Abstract
The title compound, C7H6O5·H2O, is a new polymorph of the structures reported by Jiang et al. (2000 ▶) [Acta Cryst. C56, 594–595] and Okabe et al. (2001 ▶) [Acta Cryst. E57, o764–o766]. The gallic acid molecule is essentially planar (r.m.s. deviation = 0.550 Å). An intramolecular O—H⋯O hydrogen bond occurs in the gallic acid molecule, which is linked to the water molecule by a further O—H⋯O hydrogen bond. In the crystal, the components are linked by O—H⋯O hydrogen bonds. The hydrogen-bonding pattern differs from those reported for the previous polymorphs.
Related literature
For the biological activity of gallic acid, see: Lu et al. (2006 ▶); Madlener et al. (2007 ▶). For the previously reproted polymorphs, see: Jiang et al. (2000 ▶); Okabe et al. (2001 ▶). For a related structure, see: Genç et al. (2004 ▶).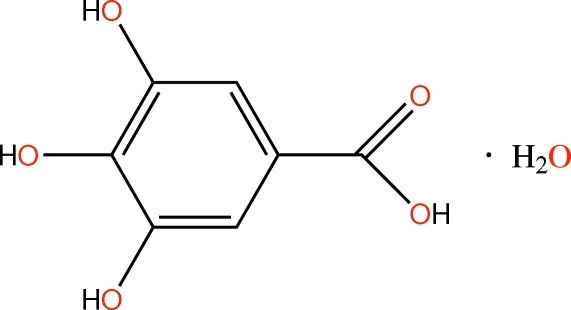
Experimental
Crystal data
C7H6O5·H2O
M r = 188.13
Monoclinic,
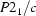
a = 9.7943 (7) Å
b = 3.6122 (2) Å
c = 21.5905 (15) Å
β = 91.268 (6)°
V = 763.66 (9) Å3
Z = 4
Mo Kα radiation
μ = 0.15 mm−1
T = 296 K
0.61 × 0.28 × 0.09 mm
Data collection
Stoe IPDS 2 diffractometer
Absorption correction: integration (X-RED32; Stoe & Cie, 2002 ▶) T min = 0.948, T max = 0.986
4558 measured reflections
1502 independent reflections
1262 reflections with I > 2σ(I)
R int = 0.042
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.104
S = 1.04
1502 reflections
127 parameters
3 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.15 e Å−3
Δρmin = −0.23 e Å−3
Data collection: X-AREA (Stoe & Cie, 2002 ▶); cell refinement: X-AREA; data reduction: X-RED32 (Stoe & Cie, 2002 ▶); program(s) used to solve structure: WinGX (Farrugia, 1997 ▶) and SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811018848/bx2352sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811018848/bx2352Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811018848/bx2352Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H3⋯O3 | 0.82 | 2.26 | 2.7006 (18) | 114 |
| O5—H6⋯O6 | 0.82 | 1.85 | 2.6542 (17) | 167 |
| O1—H2⋯O6i | 0.82 | 2.03 | 2.7539 (19) | 147 |
| O2—H3⋯O3ii | 0.82 | 2.52 | 3.1721 (19) | 137 |
| O3—H4⋯O4iii | 0.82 | 1.94 | 2.7154 (19) | 158 |
| O6—H6A⋯O2iv | 0.83 (2) | 2.03 (2) | 2.8237 (19) | 161 (3) |
| O6—H6B⋯O1v | 0.82 (2) | 2.00 (2) | 2.814 (2) | 171 (5) |
Symmetry codes: (i) 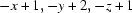 ; (ii)
; (ii) 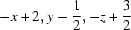 ; (iii)
; (iii) 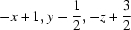 ; (iv)
; (iv) 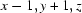 ; (v)
; (v) 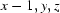 .
.
Table 2. Other crystal structures of gallic acid monohydrate (Å, °).
Acknowledgments
The authors thank the Ondokuz Mayıs University Research Fund for financial support of this project (project No. PYO.FEN.1904.09.006).
supplementary crystallographic information
Comment
Gallic acid and its derivatives are a group of naturally occurring polyphenol antioxidants which have recently been shown to have potential healty effects as an excellent free radical scavenger (Lu et al., 2006). In adition, gallic acid block DNA synthesis in leukemia cells (Madlener et al., 2007). Up to now, the different crystal structure of gallic acid monohidrate form have been reported by (Jiang et al., 2000; Okabe et al., 2001). The unit-cell parameters belong to these crystal structures have given at Table 2.
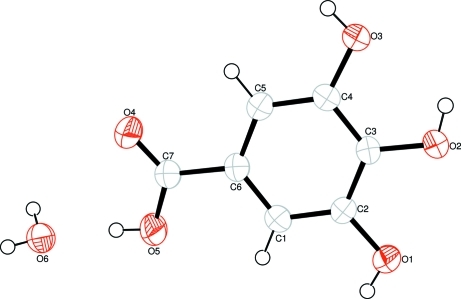
We report here the crystal structure of the title compound, C7H6O5.H2O. The asymmetric unit of title compound, C7H6O5.H2O, consists of one 3,4,5-trihydroxybenzoic acid molecule and one water molecule (Fig. 1). The C—C bond distances range from 1.378 (2) Å to 1.480 (2) Å. The longest C—C bond distance is between C6 and C7 with 1.480 Å. The C—O bond distance are range from 1.212 (2) Å to 1.3723 (19) Å. The shortest C—O bond distance is between C7 and O4 with 1.212 (2) Å. The C—O bond distance for different crystal structure was given 1.359 (2) Å by (Genç et al., 2004).
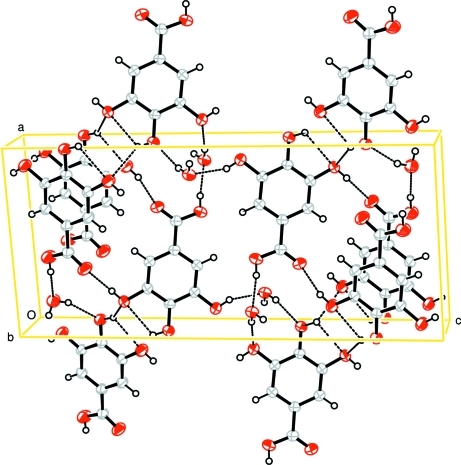
The crystal structure has two intramolecular hydrogen bonds and five intermolecular hydrogen bonds (Table 1). The O2—H3···O3 and O5—H6···O6 hydrogen bonds present in the asymmetric unit. In the crystal structure, the gallic acid molecules and water molecules are linked by the O6—H6A···O2, O6—H6B···O1, O5—H6···O6 and O1—H2···O6 hydrogen bonds. While the C—H···O hydrogen bond is present in crystal structure reported by (Jiang et al., 2000), there isn't present in our crystal structure. The via hydrogen atom of water molecule makes bifurcated hydrogen bond in crystal structure reported by (Jiang et al., 2000),but this kind hydrogen bond is not present in our crystal structure. In crystal structure reported by (Okabe et al., 2001), the hydroxyl groups make two intramolecular hydrogen bonds.
The torsion angles at ring belong to gallic acid molecule are range from 0.0 (3)° to 1.9 (3)°. Therefore, the gallic acid molecule close to planar. The tosion angles for the C2—C1—C6—C7 and C4—C5—C6—C7 are 179.51 (16)° and -179.08 (17)°, respectively. The O4—C7—O5 angle is 122.82 (15)°.
Experimental
A hot solution (60 °C) of gallic acid (0.002 mol, 0.340 g) in distilled water (approximately 20 ml) was gradually added to a hot stirring solution of magnesium sulfate heptahydrate (MgSO4.7H2O) (0.001 mol, 0.246 g) in distilled water (approximately 20 ml). The obtained mixture was stirred on a hot plate with slow evaporation to 30 ml at 60° C. The mixture was left for crystallization and in a few days, the crystals were obtained by slow evaporation from the solution at room temperature.
Refinement
H6A and H6B atoms were located in a difference map and were refined isotropically, with O—H distances in the range 0.830 (18)-0.824 (19) Å, and the H···H distance of 1.320 (2) Å. The other H atoms were positioned geometrically and refined using a riding model, with C—H = 0.930 Å, O—H = 0.820 Å, and Uiso(H) = 1.2Ueq(C) and 1.5Ueq(O).
Figures
Fig. 1.
The asymmetric unit of the title compound, showing the atomic numbering scheme. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
Packing diagram for (I), showing the three-dimensional hydrogen bonding array. Hydrogen bonds are shown as dashed lines.
Crystal data
| C7H6O5·H2O | F(000) = 392 |
| Mr = 188.13 | Dx = 1.636 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 6472 reflections |
| a = 9.7943 (7) Å | θ = 1.9–27.6° |
| b = 3.6122 (2) Å | µ = 0.15 mm−1 |
| c = 21.5905 (15) Å | T = 296 K |
| β = 91.268 (6)° | Needle, pale brown |
| V = 763.66 (9) Å3 | 0.61 × 0.28 × 0.09 mm |
| Z = 4 |
Data collection
| Stoe IPDS 2 diffractometer | 1502 independent reflections |
| Radiation source: fine-focus sealed tube | 1262 reflections with I > 2σ(I) |
| graphite | Rint = 0.042 |
| w–scan rotation | θmax = 26.0°, θmin = 1.9° |
| Absorption correction: integration (X-RED32; Stoe & Cie, 2002) | h = −9→12 |
| Tmin = 0.948, Tmax = 0.986 | k = −4→4 |
| 4558 measured reflections | l = −26→26 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.039 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.104 | w = 1/[σ2(Fo2) + (0.0466P)2 + 0.2493P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max < 0.001 |
| 1502 reflections | Δρmax = 0.15 e Å−3 |
| 127 parameters | Δρmin = −0.23 e Å−3 |
| 3 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.023 (5) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.67337 (16) | 0.6332 (5) | 0.58435 (8) | 0.0369 (4) | |
| H1 | 0.6383 | 0.7058 | 0.5459 | 0.044* | |
| C2 | 0.81149 (16) | 0.5670 (5) | 0.59282 (8) | 0.0349 (4) | |
| C3 | 0.86369 (15) | 0.4587 (5) | 0.65029 (8) | 0.0356 (4) | |
| C4 | 0.77763 (17) | 0.4238 (5) | 0.70009 (8) | 0.0395 (4) | |
| C5 | 0.63957 (17) | 0.4863 (5) | 0.69214 (8) | 0.0414 (4) | |
| H5 | 0.5814 | 0.4596 | 0.7253 | 0.050* | |
| C6 | 0.58767 (16) | 0.5895 (5) | 0.63421 (8) | 0.0370 (4) | |
| C7 | 0.43925 (16) | 0.6603 (5) | 0.62739 (8) | 0.0408 (4) | |
| O1 | 0.90229 (12) | 0.5986 (4) | 0.54561 (6) | 0.0480 (4) | |
| H2 | 0.8613 | 0.6599 | 0.5137 | 0.072* | |
| O2 | 1.00108 (11) | 0.3881 (4) | 0.65588 (6) | 0.0469 (4) | |
| H3 | 1.0200 | 0.3265 | 0.6916 | 0.070* | |
| O3 | 0.83973 (12) | 0.3290 (5) | 0.75521 (6) | 0.0546 (4) | |
| H4 | 0.7823 | 0.3169 | 0.7822 | 0.082* | |
| O4 | 0.35878 (13) | 0.5981 (5) | 0.66792 (6) | 0.0583 (4) | |
| O5 | 0.40210 (12) | 0.7959 (5) | 0.57298 (6) | 0.0567 (4) | |
| H6 | 0.3193 | 0.8292 | 0.5719 | 0.085* | |
| O6 | 0.14365 (13) | 1.0125 (5) | 0.56257 (7) | 0.0499 (4) | |
| H6A | 0.120 (3) | 1.147 (8) | 0.5916 (12) | 0.102 (11)* | |
| H6B | 0.077 (4) | 0.879 (14) | 0.555 (2) | 0.24 (3)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0338 (8) | 0.0434 (9) | 0.0333 (9) | 0.0031 (7) | −0.0022 (7) | 0.0016 (7) |
| C2 | 0.0310 (8) | 0.0418 (9) | 0.0321 (9) | 0.0009 (7) | 0.0036 (6) | 0.0008 (7) |
| C3 | 0.0264 (7) | 0.0457 (9) | 0.0346 (9) | 0.0013 (7) | −0.0009 (6) | −0.0010 (7) |
| C4 | 0.0337 (8) | 0.0530 (11) | 0.0316 (9) | 0.0013 (7) | −0.0021 (6) | 0.0037 (8) |
| C5 | 0.0322 (8) | 0.0572 (11) | 0.0349 (9) | 0.0039 (7) | 0.0042 (7) | 0.0038 (8) |
| C6 | 0.0299 (8) | 0.0444 (9) | 0.0367 (9) | 0.0041 (7) | 0.0002 (7) | −0.0001 (7) |
| C7 | 0.0319 (8) | 0.0525 (10) | 0.0381 (10) | 0.0049 (7) | 0.0003 (7) | −0.0008 (8) |
| O1 | 0.0361 (6) | 0.0738 (10) | 0.0342 (7) | 0.0052 (6) | 0.0051 (5) | 0.0085 (6) |
| O2 | 0.0286 (6) | 0.0775 (10) | 0.0345 (7) | 0.0051 (6) | −0.0026 (5) | 0.0035 (6) |
| O3 | 0.0334 (6) | 0.0984 (12) | 0.0321 (7) | 0.0070 (7) | −0.0005 (5) | 0.0138 (7) |
| O4 | 0.0335 (6) | 0.0953 (12) | 0.0465 (8) | 0.0101 (7) | 0.0074 (6) | 0.0062 (8) |
| O5 | 0.0330 (6) | 0.0882 (11) | 0.0488 (8) | 0.0137 (7) | −0.0020 (5) | 0.0158 (7) |
| O6 | 0.0358 (7) | 0.0697 (9) | 0.0440 (8) | 0.0126 (6) | −0.0016 (5) | 0.0016 (7) |
Geometric parameters (Å, °)
| C1—C2 | 1.382 (2) | C5—H5 | 0.9300 |
| C1—C6 | 1.388 (2) | C6—C7 | 1.480 (2) |
| C1—H1 | 0.9300 | C7—O4 | 1.212 (2) |
| C2—O1 | 1.372 (2) | C7—O5 | 1.317 (2) |
| C2—C3 | 1.388 (2) | O1—H2 | 0.8200 |
| C3—O2 | 1.3723 (19) | O2—H3 | 0.8200 |
| C3—C4 | 1.387 (2) | O3—H4 | 0.8200 |
| C4—O3 | 1.368 (2) | O5—H6 | 0.8200 |
| C4—C5 | 1.378 (2) | O6—H6A | 0.830 (18) |
| C5—C6 | 1.391 (2) | O6—H6B | 0.824 (19) |
| C2—C1—C6 | 118.99 (15) | C4—C5—H5 | 120.2 |
| C2—C1—H1 | 120.5 | C6—C5—H5 | 120.2 |
| C6—C1—H1 | 120.5 | C1—C6—C5 | 120.83 (15) |
| O1—C2—C1 | 122.41 (14) | C1—C6—C7 | 120.83 (15) |
| O1—C2—C3 | 117.10 (14) | C5—C6—C7 | 118.32 (15) |
| C1—C2—C3 | 120.48 (15) | O4—C7—O5 | 122.82 (15) |
| O2—C3—C4 | 121.78 (14) | O4—C7—C6 | 123.32 (16) |
| O2—C3—C2 | 118.17 (14) | O5—C7—C6 | 113.86 (15) |
| C4—C3—C2 | 120.05 (14) | C2—O1—H2 | 109.5 |
| O3—C4—C5 | 124.44 (16) | C3—O2—H3 | 109.5 |
| O3—C4—C3 | 115.54 (14) | C4—O3—H4 | 109.5 |
| C5—C4—C3 | 120.01 (15) | C7—O5—H6 | 109.5 |
| C4—C5—C6 | 119.61 (16) | H6A—O6—H6B | 105 (3) |
| C6—C1—C2—O1 | 178.97 (16) | O3—C4—C5—C6 | 178.56 (18) |
| C6—C1—C2—C3 | 0.0 (3) | C3—C4—C5—C6 | −0.9 (3) |
| O1—C2—C3—O2 | −0.5 (2) | C2—C1—C6—C5 | 1.0 (3) |
| C1—C2—C3—O2 | 178.51 (16) | C2—C1—C6—C7 | 179.51 (16) |
| O1—C2—C3—C4 | 179.56 (16) | C4—C5—C6—C1 | −0.5 (3) |
| C1—C2—C3—C4 | −1.4 (3) | C4—C5—C6—C7 | −179.08 (17) |
| O2—C3—C4—O3 | 2.4 (3) | C1—C6—C7—O4 | 173.92 (19) |
| C2—C3—C4—O3 | −177.66 (17) | C5—C6—C7—O4 | −7.5 (3) |
| O2—C3—C4—C5 | −178.04 (17) | C1—C6—C7—O5 | −5.8 (3) |
| C2—C3—C4—C5 | 1.9 (3) | C5—C6—C7—O5 | 172.79 (18) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H3···O3 | 0.82 | 2.26 | 2.7006 (18) | 114 |
| O5—H6···O6 | 0.82 | 1.85 | 2.6542 (17) | 167 |
| O1—H2···O6i | 0.82 | 2.03 | 2.7539 (19) | 147 |
| O2—H3···O3ii | 0.82 | 2.52 | 3.1721 (19) | 137 |
| O3—H4···O4iii | 0.82 | 1.94 | 2.7154 (19) | 158 |
| O6—H6A···O2iv | 0.83 (2) | 2.03 (2) | 2.8237 (19) | 161 (3) |
| O6—H6B···O1v | 0.82 (2) | 2.00 (2) | 2.814 (2) | 171 (5) |
Symmetry codes: (i) −x+1, −y+2, −z+1; (ii) −x+2, y−1/2, −z+3/2; (iii) −x+1, y−1/2, −z+3/2; (iv) x−1, y+1, z; (v) x−1, y, z.
Table 2 Other crystal structures of gallic acid monohydrate (Å, °)
| 1 | 2 | |
| Unit-cell parameters | a = 5.794 (4) | a = 14.15 (1) |
| b = 4.719 (5) | b = 3.622 (9) | |
| c = 28.688 (5) | c = 15.028 (10) | |
| β = 95.08 (3) | β = 97.52 (7) | |
| V = 781.4 (3) | V = 764 (1) | |
| Space group | Monoclinic, P21/c | Monoclinic, P2/n |
| Reference | Jiang et al. (2000) | Okabe et al. (2001) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BX2352).
References
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Genç, S., Dege, N., Çetin, A., Cansız, A., Şekerci, M. & Dinçer, M. (2004). Acta Cryst. E60, o1580–o1582.
- Jiang, R.-W., Ming, D.-S., But, P. P. H. & Mak, T. C. W. (2000). Acta Cryst. C56, 594–595. [DOI] [PubMed]
- Lu, Z., Nie, G., Belton, P. S., Tang, H. & Zhao, B. (2006). Neurochem. Int. 48, 263–274. [DOI] [PubMed]
- Madlener, S., Illmer, C., Horvath, Z., Saiko, P., Losert, A., Herbacek, I., Grusch, M., Elford, H. L., Krupitza, G., Bernhaus, A., Fritzer-Szekeres, M. & Szekeres, T. (2007). Cancer Lett. 245, 156–162. [DOI] [PubMed]
- Okabe, N., Kyoyama, H. & Suzuki, M. (2001). Acta Cryst. E57, o764–o766.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2002). X-AREA and X-RED32 Stoe & Cie, Darmstadt, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811018848/bx2352sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811018848/bx2352Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811018848/bx2352Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report