Abstract
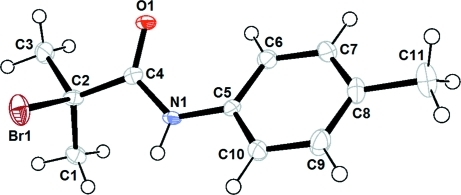
In the title molecule, C11H14BrNO, there is twist between the mean plane of the amide group and the benzene ring [C(=O)—N—C C torsion angle = −31.2 (5)°]. In the crystal, intermolecular N—H⋯O and weak C—H⋯O hydrogen bonds link molecules into chains along [100]. The methyl group H atoms are disordered over two sets of sites with equal occupancy.
Related literature
For initiators in ATRP processes (polymerization by atom transfer radical), see: Matyjaszewski & Xia (2001 ▶); Kato et al. (1995 ▶); Pietrasik & Tsarevsky (2010 ▶). For a related structure, see: Moreno-Fuquen et al. (2011 ▶). For hydrogen-bond graph sets, see: Etter (1990 ▶).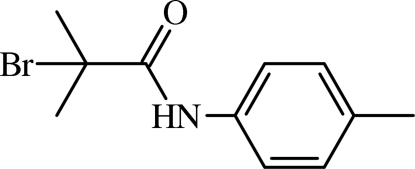
Experimental
Crystal data
C11H14BrNO
M r = 256.14
Orthorhombic,
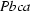
a = 10.0728 (4) Å
b = 11.2577 (4) Å
c = 20.3670 (6) Å
V = 2309.55 (14) Å3
Z = 8
Mo Kα radiation
μ = 3.53 mm−1
T = 123 K
0.25 × 0.12 × 0.05 mm
Data collection
Oxford Diffraction Xcalibur E diffractometer
Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2009 ▶) T min = 0.751, T max = 1.000
10140 measured reflections
2762 independent reflections
1819 reflections with I > 2σ(I)
R int = 0.049
Refinement
R[F 2 > 2σ(F 2)] = 0.045
wR(F 2) = 0.093
S = 1.06
2762 reflections
133 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.58 e Å−3
Δρmin = −0.46 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2009 ▶); cell refinement: CrysAlis CCD; data reduction: CrysAlis CCD; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PARST (Nardelli, 1995 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811019337/lh5254sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811019337/lh5254Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811019337/lh5254Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O1i | 0.84 (2) | 2.13 (2) | 2.937 (3) | 161 (3) |
| C1—H1C⋯O1i | 0.98 | 2.44 | 3.382 (4) | 162 |
| C10—H10⋯O1i | 0.95 | 2.57 | 3.321 (4) | 137 |
Symmetry code: (i) 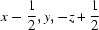 .
.
Acknowledgments
RMF is grateful to the Spanish Research Council (CSIC) for the use of a free-of-charge licence to the Cambridge Structural Database (Allen, 2002 ▶). RMF and FZ also thank the Universidad del Valle, Colombia, and the Instituto de Química de São Carlos, USP, Brazil, for partial financial support.
supplementary crystallographic information
Comment
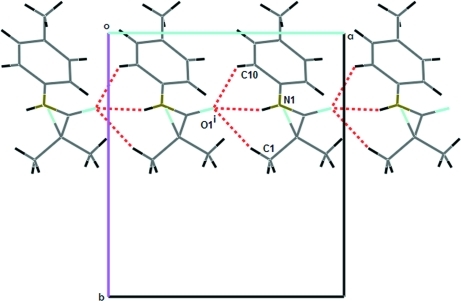
The ATRP process (polymerization by atom transfer radical) allows control of the composition and functionality in polymerization reactions (Pietrasik & Tsarevsky, 2010). The use of functional initiators in these reactions allows the synthesis of new materials. Most initiators for ATRP processes are alkyl halides (Matyjaszewski & Xia, 2001; Kato et al., 1995). As part of our work related to functional initiators in polymerization processes (Moreno-Fuquen et al., 2011) we have determined the crystal structure of the title compound (I). The molecular structure of (I) is shown in Fig. 1. There is a twist between the mean plane of the amide group and benzene ring giving a C4—N1—C5—C6 torsion angle of -31.2 (5) °. In the crystal, intermolecular N—H···O and weak C—H···O hydrogen bonds link molecules into one-dimensional chains along [100] incorporating C(4) graph motifs (Etter, 1990) (see Table 1 and Fig. 2).
Experimental
The initial reagents were purchased from Aldrich Chemical Co. and were used as received. In a 100mL round bottom flask 4-methylaniline (3.173 mmoles, 0.340 g), triethylamine (0.635 mmol, 0.064 g) were mixed, then a solution of 2-bromo isobutiryl bromide (0.685 g) in anhydrous THF (5 ml) was added drop wise, under an argon stream. The reaction was carried out in a dry bag overnight under magnetic stirring. The solid was filtered off and dichloromethane (20 ml) added to the organic phase which was washed with brine (50 ml) followed by water (10 ml). The solution was concentrated at low pressure affording colourless crystals and recrystalized from a solution of hexane and ethyl acetate (80:20). M.p. 364 (1) K.
Refinement
The H-atoms were placed geometrically with C—H= 0.95 Å for aromatic, C—H = 0.98 Å for methyl and Uiso(H) 1.2 and 1.5 times Ueq of the parent atom respectively. The methyl group H atoms are disordered over two sets of sites with equal occupancy.
Figures
Fig. 1.
An ORTEP-3 (Farrugia, 1997) plot of (I) with displacement ellipsoids drawn at the 50% probability level. H atoms are shown as spheres of arbitary radius.
Fig. 2.
Part of the crystal structure of (I), showing the formation of a one dimensional chain along [100]. Symmetry code: (i) x-1/2,+y,-z+1/2.
Crystal data
| C11H14BrNO | Dx = 1.473 Mg m−3 |
| Mr = 256.14 | Melting point: 385(1) K |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 2266 reflections |
| a = 10.0728 (4) Å | θ = 3.4–29.5° |
| b = 11.2577 (4) Å | µ = 3.53 mm−1 |
| c = 20.3670 (6) Å | T = 123 K |
| V = 2309.55 (14) Å3 | Tablet, colourless |
| Z = 8 | 0.25 × 0.12 × 0.05 mm |
| F(000) = 1040 |
Data collection
| Oxford Diffraction Xcalibur E diffractometer | 2762 independent reflections |
| Radiation source: fine-focus sealed tube | 1819 reflections with I > 2σ(I) |
| graphite | Rint = 0.049 |
| ω scans | θmax = 28.0°, θmin = 3.4° |
| Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2009) | h = −12→13 |
| Tmin = 0.751, Tmax = 1.000 | k = −10→14 |
| 10140 measured reflections | l = −26→22 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.045 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.093 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0316P)2 + 0.7321P] where P = (Fo2 + 2Fc2)/3 |
| 2762 reflections | (Δ/σ)max < 0.001 |
| 133 parameters | Δρmax = 0.58 e Å−3 |
| 0 restraints | Δρmin = −0.46 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Br1 | 0.74332 (3) | 0.28652 (3) | 0.384590 (15) | 0.02982 (13) | |
| O1 | 0.94616 (17) | 0.2851 (2) | 0.24129 (10) | 0.0200 (5) | |
| N1 | 0.7262 (2) | 0.2730 (3) | 0.21911 (12) | 0.0158 (6) | |
| C1 | 0.6864 (3) | 0.4811 (3) | 0.30022 (16) | 0.0214 (7) | |
| H1A | 0.7056 | 0.5288 | 0.2611 | 0.032* | |
| H1B | 0.6768 | 0.5335 | 0.3384 | 0.032* | |
| H1C | 0.6037 | 0.4368 | 0.2935 | 0.032* | |
| C2 | 0.7996 (3) | 0.3946 (3) | 0.31211 (15) | 0.0173 (7) | |
| C3 | 0.9232 (3) | 0.4587 (3) | 0.33541 (16) | 0.0287 (9) | |
| H3A | 0.9914 | 0.4004 | 0.3472 | 0.043* | |
| H3B | 0.9015 | 0.5073 | 0.3739 | 0.043* | |
| H3C | 0.9565 | 0.5100 | 0.3002 | 0.043* | |
| C4 | 0.8312 (3) | 0.3116 (3) | 0.25448 (14) | 0.0158 (7) | |
| C5 | 0.7307 (2) | 0.1928 (3) | 0.16557 (14) | 0.0160 (7) | |
| C6 | 0.8384 (3) | 0.1847 (3) | 0.12261 (14) | 0.0203 (7) | |
| H6 | 0.9135 | 0.2348 | 0.1281 | 0.024* | |
| C7 | 0.8347 (3) | 0.1030 (3) | 0.07213 (15) | 0.0241 (8) | |
| H7 | 0.9082 | 0.0986 | 0.0431 | 0.029* | |
| C8 | 0.7285 (3) | 0.0274 (3) | 0.06198 (15) | 0.0229 (8) | |
| C9 | 0.6210 (3) | 0.0383 (3) | 0.10438 (16) | 0.0273 (8) | |
| H9 | 0.5457 | −0.0113 | 0.0985 | 0.033* | |
| C10 | 0.6216 (3) | 0.1202 (3) | 0.15499 (15) | 0.0225 (8) | |
| H10 | 0.5463 | 0.1267 | 0.1829 | 0.027* | |
| C11 | 0.7287 (3) | −0.0636 (4) | 0.00758 (17) | 0.0338 (9) | |
| H11A | 0.6452 | −0.1082 | 0.0085 | 0.051* | 0.50 |
| H11B | 0.8033 | −0.1184 | 0.0138 | 0.051* | 0.50 |
| H11C | 0.7378 | −0.0233 | −0.0348 | 0.051* | 0.50 |
| H11D | 0.8123 | −0.0584 | −0.0168 | 0.051* | 0.50 |
| H11E | 0.6542 | −0.0482 | −0.0222 | 0.051* | 0.50 |
| H11F | 0.7197 | −0.1433 | 0.0264 | 0.051* | 0.50 |
| H1N | 0.652 (2) | 0.292 (3) | 0.2342 (15) | 0.032 (10)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0475 (2) | 0.0235 (2) | 0.01844 (18) | 0.00201 (17) | 0.00410 (15) | 0.00302 (15) |
| O1 | 0.0101 (9) | 0.0260 (14) | 0.0240 (12) | 0.0013 (10) | −0.0012 (8) | −0.0053 (11) |
| N1 | 0.0086 (12) | 0.0198 (15) | 0.0191 (13) | 0.0006 (11) | 0.0013 (9) | −0.0026 (11) |
| C1 | 0.0213 (16) | 0.017 (2) | 0.0254 (18) | 0.0021 (13) | 0.0028 (14) | −0.0020 (15) |
| C2 | 0.0131 (13) | 0.0159 (19) | 0.0228 (17) | −0.0020 (12) | 0.0012 (12) | 0.0001 (15) |
| C3 | 0.0218 (16) | 0.037 (3) | 0.0269 (19) | −0.0069 (16) | −0.0011 (14) | −0.0152 (18) |
| C4 | 0.0163 (14) | 0.0137 (18) | 0.0175 (15) | −0.0003 (12) | −0.0010 (12) | 0.0051 (14) |
| C5 | 0.0137 (14) | 0.0189 (18) | 0.0153 (15) | 0.0014 (13) | −0.0025 (11) | 0.0010 (13) |
| C6 | 0.0172 (14) | 0.025 (2) | 0.0182 (16) | −0.0014 (13) | 0.0012 (12) | 0.0021 (15) |
| C7 | 0.0217 (16) | 0.034 (2) | 0.0165 (17) | 0.0033 (15) | 0.0025 (13) | −0.0009 (16) |
| C8 | 0.0334 (19) | 0.020 (2) | 0.0155 (16) | 0.0022 (15) | −0.0021 (14) | −0.0022 (14) |
| C9 | 0.0275 (17) | 0.031 (2) | 0.0230 (18) | −0.0104 (15) | −0.0014 (14) | −0.0048 (16) |
| C10 | 0.0182 (15) | 0.030 (2) | 0.0192 (17) | −0.0032 (14) | 0.0029 (13) | −0.0042 (16) |
| C11 | 0.051 (2) | 0.029 (2) | 0.0213 (18) | −0.0049 (17) | 0.0051 (16) | −0.0064 (16) |
Geometric parameters (Å, °)
| Br1—C2 | 1.995 (3) | C6—C7 | 1.380 (4) |
| O1—C4 | 1.226 (3) | C6—H6 | 0.9500 |
| N1—C4 | 1.352 (4) | C7—C8 | 1.382 (4) |
| N1—C5 | 1.416 (4) | C7—H7 | 0.9500 |
| N1—H1N | 0.840 (17) | C8—C9 | 1.391 (4) |
| C1—C2 | 1.519 (4) | C8—C11 | 1.509 (5) |
| C1—H1A | 0.9800 | C9—C10 | 1.382 (4) |
| C1—H1B | 0.9800 | C9—H9 | 0.9500 |
| C1—H1C | 0.9800 | C10—H10 | 0.9500 |
| C2—C3 | 1.515 (4) | C11—H11A | 0.9800 |
| C2—C4 | 1.533 (4) | C11—H11B | 0.9800 |
| C3—H3A | 0.9800 | C11—H11C | 0.9800 |
| C3—H3B | 0.9800 | C11—H11D | 0.9800 |
| C3—H3C | 0.9800 | C11—H11E | 0.9800 |
| C5—C10 | 1.387 (4) | C11—H11F | 0.9800 |
| C5—C6 | 1.397 (4) | ||
| C4—N1—C5 | 126.2 (2) | C8—C7—H7 | 118.5 |
| C4—N1—H1N | 115 (2) | C7—C8—C9 | 117.1 (3) |
| C5—N1—H1N | 118 (2) | C7—C8—C11 | 121.8 (3) |
| C2—C1—H1A | 109.5 | C9—C8—C11 | 121.1 (3) |
| C2—C1—H1B | 109.5 | C10—C9—C8 | 121.2 (3) |
| H1A—C1—H1B | 109.5 | C10—C9—H9 | 119.4 |
| C2—C1—H1C | 109.5 | C8—C9—H9 | 119.4 |
| H1A—C1—H1C | 109.5 | C9—C10—C5 | 120.8 (3) |
| H1B—C1—H1C | 109.5 | C9—C10—H10 | 119.6 |
| C3—C2—C1 | 111.2 (3) | C5—C10—H10 | 119.6 |
| C3—C2—C4 | 111.1 (2) | C8—C11—H11A | 109.5 |
| C1—C2—C4 | 115.1 (2) | C8—C11—H11B | 109.5 |
| C3—C2—Br1 | 107.0 (2) | H11A—C11—H11B | 109.5 |
| C1—C2—Br1 | 107.20 (19) | C8—C11—H11C | 109.5 |
| C4—C2—Br1 | 104.7 (2) | H11A—C11—H11C | 109.5 |
| C2—C3—H3A | 109.5 | H11B—C11—H11C | 109.5 |
| C2—C3—H3B | 109.5 | C8—C11—H11D | 109.5 |
| H3A—C3—H3B | 109.5 | H11A—C11—H11D | 141.1 |
| C2—C3—H3C | 109.5 | H11B—C11—H11D | 56.3 |
| H3A—C3—H3C | 109.5 | H11C—C11—H11D | 56.3 |
| H3B—C3—H3C | 109.5 | C8—C11—H11E | 109.5 |
| O1—C4—N1 | 123.0 (3) | H11A—C11—H11E | 56.3 |
| O1—C4—C2 | 120.8 (3) | H11B—C11—H11E | 141.1 |
| N1—C4—C2 | 116.2 (2) | H11C—C11—H11E | 56.3 |
| C10—C5—C6 | 118.7 (3) | H11D—C11—H11E | 109.5 |
| C10—C5—N1 | 118.1 (3) | C8—C11—H11F | 109.5 |
| C6—C5—N1 | 123.3 (3) | H11A—C11—H11F | 56.3 |
| C7—C6—C5 | 119.3 (3) | H11B—C11—H11F | 56.3 |
| C7—C6—H6 | 120.4 | H11C—C11—H11F | 141.1 |
| C5—C6—H6 | 120.4 | H11D—C11—H11F | 109.5 |
| C6—C7—C8 | 122.9 (3) | H11E—C11—H11F | 109.5 |
| C6—C7—H7 | 118.5 | ||
| C5—N1—C4—O1 | 4.0 (5) | C10—C5—C6—C7 | −1.6 (5) |
| C5—N1—C4—C2 | −177.5 (3) | N1—C5—C6—C7 | 179.3 (3) |
| C3—C2—C4—O1 | 14.6 (4) | C5—C6—C7—C8 | −0.4 (5) |
| C1—C2—C4—O1 | 142.0 (3) | C6—C7—C8—C9 | 1.6 (5) |
| Br1—C2—C4—O1 | −100.5 (3) | C6—C7—C8—C11 | −178.2 (3) |
| C3—C2—C4—N1 | −163.9 (3) | C7—C8—C9—C10 | −1.0 (5) |
| C1—C2—C4—N1 | −36.5 (4) | C11—C8—C9—C10 | 178.8 (3) |
| Br1—C2—C4—N1 | 81.0 (3) | C8—C9—C10—C5 | −0.9 (5) |
| C4—N1—C5—C10 | 149.7 (3) | C6—C5—C10—C9 | 2.2 (5) |
| C4—N1—C5—C6 | −31.2 (5) | N1—C5—C10—C9 | −178.6 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1i | 0.84 (2) | 2.13 (2) | 2.937 (3) | 161 (3) |
| C1—H1C···O1i | 0.98 | 2.44 | 3.382 (4) | 162 |
| C10—H10···O1i | 0.95 | 2.57 | 3.321 (4) | 137 |
Symmetry codes: (i) x−1/2, y, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LH5254).
References
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Etter, M. (1990). Acc. Chem. Res. 23, 120–126.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Kato, M., Kamigaito, M., Sawamoto, M. & Higashimura, T. (1995). Macromolecules, 28, 1721–1723.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Matyjaszewski, K. & Xia, J. (2001). Chem. Rev. 101, 2921–2990. [DOI] [PubMed]
- Moreno-Fuquen, R., Quintero, D. E., Zuluaga, F., Haiduke, R. L. A. & Kennedy, A. R. (2011). Acta Cryst. E67, o659. [DOI] [PMC free article] [PubMed]
- Nardelli, M. (1995). J. Appl. Cryst. 28, 659.
- Oxford Diffraction (2009). CrysAlis CCD and CrysAlis RED Oxford Diffraction Ltd, Yarnton, England.
- Pietrasik, J. & Tsarevsky, N. V. (2010). Eur. Polym. J. 46, 2333–2340.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811019337/lh5254sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811019337/lh5254Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811019337/lh5254Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report