferase activity with a polymorphism in the promoter region of the UGT1A1 gene. J. Hepatol. 33: 348–351 [DOI] [PubMed] [Google Scholar]
- Sainz M. B., Grotewold E., Chandler V. L., 1997. Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley B. W., 1996. Flavonoid biosynthesis: ‘new’ functions for an ‘old’ pathway. Trends Plant Sci. 1: 377–382 [Google Scholar]
- Shirley B. W., Kubasek W. L., Storz G., Bruggemann E., Koornneef M., et al. , 1995. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Snook M. E., Widstrom N. W., Wiseman B. R., Gueldner R. C., Wilson R. L., et al. 1994. New flavone C-glycosides from corn (Zea mays L.) for the control of the corn earworm (Helicoverpa zea), pp. 122–135 in Bioregulators for crop protection and pest control. Edited by Hedin P. A. Symposium Series 557 of the American Chemical Society, Washington, DC [Google Scholar]
- Snyder B. A., Nicholson R. L., 1990. Synthesis of phytoalexins in sorghum as a site-specific response to fungal ingress. Science 248: 1637–1639 [DOI] [PubMed] [Google Scholar]
- Stafford H. A., 1990. Flavonoid Metabolism. CRC Press, Boca Raton, FL [Google Scholar]
- Styles E. D., Ceska O., 1989. Pericarp flavonoids in genetic strains of Zea mays. Maydica 34: 227–237 [Google Scholar]
- Taylor L. P., Grotewold E., 2005. Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 8: 317–323 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J., 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda K., Yang D., Yamanaka N., Watanabe S., Harada K., et al. , 2002. A single-base deletion in soybean flavonoid 3′-hydroxylase gene is associated with gray pubescence color. Plant Mol. Biol. 50: 187–196 [DOI] [PubMed] [Google Scholar]
- Tuerck J. A., Fromm M. E., 1994. Elements of the maize A1 promoter required for transactivation by the anthocyanin B/C1 or phlobaphene P regulatory genes. Plant Cell 6: 1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces M. D., Legendre M., Caldara M., Hagihara M., Verstrepen K. J., 2009. Unstable tandem repeats in promoters confer transcriptional evolvability. Science 324: 1213–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroh Bi I., McMullen M. D., Sanchez-Villeda H., Schroeder S., Gardiner J., et al. , 2006. Single nucleotide polymorphisms and insertion-deletions for genetic markers and anchoring the maize fingerprint contig physical map. Crop Sci. 46: 12–21 [Google Scholar]
- Wang H., Cao G., Prior R. L., 1997. Oxygen radical absorbing capacity of anthocyanins. J. Agricult. Food Chem. 45: 304–309 [Google Scholar]
- Winkel-Shirley B., 2001. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarudnaya K. I., 1950. A chromatographic study of anthocyanins and related substances in various genotypes of maize. Ph.D. Thesis, University of Missouri, Columbia, MO [Google Scholar]