Abstract
Background:
Apolipoprotein E (ApoE) plays a role in the regulation of lipid metabolism in humans. ApoE, a 229-amino-acid polypeptide, is classified into three major isoforms (E2, E3, and E4) according to the differences in amino acids at positions 112 and 158. In the normal population, ApoE3 isoform is the most prevalent, and ApoE2 or E4 is frequently associated with hyperlipoproteinemia. The objective of this work was to investigate the relationship between ApoE gene polymorphism and coronary heart disease (CHD) in Gaza Strip and investigate the association between serum lipid levels and CHD.
Material and Methods:
The study population consisted of 137 subjects including 69 CHD cases (45 male, 24 female) and 68 healthy subjects (33 male and 35 female).
Results:
The ApoE3/E3 genotype was the most common in the control and the CHD groups. ApoE2/E3 and ApoE3/E4 were the next most common genotypes. The frequencies of ApoE alleles in the CHD subjects were 0.826 for E3, 0.137 for E4, and 0.0362 for E2. These frequencies are comparable to those found in the control group which were 0.875 for the E3, 0.073 for E4, and 0.0515 for E2. No statistically significant differences in ApoE genotypes were found between the patients and the control groups. Moreover, there was no significant difference between the mean of triglyceride (TG) and HDL levels among different ApoE genotypes. However, there was a significant difference in the mean of LDL and ApoE genotypes where the mean of LDL was 218.17 mg/dl in ApoE4, 149.67 mg/dl in ApoE2, and 184.52 mg/dl in ApoE3. A significant difference was also evident between the mean of LDL levels in the CHD and the control group where the mean of LDL was 126 mg/dl in CHD and 111.47 mg/dl in the control group. Our study indicated that there was no significant difference between the mean of cholesterol and TG levels of the CHD and the control groups.
Conclusions:
To our knowledge, this is the first study in Gaza Strip investigating the relation between ApoE genotypes and CHD. Further investigations are needed to link other genetic factors to CHD.
Keywords: Apolipoprotien E, Polymerase chain reaction- restriction fragment length polymorphism, coronary heart disease, gene polymorphism
INTRODUCTION
Apolipoprotein E (ApoE) is a polymorphic protein codified by three alleles in the same locus. Variability in ApoE binding to ApoE or LDL receptors results in different levels of lipids and lipoproteins and risk of coronary heart disease (CHD) among individuals predisposed to CHD.[1] Cardiovascular diseases (CVDs), including CHD and stroke, are the most important causes of mortality and morbidity. The known risk factors affecting CHD will inevitably affect the relation of ApoE to CHD. Several studies have indicated that the classic cardiovascular risk factors including smoking, high blood pressure, low HDL cholesterol, and in most studies, also high total cholesterol are important predictors of CHD events in elderly as well as middle-aged subjects. Lately, new cardiovascular risk factors such as apolipoproteins, including ApoE, have been a focus of keen investigation.[2] ApoE is an essential part of lipoprotein metabolism. It is present in lipoprotein particles and mediates lipoprotein binding to the LDL and lipoprotein remnant receptors. Defects in the ApoE protein could diminish its ability to bind to the receptors, which then leads to an elevated blood cholesterol level.[3] The gene coding for ApoE is located on chromosome 19 near the genes for Apo C-I and C-II. Three different alleles, E2, E3, and E4, account for ApoE polymorphism and determine the six genotypes E2/2, E2/3, E2/4, E3/3, E4/3, and E4/4.[2] ApoE polymorphisms have been associated with variations in the blood cholesterol level and with the risk of atherosclerosis and premature CVDs.[4]
ApoE polymorphism has functional effects on lipoprotein metabolism mediated through the hepatic binding, uptake, and catabolism of lipid particles, i.e., chylomicrons, chylomicron remnants, VLDL, and HDL subspecies. ApoE contributes to variability in normal cholesterol levels in populations.[5]
The major effect of genetic variation at this locus is its influence on cholesterol levels, one of the major risk factors for CVDs, particularly coronary artery disease. With reference to cholesterol effects from the E3 allele, E4 is associated with higher total and low density lipoprotein cholesterol levels and E2 with lower levels. The cholesterol-lowering effect of E2 tends to be greater than the cholesterol-raising effect of E4. In total, the contribution of this gene to cholesterol variability based on a variety of populations that have been studied is no more than 10%. Diet and other genes contribute to each individual's cholesterol level, as well as to the population's average level. Many studies have looked at interactions with variants of this gene as possible modifiers of other cardiovascular risks, such as high- and low-fat diets and active versus sedentary lifestyles. Interactions with lipid-lowering medications have also been investigated in relation to ApoE.[6]
ApoE facilitates the binding of triglyceride (TG)-richlipoprotein remnants to receptors that determine their clearance. The isoforms vary in their receptor-binding activity, with ApoE4 having the greatest receptor binding and ApoE2 havinga decreased affinity. Individuals with ApoE2 have higher levelsof TGs, and ApoE2 homozygotes have greatly increasedconcentrations of remnants, which frequently results in a formof dyslipidemia called type III or dysbetalipoproteinemia. Individualswith E4, conversely, tend to have higher concentrations of LDLcholesterol; this increase is due, in part, to more efficientabsorption of dietary cholesterol and perhaps down-regulationof the LDL receptor. Thus, ApoE is an important candidate genefor CVD.[7]
Approximately, 1% of individuals in the general population are ApoE2/2 homozygotes. However, only a small percentage (2–5%) of these ApoE2/2 homozygotes develop type III hyperlipoproteinemia. Utermann et al. suggested that homozygosityfor ApoE2 is a necessary but insufficient genetic influenceand that additional genetic or environmental factors are requiredfor the expression of type III hyperlipoproteinemia in ApoE2/2 homozygotes. Until 1979, no other abnormalities in other geneshad been identified.[8] High levels of total and LDL cholesterol and low levels of HDLcholesterol predispose individuals to the development of atherosclerosis. Serum levels of these lipids are partly determined by the ApoE genotype. With reference to ApoE3 homozygotes, ApoE2 is associated with lower levels of total and LDL cholesteroland with higher levels of HDL cholesterol, while ApoE4 hasopposite effects. ApoE plays a pivotal role in thetransport of lipoproteins and is involved in numerous processesin the arterial wall.[9]
The risk of atherosclerosis and coronary heart disease inmiddle ageappears to be higher in the presence ofthe ApoE4 allele, possibly because of altered lipid metabolism. Not all studies, especially those of elderly subjects or survivorsof heart attack, however, find an association between ApoE4 and CHD.[10]
The ApoE4 allele (Arg-112 and Arg-158) hasbeen linked to atherosclerosis. Individuals with E4/E4 genotypeare at a higher risk of developing the disease.[11]
The following were the objectives of this study
To investigate the association between ApoE gene polymorphism and CHD
To investigate the association between serum lipid levels and CHD
To evaluate the effect of ApoE gene polymorphism on serum lipid levels.
MATERIALS AND METHODS
Study population
This study is a retrospective case-control study in which ApoE genotyping was performed on 137 individuals whowere randomly selected. Sixty-nine subjects (24 female and 45 male) with coronary artery disease from Nasser Hospital in Khan Younis and 68 normal subjects were included in this study (35 female and 33 male).
Sample collection
Blood samples were collected from the patients and the control subjectsafter a 12- to 16-h overnight fasting. Each sample was collected into two tubes, one EDTA tube and one glass tube; the sample in the glass tube was separated serum was used for lipid profiling. The EDTA sample kept at 4°C was used within 24 h for DNA extraction and subsequent PCR analysis.
Data analysis
The data was entered, stored, and analyzed by a personal computer using the SPSS 8.0 statistical package. Differences in proportions were assessed by ANOVA, t-test, and chi-square test; P-values < 0.05 were considered statistically significant.
Lipid profiling
Serum cholesterol determination
10 µl Sample /standard was mixed1000 µl with reagent Cholesterolkit (DiaSys , Germany) . The contents were mixed and incubated for 20 min at 20–25°C; then the absorbance of each tube was read at wavelength 500 nm against blank. The cholesterol content was then calculated by applying the following equation:

And the Triglyceride the same method but used the triglyceride kit (DiaSys , Germany) and in both method used the normal and high Cholesterol control (DiaSys , Germany) , normal and high Triglyceride control (DiaSys , Germany).
HDL-C measurement
HDL-cholesterol was measured after chylomicrons, VLDL, and LDL, were precipitated by adding phosphotungestic acid to the sample. Centrifugation afterward left only HDL in the supernatant. The HDL-C content was determined enzymatically using Ecoline S+ cholesterol as described below.
Assay procedure
The200-µl serum samples were mixed with 500 µl of the precipitation reagent (phosphotungestic acid and magnesium chloride) and the mixture was incubated for 15 min at room temperature; then the mixture was centrifuged for 20 min at a speed of 2500 rpm; 0.1 ml of the clear supernatant was then transferred to the reaction solution for the determination of cholesterol.
The supernatant after the centrifugation should be clear; serum or plasma with TG contents ≥1000 mg/dl tends to produce a turbid supernatant or floating precipitates. In such case, the sample should be diluted 1:1 with the NaCl solution (0.9%) and then precipitation should be performed and the final result should be multiplied by 2.
LDL calculation
LDL can be calculated from the parameters determined above using the following formula:
LDL = Total cholesterol − (HDL + Triglyceride) 5
ApoE genotyping
DNA extraction
The human genomic DNA was isolated from the human blood sample by using the Promega kit(Promega, WI, USA). Then the quality of the isolated DNA was determined by running each sample on ethidium bromide-stained 1.0% agarose gel.
PCR amplification of the ApoE gene
The primers used in the PCR forward Sequence “5 to “3 (TCCAAGGAGCTGCAGGCGGCGCA) reverse Sequence “5 to “3 (GCCCCGGCCTGGTACACTGCCA)
Each PCR reaction mixture contained (~200 ng) of the prepared DNA template, 1× PCR buffer, 3.0 mM MgCl2 , 2.0 mM of each of the primers, and 200 mM of each deoxynucleotide triphosphate (dNTP); 1.25 U was the final concentration of Taq DNA polymerase (Promega) and DMSO (5%) in a thin-walled microfuge tube. The microfuge tubes were then placed in a thermal cycler and PCR amplification was done according to the following steps:
Step 1. Denaturation for 3 min at 95ºC
Step 2. 40 cycles of
2.1. Melting for 60 s at 95°C
2.2. Annealing for 60 s at 58°C
2.3. Extension for 90 s at 72°C
Step 3. Final elongation for 10 min at 72°C.
Upon the completion of PCR, the products were analyzed by electrophoresis on 2% ethidium bromide-stained agarose gel; then the 218-bp amplicons were stored at 4°C until analysis and digestion.
Restriction fragment length polymorphism analysis
Genotyping for ApoE was performed by digesting the PCR product with restriction enzymes AflIII (5.000 U/ml) and HaeII (20.000 U/ml), 10× buffer, and 0.2 µl BSA. The contents were incubated for 24 h at 37°C. The digests were resolved on ethidium bromide-stained agarose gel and the results were documented by photography and separation of the resulting DNA fragments on agarose gel [Figure 1].
Figure 1.
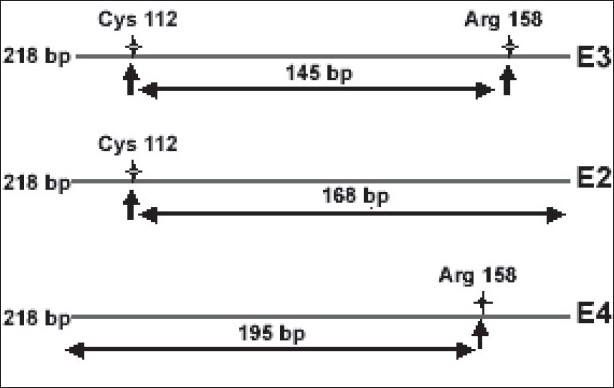
A schematic diagram illustrating ApoE RFLP analysis
RESULTS
PCR results
The amplicon (PCR product) generated from the ApoE gene should yield a 218-bp-long double-stranded DNA fragment. A negative control (with water instead of the DNA template) was included in each reaction. The size of the amplicon was estimated by comparing it with a DNA molecular size marker (100 bp ladder DNA) run on the same gel.
Amplification product
A representative photograph of the ApoE PCR amplification product is illustrated in Figure 2. Lane 1 in the figure shows the 100 bp ladder, lane 4 contains a negative control, and the other lanes show the 218 bp ApoE amplicon.
Figure 2.
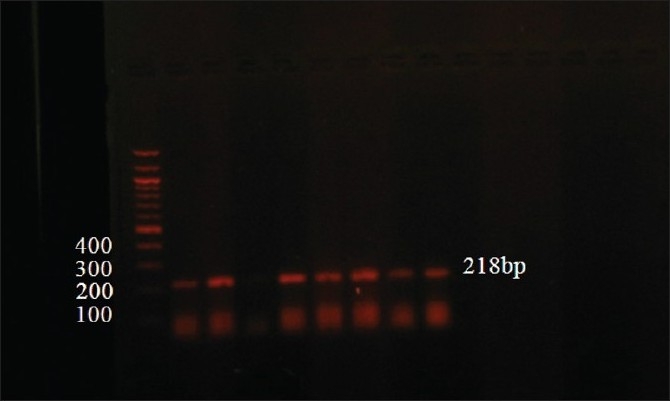
A photograph of the ApoE amplification product. Lane 1 shows the 100 bp ladder, lane 4 negative control, and the rest of lanes show the 218 bp ApoE PCR product
RFLP result
Figure 3 illustrates a representative ApoE genotyping by AflIII (A) and HaeII (H) digestion of the 218 bp amplified fragment.
Figure 3.
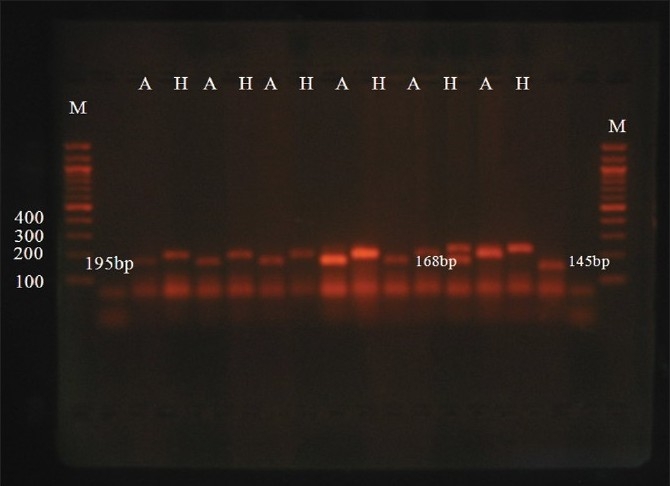
A representative photograph of AflIII (A) and HaeII (H) digestion of the ApoE 218 bp amplified fragment. M indicates the 100 bp size marker. The digests were run on ethidium bromide-stained 3% agarose gel; E3 = 145 bp, E2 = 168 bp, E4 = 195 bp
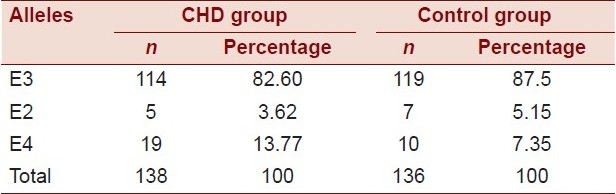
Genotype frequencies
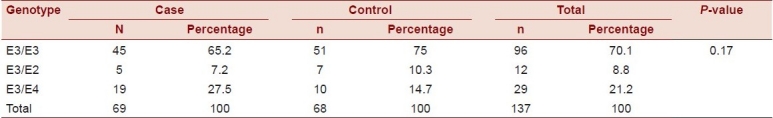
The ApoE allele frequencies in the control subjects were 5.15% for the E2 allele, 87.5% for the E3 allele, and 7.3% for the E4 allele. The frequencies in the CHD group were 3.62% for the E2 allele, 82.6% for the E3 allele, and 13.77% for the E4 allele [Table 1].
Table 1.
Frequency of ApoE alleles among the case and the control groups
Distribution of subjects according to ApoE genotypes
The distribution of the subjects according to ApoE genotypes was as follows: 70.1% E3/E3, 21.2% E3/E4, and 8.8% E3/E2
Relationship between CHD and ApoE gene polymorphism
The results showed that there was no statistically significant relation between CHD and ApoE gene polymorphism (χ2 = 3.4, P-value = 0.17), where the presence of the E3/E3 genotype was the most frequent among cases and controls (65.2% and 75%, respectively) as shown in Table 2.
Table 2.
Distribution of the ApoE genotypes among the cases and the control groups
Relationship between the ApoE genotype and gender
The results showed that there was no statistically significant relation between the ApoE genotype and gender (χ2 = 3.39, P-value = 0.18; Table 3).
Table 3.
Relationship between the ApoE gene and gender

Relationship between CHD and lipid profile
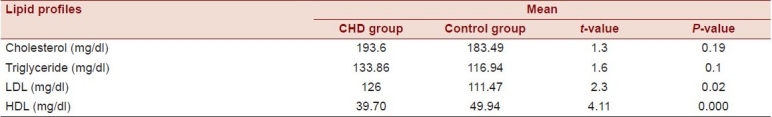
There was no statistically significant difference between the means of cholesterol and TG levels between the CHD and the control groups (t= 1.3, P-value = 0.19 and t= 1.6, P-value = 0.1, respectively). On the other hand, there were significant differences between the mean of LDL and HDL levels between cases and controls (P-value = 0.02 and P < 0.05, respectively).
Relationship between the lipid profile and ApoE genotypes
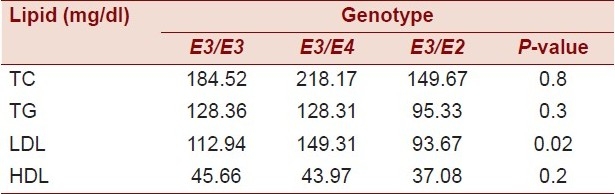
The relationship between the lipid profile and ApoE is provided in Table 4.
Table 4.
Relationship between CHD and lipid profile
As shown in the table, there was no significant difference between the mean of cholesterol, TGs, and HDL regardless of the ApoE genotype. On the other hand, there was a significant difference between the ApoE genotype and the mean of LDL.
Table 5.
Plasma lipid profile according to the ApoE genotype
CHD and age
The results of the study showed that there was a statistically significant difference between the means of age of cases and controls (P-value < 0.05) where the mean age of cases was 57.65 years, while that of the controls was 44.01.
CHD and smoking
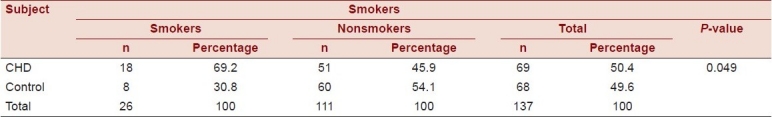
The results of the study showed that there was a statistically significant difference between the smokers and nonsmokers in cases and controls (χ2 = 4.56, P-value = 0.049) as shown in Table 6.
Table 6.
Relationship between CHD and smokers
DISCUSSION
According to the results obtained from PCR-RFLP, the ApoE allele frequencies in the control group were 0.0515 for the E2 allele, 0.875 for the E3 allele, and 0.073 for the E4 allele. This distribution is in agreement with the distribution in the control group reported in a Saudi study: 0.888 for E3, 0.050 for E2, and 0.062 for E4.[12] The frequencies of the ApoE alleles in the CHD subjects were 0.0362 for the E2 allele, 0.826 for the E3 allele, and 0.1377 for the E4.
Relation between ApoE gene polymorphism and CHD
According to our results, it was found that 27.5% (19 of 69) had E3/E4 in the CHD group as compared to 14.7% (10/68) of the control group. When compared to other studies, this percentage is close to that reported in Saudi Arabia where 20.8% of the CHD subjects had E3/E4 as compared to 12.5% from the control group.[12]Kolovou has shown that 21.8% of the CHD and 16.3% of the control group had the E3/E4 genotype.[13] The present study however indicated that there is no statistically significant relation between CHD and ApoE genes (χ2 = 3.4, P-value = 0.17). These results are in agreement with different studies such as the Saudi and the Taiwanese studies which recorded that no statistical difference in genotype or allelic distribution was found between the patients of CHD and the control groups.[12,13] These results can be explained by the fact that other genetic or environmentalrisk factors may interact with ApoE4 in determining the CHD risk. We did not find a significant differencefor the ApoE gene and genotype frequencies between patientsand controls, suggesting that the E4 allele is not a strong riskfactor alone for CHD in Gaza Strip. For the E4 allele, a significantly increased frequency in patientscompared to healthy controls has been described in some butnot all studies and the E4 allele could be a risk factor in associationwith other genetic or environmental factors.[14]
CVD risk is influenced by several well-establishedrisk factors, such as body mass index (BMI), an indicator of overweight and obesity, blood lipids, diabetes, and blood pressure. These are intermediate phenotypes, correlated among themselves and having their own genetic and environmental determinants,including diet, nutrition, hormones, smoking, alcohol intake, and physical activity.[15]
Serum lipid profile and the ApoE genotypes
As we noted from our results, the mean of cholesterol level (218.17 mg/dl) was higher in subjects with the E4 genotype as compared to those with E3 (184.5 mg/dl), while the E2 genotype had the lowest level (149.6 mg/dl). These results confirm the effect of variation in the ApoE genotype on lipoproteinlevels. The E4 allele has been shown to increase intestinal cholesterol absorption,to affect LDL synthesis in the liver, and to be associated withhigher levels of total cholesterol and LDL-C anda higher prevalence of atherosclerosis.
CHD and lipid profile
Our study results showed that there is a significant increase in LDL in the case group as compared to the control group. No statistically significant differences were evident between the mean of cholesterol between the case group and the control group. It has been suggested that LDL-C indicates the cardiovascular riskin individuals better than total cholesterol. This result is in agreement with other authors who reported that the most common lipid disorder was high TC and high LDL-C in CHD, which was much more prevalent than high TG. A high concentration of LDL must be considered the principal risk factor because in the absence of elevation of LDL-C, atherosclerosis develops slowly and usually remains subclinical. This is true even when the other independent risk factors are present.
CONCLUSION AND RECOMMENDATIONS
The present study focused on the detection of ApoE genotypes in Gaza Strip and the relationship between those genotypes and lipid profile in 69 cases of CHD from Nasser and AlShifa Hospitals as compared to 68 healthy subjects. The results of this study can be summarized as follows:
In Gaza Strip, the ApoE3/E3 genotype was the most common in the control and the CHD groups. ApoE2/E3 and ApoE4/E3 were the next most common genotypes. Other genotypes such as ApoE2/E2, ApoE4/E4, or ApoE2/E4 were not encountered in the examined subjects.
The frequencies of ApoE alleles in the CHD subjects were as follows: 0.826 for E3, 0.137 for E4, and 0.0362 for E2. These frequencies are comparable to those found in the control group where we obtained the following results: 0.875 for E3, 0.073 for E4, and 0.0515 for E2.
No statistically significant differences in ApoE genotypes were found between the patients and the control groups.
The distribution of the ApoE genotypes in the Gaza Strip population was similar to that of other Asian populations.
Our study indicated that there was no significant difference between the mean of cholesterol and TG levels of the CHD and control groups.
There was a significant difference between the mean of LDL and HDL levels between the CHD and the control group.
There was no significant difference between the mean of TGs and HDL between the different ApoE genotypes. However, there was a significant difference in the mean of LDL and ApoE genotypes.
Smoking, hypertension, and advanced age are important risk factors for CHD.
Acknowledgments
This work has been carried out in the medical technology laboratories at the Islamic University of Gaza, Palestine. We would like to thank the following people for their support, generous advices, and help: Professor Fadel A. Sharif, for his great knowledge and tremendous intelligence; Dr. Abbood Kishawee, for his support and help; Mr. Nasser Abu Shaaban, for his helpful and friendly support during the laboratory work. We would like to extend our thanks to the staff of the Medical Sciences Department, College of Science and Technology, Khan Younis, for their support and help, and to all the staff at Nasser Hospital (nurses, doctors, and lab technicians), specially of the Cardiology Department. We are deeply grateful to Dr. Amjad Al Shanty for his helpful guidance in the statistical analysis.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Hsieh WJ, Sing-Ka L, Ming-Shien W, Tsuen KJ. Characterization of apolipoprotein E genetic variations in Taiwanese: Association with coronary heart disease and plasma lipid levels. Hum Biol. 2002;5:1–3. doi: 10.1353/hub.2002.0012. [DOI] [PubMed] [Google Scholar]
- 2.Kuusisto J, Mykkänen L, Kervinen K, Kesäniemi YA, Laakso M. Apolipoprotein E4 phenotype is not an important risk factor for coronary heart disease or stroke in elderly subjects. Arterioscler Thromb Vasc Biol. 1995;15:1280–6. doi: 10.1161/01.atv.15.9.1280. [DOI] [PubMed] [Google Scholar]
- 3.Ghebranious N, Ivacic L, Mallum J, Dokken C. Detection of ApoE2, E3 and E4 alleles using maldi-tof mass spectrometry and the homogeneous mass-extend technology. Mole Diagn. 2005;33:1–6. doi: 10.1093/nar/gni155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendez AP, Mayeux R, Ngai C, Shea S, Berglund L. Association of ApoE polymorphism with plasma lipid levels in a multiethnic elderly population. Arterioscler Thromb Vasc Biol. 1997;17:3534–41. doi: 10.1161/01.atv.17.12.3534. [DOI] [PubMed] [Google Scholar]
- 5.Slooter AJ, Bots ML, Havekes LM, Iglesias AD, Cruts M, Grobbee DE, et al. Apolipoprotein E and carotid artery atherosclerosis. J Vasc Biol. 2001;32:19–47. doi: 10.1161/hs0901.095377. [DOI] [PubMed] [Google Scholar]
- 6.Lahoz C, Schaefer EJ, Cupples LA, Wilson PW, Levy D, Osgood D, et al. Apolipoprotein E genotype and cardiovascular disease in the Framingham Heart Study. Atherosclerosis. 2001;154:529–66. doi: 10.1016/s0021-9150(00)00570-0. [DOI] [PubMed] [Google Scholar]
- 7.Kataoka S, Robbins DC, Cowan LD, Go O, Yeh JL, Devereux RB, et al. Apolipoprotein E polymorphism in American Indians and its relation to plasma lipoproteins and diabetes. The Strong Heart Study. Arterioscler Throm Vasc Biol. 1996;16:918–34. doi: 10.1161/01.atv.16.8.918. [DOI] [PubMed] [Google Scholar]
- 8.Utermann G, Vogelberg H, Steinmetz A, Schoenborn W, Pruin N, Jaeschke M, et al. Polymorphism of apolipoprotein E, II: Genetics of hyperlipoproteinemia type III. Clin Genet. 1979;15:37–62. [PubMed] [Google Scholar]
- 9.Basun H, Corder EH, Guo Z, Lannfelt L, Corder LS, Manton KG, et al. Apolipoprotein E polymorphism and stroke in a population sample aged 75 years or more. Am Heart Assoc. 1996;27:1310–5. doi: 10.1161/01.str.27.8.1310. [DOI] [PubMed] [Google Scholar]
- 10.Zivelin A, Rosenberg N, Peretz H, Amit Y, Kornbrot , Seligsohn U. Improved method for genotyping apolipoprotein E polymorphisms by a PCR-based assay simultaneously utilizing two distinct restriction enzymes. Am Assoc Clin Chem. 1997;43:1657–9. [PubMed] [Google Scholar]
- 11.Mooijaart SP, Berbee JF, Heemst D, Havekes LM, Craen JM, Slagboom PE, et al. ApoE plasma levels and risk of cardiovascular mortality in old age: A peer reviewed open access. J Pub Lib Sci. 2006;6:1–20. doi: 10.1371/journal.pmed.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dzimiri N, Meyer FB, Hussain SS, Basco C, Afrane B, Halees Z. Relevance of apolipoprotein E polymorphism for coronary artery disease in the Saudi population. Biol Med Res Cardiovasc Dis. 1999;123:1241–5. doi: 10.5858/1999-123-1241-ROAEPF. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh WJ, Sing-Ka L, Ming-Shien W, Tsuen KJ. Characterization of apolipoprotein E genetic variations in Taiwanese: Association with coronary heart disease and plasma lipid levels. Hum Biol. 2002;5:1–3. doi: 10.1353/hub.2002.0012. [DOI] [PubMed] [Google Scholar]
- 14.Rossouw JE. Hormones, genetic factors, and gender differences in cardiovascular disease. Cardiovasc Res. 2002;53:550–7. doi: 10.1016/s0008-6363(01)00478-3. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein MS, Costanza MC, James RW, Morris MA, Cambien F, Raoux S, et al. Physical activity may modulate effects of ApoE genotype on lipid profile. Arterioscler Throm Vasc Biol. 2002;22:133–40. doi: 10.1161/hq0102.101819. [DOI] [PubMed] [Google Scholar]


