Abstract
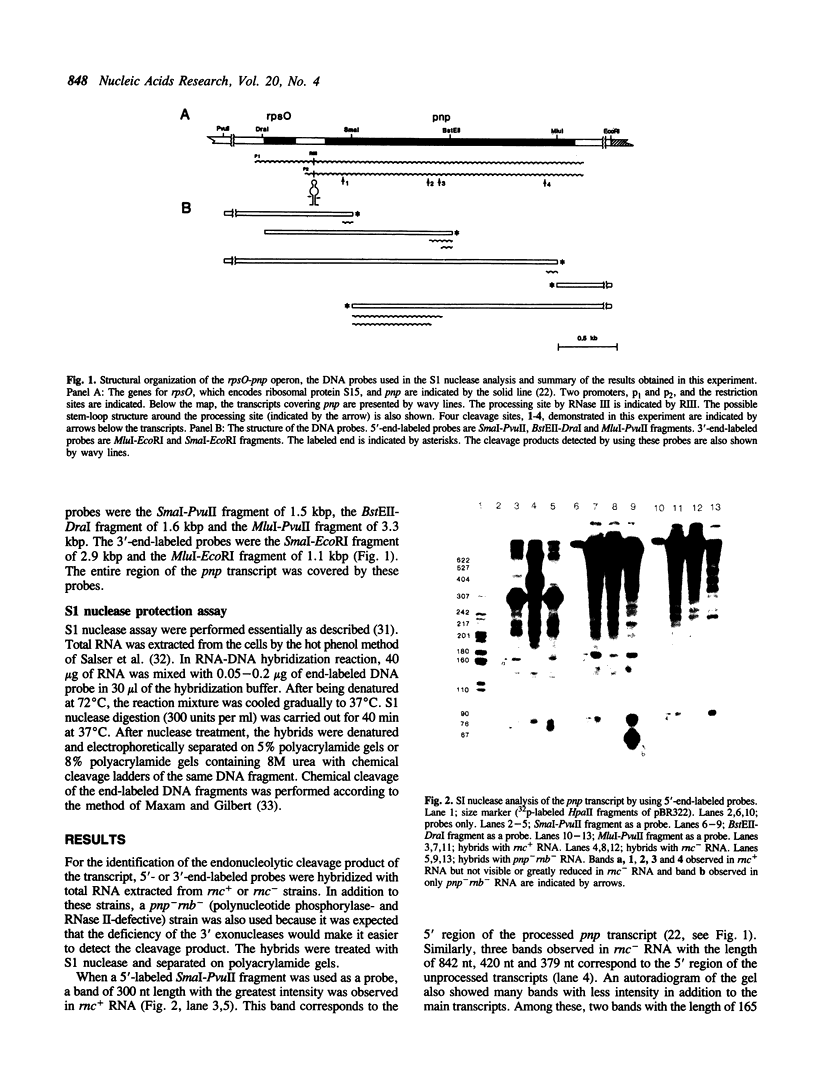
The primary transcript of pnp, the gene encoding polynucleotide phosphorylase in Escherichia coli, is processed in the 5' end region by ribonuclease III (RNase III). The unprocessed transcript shows enhanced stability compared with the processed transcript. We report here that, unlike the processed transcript, the unprocessed pnp transcript did not accept endonucleolytic attack at, at least, five cleavage sites. Sequencing analysis of the four cleavage products shows no sequence specific to all these sites, but AU rich stretches were observed at three sites.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apirion D., Gitelman D. R. Decay of RNA in RNA processing mutants of Escherichia coli. Mol Gen Genet. 1980 Jan;177(2):339–343. doi: 10.1007/BF00267448. [DOI] [PubMed] [Google Scholar]
- Apirion D., Watson N. Mapping and characterization of a mutation in Escherichia coli that reduces the level of ribonuclease III specific for double-stranded ribonucleic acid. J Bacteriol. 1975 Oct;124(1):317–324. doi: 10.1128/jb.124.1.317-324.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P., Kushner S. R. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell J. C., Régnier P., Chen S. M., Nakamura Y., Grunberg-Manago M., Court D. L. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989 Nov;8(11):3401–3407. doi: 10.1002/j.1460-2075.1989.tb08504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Blumer K. J., Steege D. A. mRNA processing in Escherichia coli: an activity encoded by the host processes bacteriophage f1 mRNAs. Nucleic Acids Res. 1984 Feb 24;12(4):1847–1861. doi: 10.1093/nar/12.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Z. F., Gross C. A., Watanabe K. K., Burgess R. R. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983 Feb;32(2):335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Purification and characterization of ribonuclease M and mRNA degradation in Escherichia coli. Eur J Biochem. 1989 May 1;181(2):363–370. doi: 10.1111/j.1432-1033.1989.tb14733.x. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Subbarao M. N., Kennell D. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J Mol Biol. 1986 Nov 20;192(2):257–274. doi: 10.1016/0022-2836(86)90363-3. [DOI] [PubMed] [Google Scholar]
- Claverie-Martin F., Diaz-Torres M. R., Yancey S. D., Kushner S. R. Cloning of the altered mRNA stability (ams) gene of Escherichia coli K-12. J Bacteriol. 1989 Oct;171(10):5479–5486. doi: 10.1128/jb.171.10.5479-5486.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory S. A., Belasco J. G. The ompA 5' untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J Bacteriol. 1990 Aug;172(8):4472–4481. doi: 10.1128/jb.172.8.4472-4481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski K., Roch J. M., Prentki P., Krisch H. M. The stability of bacteriophage T4 gene 32 mRNA: a 5' leader sequence that can stabilize mRNA transcripts. Cell. 1985 Dec;43(2 Pt 1):461–469. doi: 10.1016/0092-8674(85)90176-x. [DOI] [PubMed] [Google Scholar]
- Lundberg U., von Gabain A., Melefors O. Cleavages in the 5' region of the ompA and bla mRNA control stability: studies with an E. coli mutant altering mRNA stability and a novel endoribonuclease. EMBO J. 1990 Sep;9(9):2731–2741. doi: 10.1002/j.1460-2075.1990.tb07460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the product of the ams gene in vivo and in vitro. J Bacteriol. 1991 Apr;173(8):2488–2497. doi: 10.1128/jb.173.8.2488-2497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988 Mar 25;52(6):893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Carpousis A. J., Krisch H. M. Escherichia coli RNase E has a role in the decay of bacteriophage T4 mRNA. Genes Dev. 1990 May;4(5):873–881. doi: 10.1101/gad.4.5.873. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Krisch H. M., Higgins C. F. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990 Dec;4(12):2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Prentki P., Belin D., Krisch H. M. Processing of unstable bacteriophage T4 gene 32 mRNAs into a stable species requires Escherichia coli ribonuclease E. EMBO J. 1988 Nov;7(11):3601–3607. doi: 10.1002/j.1460-2075.1988.tb03238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Higgins C. F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987 Dec 24;51(6):1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature. 1984 Nov 1;312(5989):75–77. doi: 10.1038/312075a0. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Lundberg U., von Gabain A. In vivo and in vitro identity of site specific cleavages in the 5' non-coding region of ompA and bla mRNA in Escherichia coli. EMBO J. 1988 Jul;7(7):2269–2275. doi: 10.1002/j.1460-2075.1988.tb03067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S., Reeh S. Functional mRNA half lives in E. coli. Mol Gen Genet. 1978 Nov 9;166(3):329–336. doi: 10.1007/BF00267626. [DOI] [PubMed] [Google Scholar]
- Petersen C. Multiple determinants of functional mRNA stability: sequence alterations at either end of the lacZ gene affect the rate of mRNA inactivation. J Bacteriol. 1991 Apr;173(7):2167–2172. doi: 10.1128/jb.173.7.2167-2172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5' end. EMBO J. 1987 Jul;6(7):2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M. K., Singh B., Ray B. K., Apirion D. Maturation of 5-S rRNA: ribonuclease E cleavages and their dependence on precursor sequences. Eur J Biochem. 1983 Mar 1;131(1):119–127. doi: 10.1111/j.1432-1033.1983.tb07238.x. [DOI] [PubMed] [Google Scholar]
- Régnier P., Grunberg-Manago M., Portier C. Nucleotide sequence of the pnp gene of Escherichia coli encoding polynucleotide phosphorylase. Homology of the primary structure of the protein with the RNA-binding domain of ribosomal protein S1. J Biol Chem. 1987 Jan 5;262(1):63–68. [PubMed] [Google Scholar]
- Salser W., Gesteland R. F., Bolle A. In vitro synthesis of bacteriophage lysozyme. Nature. 1967 Aug 5;215(5101):588–591. doi: 10.1038/215588a0. [DOI] [PubMed] [Google Scholar]
- Schmeissner U., McKenney K., Rosenberg M., Court D. Removal of a terminator structure by RNA processing regulates int gene expression. J Mol Biol. 1984 Jun 15;176(1):39–53. doi: 10.1016/0022-2836(84)90381-4. [DOI] [PubMed] [Google Scholar]
- Takata R., Izuhara M., Hori K. Differential degradation of the Escherichia coli polynucleotide phosphorylase mRNA. Nucleic Acids Res. 1989 Sep 25;17(18):7441–7451. doi: 10.1093/nar/17.18.7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata R., Mukai T., Hori K. RNA processing by RNase III is involved in the synthesis of Escherichia coli polynucleotide phosphorylase. Mol Gen Genet. 1987 Aug;209(1):28–32. doi: 10.1007/BF00329832. [DOI] [PubMed] [Google Scholar]
- Tomcsányi T., Apirion D. Processing enzyme ribonuclease E specifically cleaves RNA I. An inhibitor of primer formation in plasmid DNA synthesis. J Mol Biol. 1985 Oct 20;185(4):713–720. doi: 10.1016/0022-2836(85)90056-7. [DOI] [PubMed] [Google Scholar]