Abstract
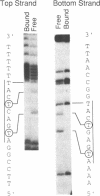
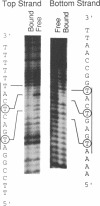
We describe a novel uracil interference method for examining protein contacts with the 5-methyl group of thymines. The protein of interest is incubated with target DNA containing randomly distributed deoxyuracil substitutions that is generated by carrying out the polymerase chain reaction in the presence of a mixture of TTP and dUTP. After separating DNA-protein complexes away from unbound DNA, the locations of deoxyuracil residues that either do or do not interfere with DNA-binding are determined by cleavage with uracil-N-glycosylase followed by piperidine. Using this uracil interference assay, we show that the methyl groups of the four core thymines, but not the two peripheral thymines, of the optimal binding site (ATG-ACTCAT) are important for high affinity binding of GCN4. Similar, but not identical, results are obtained using KMnO4 interference, another method used for studying protein-DNA interactions involving thymine residues. These observations strongly suggest that GCN4 directly contacts the 5-methyl groups of the four core thymines that lie in the major groove of the target DNA. Besides providing specific structural information about protein-DNA complexes, uracil interference should also be useful for identifying DNA-binding proteins and their target sites in eukaryotic promoter regions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abate C., Luk D., Curran T. Transcriptional regulation by Fos and Jun in vitro: interaction among multiple activator and regulatory domains. Mol Cell Biol. 1991 Jul;11(7):3624–3632. doi: 10.1128/mcb.11.7.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D. R., Curran T. Analysis of dimerization and DNA binding functions in Fos and Jun by domain-swapping: involvement of residues outside the leucine zipper/basic region. Oncogene. 1990 Jun;5(6):929–939. [PubMed] [Google Scholar]
- Curran T., Franza B. R., Jr Fos and Jun: the AP-1 connection. Cell. 1988 Nov 4;55(3):395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- Delort A. M., Neumann J. M., Molko D., Hervé M., Téoule R., Tran Dinh S. Influence of uracil defect on DNA structure: 1H NMR investigation at 500 MHz. Nucleic Acids Res. 1985 May 10;13(9):3343–3355. doi: 10.1093/nar/13.9.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Ampe C., Steitz T. A., Crothers D. M. Molecular characterization of the GCN4-DNA complex. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6034–6038. doi: 10.1073/pnas.87.16.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeddel D. V., Yansura D. G., Caruthers M. H. How lac repressor recognizes lac operator. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3578–3582. doi: 10.1073/pnas.75.8.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarie R. Thymine methyls and DNA-protein interactions. Nucleic Acids Res. 1987 Dec 10;15(23):9975–9983. doi: 10.1093/nar/15.23.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991 Jul 26;66(2):317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- O'Neil K. T., Hoess R. H., DeGrado W. F. Design of DNA-binding peptides based on the leucine zipper motif. Science. 1990 Aug 17;249(4970):774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- Ogata R., Gilbert W. Contacts between the lac repressor and the thymines in the lac operator. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4973–4976. doi: 10.1073/pnas.74.11.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A. R., Brandl C. J., Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol Cell Biol. 1989 Jul;9(7):2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu W. T., Struhl K. The leucine zipper symmetrically positions the adjacent basic regions for specific DNA binding. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):6901–6905. doi: 10.1073/pnas.88.16.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risse G., Jooss K., Neuberg M., Brüller H. J., Müller R. Asymmetrical recognition of the palindromic AP1 binding site (TRE) by Fos protein complexes. EMBO J. 1989 Dec 1;8(12):3825–3832. doi: 10.1002/j.1460-2075.1989.tb08560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers J. W., Vincent A. C., Struhl K. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol Cell Biol. 1990 Oct;10(10):5077–5086. doi: 10.1128/mcb.10.10.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truss M., Chalepakis G., Beato M. Contacts between steroid hormone receptors and thymines in DNA: an interference method. Proc Natl Acad Sci U S A. 1990 Sep;87(18):7180–7184. doi: 10.1073/pnas.87.18.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M. A., Ellenberger T., Wobbe C. R., Lee J. P., Harrison S. C., Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990 Oct 11;347(6293):575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]