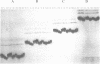
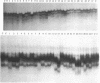
Abstract
Single-strand conformation polymorphism (SSCP) is a simple method for detecting the presence of mutations in a segment of DNA, but the fraction of all mutations detected is unclear. We have evaluated SSCP for the detection of single-base mutations in the factor IX gene. Multiple conditions were examined including electrophoresis temperature, electrophoresis buffer concentration, acrylamide to bis-acrylamide ratio, and water-cooled versus fan-cooled gel apparatuses. Depending on conditions, 10-11 of 12 known mutations were detected in a 183 bp segment whereas only 11-14 of 22 known mutations were detected in a 307 bp segment. We hypothesized that single stranded RNA should have a larger repertoire of secondary structure because shorter hairpins form stable duplexes and the 2' hydroxyl group is available for sugar-base and sugar-sugar hydrogen bonds. By incorporating phage promoter sequences into PCR primers, RNA-SSCP (rSSCP) could be compared directly with standard DNA SSCP. rSSCP was generally superior to SSCP, especially for the 307 bp segment. In addition, the abundance of transcript produced as a result of rSSCP allows the rapid, nonradioactive detection of mutations by staining the gel with ethidium bromide. To gauge the utility of the method in a prospective manner, a blinded study was performed in which SSCP, rSSCP, and direct genomic sequencing were compared in 28 patients with hemophilia B. A total of 2.6 kb of factor IX genomic sequence was examined in nine regions ranging from 180 to 497 nucleotides of factor IX sequence. Sequence changes at 20 different sites were detected by direct genomic sequencing; 70% of these were detected by rSSCP while only 35% were detected by SSCP.
Full text
PDF


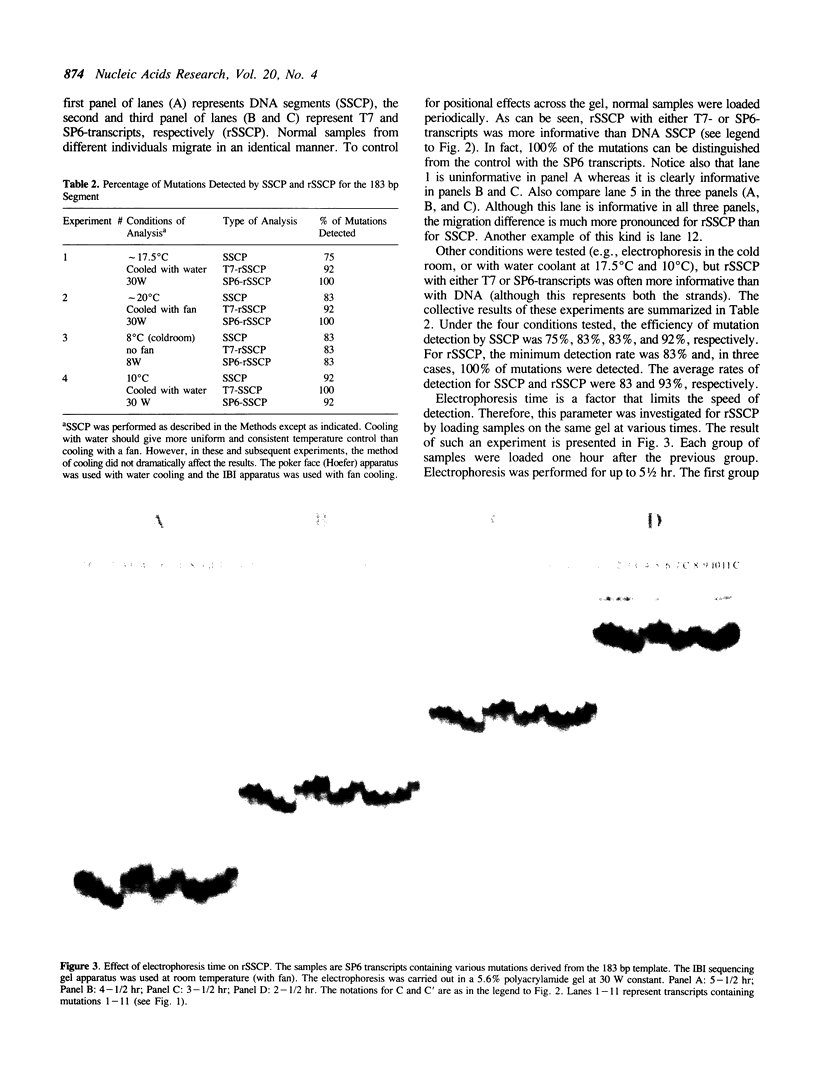

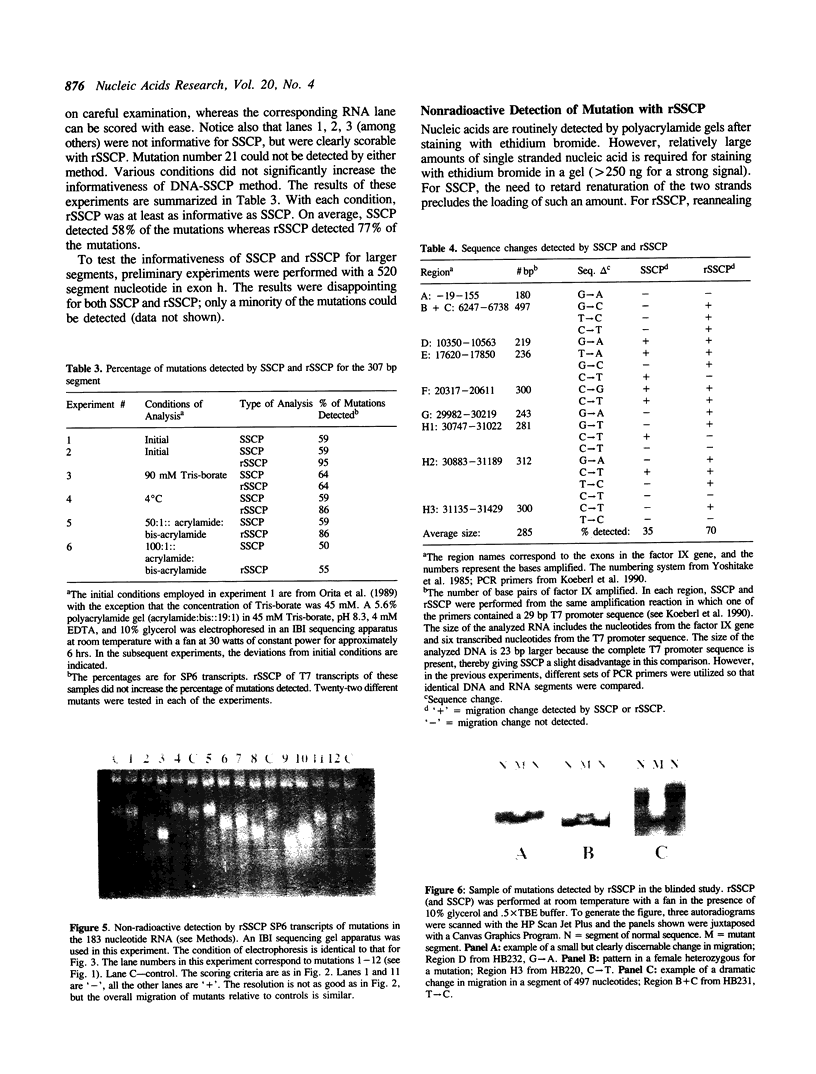


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ainsworth P. J., Surh L. C., Coulter-Mackie M. B. Diagnostic single strand conformational polymorphism, (SSCP): a simplified non-radioisotopic method as applied to a Tay-Sachs B1 variant. Nucleic Acids Res. 1991 Jan 25;19(2):405–406. doi: 10.1093/nar/19.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolos A. M., Dean M., Lucas-Derse S., Ramsburg M., Brown G. L., Goldman D. Population and pedigree studies reveal a lack of association between the dopamine D2 receptor gene and alcoholism. JAMA. 1990 Dec 26;264(24):3156–3160. [PubMed] [Google Scholar]
- Bottema C. D., Koeberl D. D., Ketterling R. P., Bowie E. J., Taylor S. A., Lillicrap D., Shapiro A., Gilchrist G., Sommer S. S. A past mutation at isoleucine 397 is now a common cause of moderate/mild haemophilia B. Br J Haematol. 1990 Jun;75(2):212–216. doi: 10.1111/j.1365-2141.1990.tb02651.x. [DOI] [PubMed] [Google Scholar]
- Bottema C. D., Koeberl D. D., Sommer S. S. Direct carrier testing in 14 families with haemophilia B. Lancet. 1989 Sep 2;2(8662):526–529. doi: 10.1016/s0140-6736(89)90653-3. [DOI] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., Prockop D. J. Detection of single-base mutations by reaction of DNA heteroduplexes with a water-soluble carbodiimide followed by primer extension: application to products from the polymerase chain reaction. Nucleic Acids Res. 1990 Jul 11;18(13):3933–3939. doi: 10.1093/nar/18.13.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., Rooney J. E., Hosomi S., Zeiger A. R., Prockop D. J. Detection and location of single-base mutations in large DNA fragments by immunomicroscopy. Genomics. 1989 May;4(4):530–538. doi: 10.1016/0888-7543(89)90276-0. [DOI] [PubMed] [Google Scholar]
- Koeberl D. D., Bottema C. D., Buerstedde J. M., Sommer S. S. Functionally important regions of the factor IX gene have a low rate of polymorphism and a high rate of mutation in the dinucleotide CpG. Am J Hum Genet. 1989 Sep;45(3):448–457. [PMC free article] [PubMed] [Google Scholar]
- Koeberl D. D., Bottema C. D., Ketterling R. P., Bridge P. J., Lillicrap D. P., Sommer S. S. Mutations causing hemophilia B: direct estimate of the underlying rates of spontaneous germ-line transitions, transversions, and deletions in a human gene. Am J Hum Genet. 1990 Aug;47(2):202–217. [PMC free article] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Hayashi K., Sekiya T. Detection of aberrations of the p53 alleles and the gene transcript in human tumor cell lines by single-strand conformation polymorphism analysis. Cancer Res. 1991 Jul 1;51(13):3356–3361. [PubMed] [Google Scholar]
- Myers R. M., Lumelsky N., Lerman L. S., Maniatis T. Detection of single base substitutions in total genomic DNA. Nature. 1985 Feb 7;313(6002):495–498. doi: 10.1038/313495a0. [DOI] [PubMed] [Google Scholar]
- Orita M., Sekiya T., Hayashi K. DNA sequence polymorphisms in Alu repeats. Genomics. 1990 Oct;8(2):271–278. doi: 10.1016/0888-7543(90)90282-y. [DOI] [PubMed] [Google Scholar]
- Orita M., Suzuki Y., Sekiya T., Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989 Nov;5(4):874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- Poduslo S. E., Dean M., Kolch U., O'Brien S. J. Detecting high-resolution polymorphisms in human coding loci by combining PCR and single-strand conformation polymorphism (SSCP) analysis. Am J Hum Genet. 1991 Jul;49(1):106–111. [PMC free article] [PubMed] [Google Scholar]
- Sarkar G., Koeberl D. D., Sommer S. S. Direct sequencing of the activation peptide and the catalytic domain of the factor IX gene in six species. Genomics. 1990 Jan;6(1):133–143. doi: 10.1016/0888-7543(90)90458-7. [DOI] [PubMed] [Google Scholar]
- Sarkar G., Sommer S. S. Access to a messenger RNA sequence or its protein product is not limited by tissue or species specificity. Science. 1989 Apr 21;244(4902):331–334. doi: 10.1126/science.2565599. [DOI] [PubMed] [Google Scholar]
- Yoshitake S., Schach B. G., Foster D. C., Davie E. W., Kurachi K. Nucleotide sequence of the gene for human factor IX (antihemophilic factor B). Biochemistry. 1985 Jul 2;24(14):3736–3750. doi: 10.1021/bi00335a049. [DOI] [PubMed] [Google Scholar]