Abstract
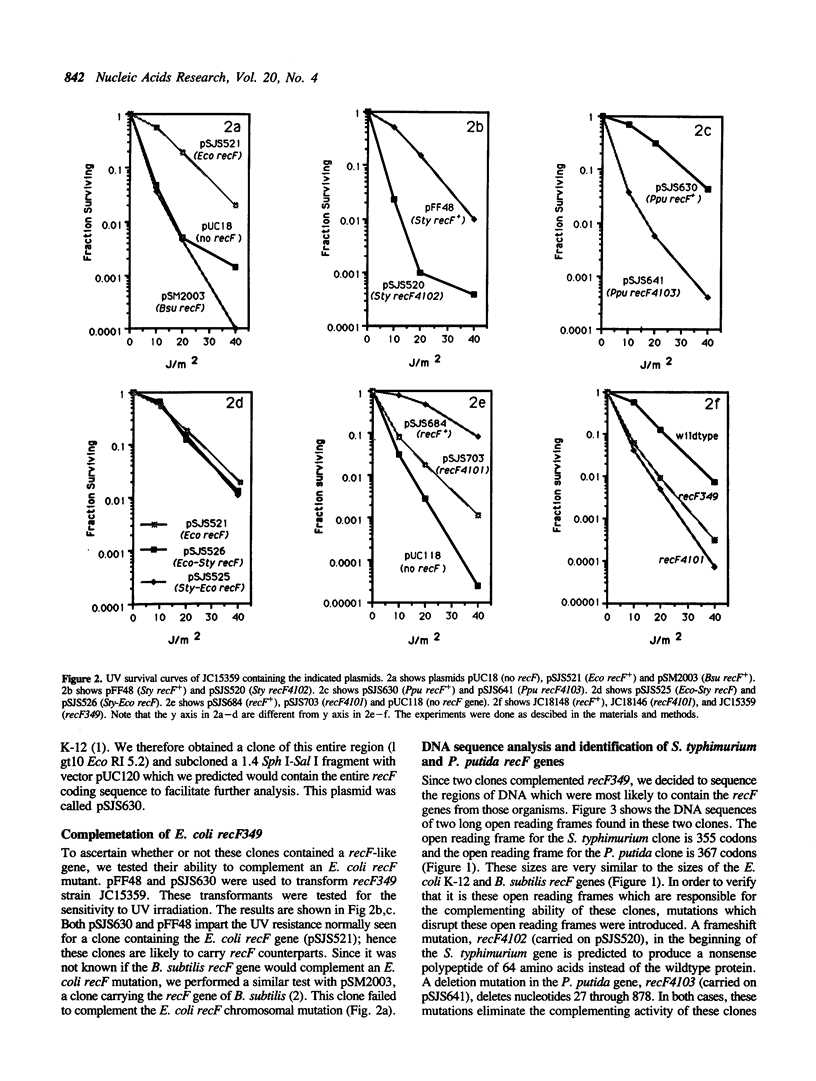
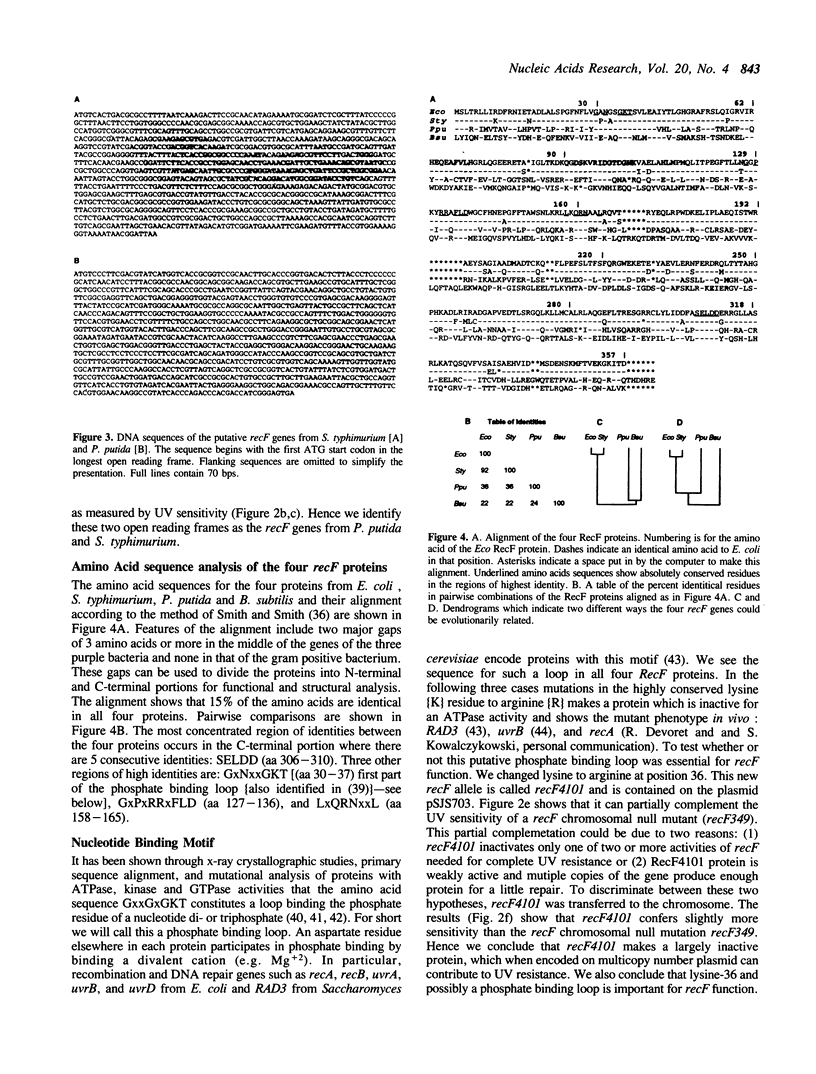
We have compared the recF genes from Escherichia coli K-12, Salmonella typhimurium, Pseudomonas putida, and Bacillus subtilis at the DNA and amino acid sequence levels. To do this we determined the complete nucleotide sequence of the recF gene from Salmonella typhimurium and we completed the nucleotide sequence of recF gene from Pseudomonas putida begun by Fujita et al. (1). We found that the RecF proteins encoded by these two genes contain respectively 92% and 38% amino acid identity with the E. coli RecF protein. Additionally, we have found that the S. typhimurium and P. putida recF genes will complement an E. coli recF mutant, but the recF gene from Bacillus subtilis [showing about 20% identity with E. coli (2)] will not. Amino acid sequence alignment of the four proteins identified four highly conserved regions. Two of these regions are part of a putative phosphate binding loop. In one region (position 36), we changed the lysine codon (which is essential for ATPase, GTPase and kinase activity in other proteins having this phosphate binding loop) to an arginine codon. We then tested this mutation (recF4101) on a multicopy plasmid for its ability to complement a recF chromosomal mutation and on the E. coli chromosome for its effect on sensitivity to UV irradiation. The strain with recF4101 on its chromosome is as sensitive as a null recF mutant strain. The strain with the plasmid-borne mutant allele is however more UV resistant than the null mutant strain. We conclude that lysine-36 and possibly a phosphate binding loop is essential for full recF activity. Lastly we made two chimeric recF genes by exchanging the amino terminal 48 amino acids of the S. typhimurium and E. coli recF genes. Both chimeras could complement E. coli chromosomal recF mutations.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso J. C., Tailor R. H., Lüder G. Characterization of recombination-deficient mutants of Bacillus subtilis. J Bacteriol. 1988 Jul;170(7):3001–3007. doi: 10.1128/jb.170.7.3001-3007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengod M. E., García-Sogo M., Lambíes E. Transcriptional organization of the dnaN and recF genes of Escherichia coli K-12. J Biol Chem. 1988 Aug 25;263(24):12109–12114. [PubMed] [Google Scholar]
- Armengod M. E., Lambíes E. Overlapping arrangement of the recF and dnaN operons of Escherichia coli; positive and negative control sequences. Gene. 1986;43(3):183–196. doi: 10.1016/0378-1119(86)90206-4. [DOI] [PubMed] [Google Scholar]
- Blanar M. A., Sandler S. J., Armengod M. E., Ream L. W., Clark A. J. Molecular analysis of the recF gene of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4622–4626. doi: 10.1073/pnas.81.15.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesla Z., O'Brien P., Clark A. J. Genetic analysis of UV mutagenesis of the Escherichia coli glyU gene. Mol Gen Genet. 1987 Apr;207(1):1–8. doi: 10.1007/BF00331483. [DOI] [PubMed] [Google Scholar]
- Cohen A., Laban A. Plasmidic recombination in Escherichia coli K-12: the role of recF gene function. Mol Gen Genet. 1983;189(3):471–474. doi: 10.1007/BF00325911. [DOI] [PubMed] [Google Scholar]
- Engstrom J., Wong A., Maurer R. Interaction of DNA polymerase III gamma and beta subunits in vivo in Salmonella typhimurium. Genetics. 1986 Jul;113(3):499–515. doi: 10.1093/genetics/113.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R., Kolodner R. Gene conversion in Escherichia coli: the recF pathway for resolution of heteroduplex DNA. J Bacteriol. 1989 Jun;171(6):3046–3052. doi: 10.1128/jb.171.6.3046-3052.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M. Q., Yoshikawa H., Ogasawara N. Structure of the dnaA region of Pseudomonas putida: conservation among three bacteria, Bacillus subtilis, Escherichia coli and P. putida. Mol Gen Genet. 1989 Feb;215(3):381–387. doi: 10.1007/BF00427033. [DOI] [PubMed] [Google Scholar]
- Griffin T. J., 4th, Kolodner R. D. Purification and preliminary characterization of the Escherichia coli K-12 recF protein. J Bacteriol. 1990 Nov;172(11):6291–6299. doi: 10.1128/jb.172.11.6291-6299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii Z., Clark A. J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973 Oct 25;80(2):327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- Lavery P. E., Kowalczykowski S. C. Properties of recA441 protein-catalyzed DNA strand exchange can be attributed to an enhanced ability to compete with SSB protein. J Biol Chem. 1990 Mar 5;265(7):4004–4010. [PubMed] [Google Scholar]
- Madiraju M. V., Clark A. J. Effect of RecF protein on reactions catalyzed by RecA protein. Nucleic Acids Res. 1991 Nov 25;19(22):6295–6300. doi: 10.1093/nar/19.22.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju M. V., Templin A., Clark A. J. Properties of a mutant recA-encoded protein reveal a possible role for Escherichia coli recF-encoded protein in genetic recombination. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6592–6596. doi: 10.1073/pnas.85.18.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4185–4189. doi: 10.1073/pnas.84.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura C. S., Holbrook S. R., Ames G. F. Structural model of the nucleotide-binding conserved component of periplasmic permeases. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):84–88. doi: 10.1073/pnas.88.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya S., Ogasawara N., Yoshikawa H. Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. III. Nucleotide sequence of some 10,000 base pairs in the origin region. Nucleic Acids Res. 1985 Apr 11;13(7):2251–2265. doi: 10.1093/nar/13.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., Yoshikawa H. Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames. Nucleic Acids Res. 1985 Apr 11;13(7):2267–2279. doi: 10.1093/nar/13.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., von Meyenburg K., Hansen F. G., Yoshikawa H. Conservation of genes and their organization in the chromosomal replication origin region of Bacillus subtilis and Escherichia coli. EMBO J. 1985 Dec 1;4(12):3345–3350. doi: 10.1002/j.1460-2075.1985.tb04087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream L. W., Clark A. J. Cloning and deletion mapping of the recF dnaN region of the Escherichia coli chromosome. Plasmid. 1983 Sep;10(2):101–110. doi: 10.1016/0147-619x(83)90062-8. [DOI] [PubMed] [Google Scholar]
- Ream L. W., Margossian L., Clark A. J., Hansen F. G., von Meyenburg K. Genetic and physical mapping of recF in Escherichia coli K-12. Mol Gen Genet. 1980;180(1):115–121. doi: 10.1007/BF00267359. [DOI] [PubMed] [Google Scholar]
- Roca A. I., Cox M. M. The RecA protein: structure and function. Crit Rev Biochem Mol Biol. 1990;25(6):415–456. doi: 10.3109/10409239009090617. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y., Mizukami T. A temperature-sensitive Escherichia coli mutant defective in DNA replication: dnaN, a new gene adjacent to the dnaA gene. Mol Gen Genet. 1980;178(3):541–553. doi: 10.1007/BF00337859. [DOI] [PubMed] [Google Scholar]
- Sandler S. J., Clark A. J. Factors affecting expression of the recF gene of Escherichia coli K-12. Gene. 1990 Jan 31;86(1):35–43. doi: 10.1016/0378-1119(90)90111-4. [DOI] [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990 Nov;15(11):430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Seeley T. W., Grossman L. Mutations in the Escherichia coli UvrB ATPase motif compromise excision repair capacity. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6577–6581. doi: 10.1073/pnas.86.17.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovgaard O., Hansen F. G. Comparison of dnaA nucleotide sequences of Escherichia coli, Salmonella typhimurium, and Serratia marcescens. J Bacteriol. 1987 Sep;169(9):3976–3981. doi: 10.1128/jb.169.9.3976-3981.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. F., Smith T. F. Automatic generation of primary sequence patterns from sets of related protein sequences. Proc Natl Acad Sci U S A. 1990 Jan;87(1):118–122. doi: 10.1073/pnas.87.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Higgins D., Prakash L., Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988 Oct;7(10):3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms B., Wackernagel W. Regulatory role of recF in the SOS response of Escherichia coli: impaired induction of SOS genes by UV irradiation and nalidixic acid in a recF mutant. J Bacteriol. 1987 Apr;169(4):1731–1736. doi: 10.1128/jb.169.4.1731-1736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert M. R. Altered induction of the adaptive response to alkylation damage in Escherichia coli recF mutants. J Bacteriol. 1989 Jan;171(1):99–103. doi: 10.1128/jb.171.1.99-103.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert M. R., Hartke M. A. Suppression of Escherichia coli recF mutations by recA-linked srfA mutations. J Bacteriol. 1984 Feb;157(2):498–506. doi: 10.1128/jb.157.2.498-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert M. R., Margossian L. J., Clark A. J. Two-component suppression of recF143 by recA441 in Escherichia coli K-12. J Bacteriol. 1984 Nov;160(2):702–705. doi: 10.1128/jb.160.2.702-705.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. P., Kaguni J. M. Transcriptional repression of the dnaA gene of Escherichia coli by dnaA protein. Mol Gen Genet. 1987 Oct;209(3):518–525. doi: 10.1007/BF00331158. [DOI] [PubMed] [Google Scholar]
- Willetts N. S., Clark A. J., Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969 Jan;97(1):244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]


