Abstract
The title compound, C22H21NO2, was synthesized from 4-methoxy-2-methylaniline and 2-hydroxy-1,2-diphenylethanone. In the title compound, the C—C—C—N—C backbone adopts an all-trans conformation. The crystal structure is stabilized by weak intermolecular C—H⋯O hydrogen-bond interactions.
Related literature
For the synthesis and similar structures, see: Au & Tafeenko (1986 ▶, 1987 ▶); Batsanov et al., (2006 ▶). For general background to these structures, see: Batsanov et al. (2006 ▶); Abdulla et al. (1985 ▶). For bond-length data, see: Allen et al. (1987 ▶). For geometrical analysis, see: Bruno et al. (2002 ▶).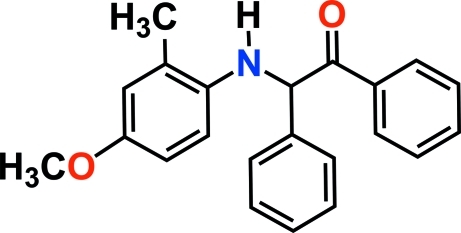
Experimental
Crystal data
C22H21NO2
M r = 331.40
Monoclinic,

a = 12.570 (12) Å
b = 8.009 (8) Å
c = 18.091 (17) Å
β = 100.544 (15)°
V = 1791 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.08 mm−1
T = 173 K
0.21 × 0.19 × 0.15 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2008 ▶) T min = 0.984, T max = 0.988
28799 measured reflections
4122 independent reflections
2935 reflections with I > 2σ(I)
R int = 0.055
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.112
S = 1.05
4122 reflections
226 parameters
H-atom parameters constrained
Δρmax = 0.27 e Å−3
Δρmin = −0.25 e Å−3
Data collection: SMART (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: OLEX2 (Dolomanov et al., 2009 ▶); software used to prepare material for publication: SHELXTL (Sheldrick, 2008 ▶), OLEX2, publCIF (Westrip, 2010 ▶) and Mercury (Macrae et al., 2006 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811015960/hg5030sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811015960/hg5030Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811015960/hg5030Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C11—H11A⋯O1i | 0.95 | 2.48 | 3.352 (4) | 153 |
Symmetry code: (i)  .
.
Acknowledgments
This work was supported by the Mersin University Research Fund [Project No. BAP-SBE FK (SZ) 2008–8 YL].
supplementary crystallographic information
Comment
A few 1-arylanilinoethanone derivatives have been structurally charactized because of their importance in synthesis or because of their interesting charge-transfer properties (Abdulla et al., 1985). We found only four structurally similar compounds (Au & Tafeenko, 1987; Au & Tafeenko, 1986; Batsanov et al., 2006) in the Cambridge Structural Database (CSD CONQUEST 1.11, B4; Bruno et al., 2002).
Compound I was synthesized by the HCl acid-catalyzed reaction of a benzoin derivative to 4-methoxy-2-methylaniline (Scheme 1) resulting in the title compound, 2-((4-methoxy-2-methylphenyl)amino)-1,2-diphenylethanone, I, Figure 1, its structure was determined by X-ray crystallography.
The molecular structure of the title compound contains two phenyl rings and one substituted aniline ring connected by a –C(O)—C– linker. The dihedral angle between the two phenyl rings is 87.78 (7) ° and the dihedral angle between the substitute aniline and phenyl rings are 55.30 (7) and 84.57 (7) °, respectively. In adition, in the title compound, the C2—C1—C8—N1—C15 backbone adopts an all-trans conformation (Au & Tafeenko, 1987; Au & Tafeenko, 1986; Batsanov et al., 2006).
The C—N bond lengths C8—N1 and C15—N1 are shorter than the normal C—N single-bond length of about 1.48 Å. The shortening of these C—N bonds reveals the effects of some conjugation in this part of the molecule. All other bond lengths fall within the expected ranges (Allen et al., 1987).
Intramolecular hydrogen bonding N1–H1A···O1 with N–H 0.88 Å, H···O 2.22 Å, N–H···O 107 ° results in the formation of a five membered ring in the O1—C1—C8—N1–H1A plane. The crystal packing is dominated by weak intermolecular C11—H11A···O1 (x, 1 + y, z) hydrogen bonds, with H···O = 2.48 Å and a C—H···O angle of 153 ° (Figure 2).
Experimental
A mixture of 4-methoxy-2-methylaniline (15 mmol), 2-hydroxy-1,2-diphenylethanone (5 mmol) and 1 ml conc. HCl in 20 ml of ethanol were refluxed for 5 h (Figure 3). After reaction was complete, the mixture was allowed to cool to room temperature, poured into cold water (20 ml) and finally extracted with CH2Cl2 (3x15 ml). The organic layer was dried over magnesium sulfate and the solvent removed under reduced pressure to yield a crude product that was purified by recrystallization in ethyl acetate. 2-((4-methoxy-2-methylphenyl)amino)-1,2-diphenylethanone: Yield: 1.20 g, 46%. M.p.: 110–112 °C. 1H NMR (DMSO-d6) δ: 8.18–8.16 (d, 2H, Ar—H (C3, C7)), 7.62 (t, 1H, Ar—H (C5)), 7.56–7.49 (m, 4H, Ar—H (C4, C6, C11, C13)), 7.30–7.26 (m, 2H, Ar—H (C10, C14)), 7.16 (t, 1H, Ar—H (C12)), 6.70–6.68 (d, J=4.8 Hz, 1H, Ar—H (C19)), 6.67 (s, 1H, Ar—H (C17)), 6.57–6.55 (d, J=8 Hz, 1H, Ar—H (C20)), 6.46–6.44 (d, J=8 Hz, 1H, Ar—H (C8)), 5.19–5.17 (d, J=8 Hz, 1H, NH), 3.60 (s, 3H, OCH3), 2.22 (s, 3H, CH3). 13C NMR (400 MHz, p.p.m.) δ: 17.5 (CH3), 55.1 (OCH3), 61.5 (C8), 111.2 (C19), 112.5 (C20), 116.4 (C17), 123.9 (C16), 127.6, 128.1, 128.6, 128.7, 128.8 (C), 133.7 (C5), 134.7 (C2), 138.1 (C9), 138.3 (C15), 151.1 (C18), 197.6 (C9). Anal. Calc. for C22H21NO2: C, 79.73; H, 6.39; N, 4.23%. Found: C, 79.70; H, 6.21; N, 4.19%.
Refinement
H atom positions were clearly derived from difference Fourier maps and refined using a riding model, fixing the bond lengths at 0.98 and 0.95 Å for CH3 and CH(aromatic), respectively. The displacement parameters of the H atoms were constrained with Uiso(H) = 1.2Ueq (C) or 1.5Ueq (methyl C).
Figures
Fig. 1.
The molecular structure of (I), showing ellipsoids at the 50% probability level.
Fig. 2.
The molecular packing of (I). The hydrogen bonds are shown as dashed lines.
Fig. 3.
Synthesis of the title compound.
Crystal data
| C22H21NO2 | F(000) = 704 |
| Mr = 331.40 | Dx = 1.229 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4852 reflections |
| a = 12.570 (12) Å | θ = 2.3–24.5° |
| b = 8.009 (8) Å | µ = 0.08 mm−1 |
| c = 18.091 (17) Å | T = 173 K |
| β = 100.544 (15)° | Block, yellow |
| V = 1791 (3) Å3 | 0.21 × 0.19 × 0.15 mm |
| Z = 4 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 4122 independent reflections |
| Radiation source: fine-focus sealed tube | 2935 reflections with I > 2σ(I) |
| graphite | Rint = 0.055 |
| φ and ω scans | θmax = 27.5°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −16→16 |
| Tmin = 0.984, Tmax = 0.988 | k = −10→10 |
| 28799 measured reflections | l = −23→23 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.112 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0413P)2 + 0.4229P] where P = (Fo2 + 2Fc2)/3 |
| 4122 reflections | (Δ/σ)max = 0.004 |
| 226 parameters | Δρmax = 0.27 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.70413 (11) | 0.71666 (18) | 0.13255 (8) | 0.0336 (3) | |
| C2 | 0.65511 (11) | 0.79413 (17) | 0.05905 (7) | 0.0314 (3) | |
| C3 | 0.70228 (12) | 0.92642 (19) | 0.02686 (8) | 0.0380 (3) | |
| H3A | 0.7667 | 0.9764 | 0.0533 | 0.046* | |
| C4 | 0.65510 (14) | 0.9856 (2) | −0.04399 (8) | 0.0453 (4) | |
| H4A | 0.6879 | 1.0748 | −0.0662 | 0.054* | |
| C5 | 0.56034 (14) | 0.9144 (2) | −0.08206 (8) | 0.0436 (4) | |
| H5A | 0.5286 | 0.9546 | −0.1305 | 0.052* | |
| C6 | 0.51179 (13) | 0.7853 (2) | −0.05009 (8) | 0.0420 (4) | |
| H6A | 0.4460 | 0.7385 | −0.0761 | 0.050* | |
| C7 | 0.55897 (12) | 0.72392 (18) | 0.01989 (8) | 0.0361 (3) | |
| H7A | 0.5260 | 0.6339 | 0.0414 | 0.043* | |
| C8 | 0.78651 (11) | 0.81396 (18) | 0.19059 (8) | 0.0339 (3) | |
| H8A | 0.8385 | 0.8744 | 0.1644 | 0.041* | |
| C9 | 0.72052 (11) | 0.94044 (17) | 0.22728 (7) | 0.0297 (3) | |
| C10 | 0.72302 (12) | 1.11045 (18) | 0.21167 (8) | 0.0370 (3) | |
| H10A | 0.7685 | 1.1508 | 0.1790 | 0.044* | |
| C11 | 0.65930 (13) | 1.22179 (18) | 0.24361 (9) | 0.0421 (4) | |
| H11A | 0.6615 | 1.3376 | 0.2327 | 0.051* | |
| C12 | 0.59250 (12) | 1.16418 (19) | 0.29135 (8) | 0.0396 (4) | |
| H12A | 0.5489 | 1.2403 | 0.3129 | 0.048* | |
| C13 | 0.58976 (12) | 0.99566 (19) | 0.30736 (8) | 0.0378 (3) | |
| H13A | 0.5441 | 0.9557 | 0.3399 | 0.045* | |
| C14 | 0.65372 (11) | 0.88458 (18) | 0.27584 (8) | 0.0334 (3) | |
| H14A | 0.6519 | 0.7691 | 0.2875 | 0.040* | |
| C15 | 0.91952 (11) | 0.72896 (19) | 0.30593 (8) | 0.0330 (3) | |
| C16 | 0.95263 (11) | 0.59902 (19) | 0.35825 (8) | 0.0346 (3) | |
| C17 | 1.02943 (11) | 0.6351 (2) | 0.42183 (8) | 0.0393 (4) | |
| H17A | 1.0524 | 0.5485 | 0.4570 | 0.047* | |
| C18 | 1.07393 (12) | 0.7936 (2) | 0.43580 (8) | 0.0412 (4) | |
| C19 | 1.04216 (12) | 0.9204 (2) | 0.38448 (8) | 0.0409 (4) | |
| H19A | 1.0725 | 1.0289 | 0.3929 | 0.049* | |
| C20 | 0.96492 (11) | 0.88712 (19) | 0.32004 (8) | 0.0375 (3) | |
| H20A | 0.9429 | 0.9745 | 0.2851 | 0.045* | |
| C21 | 0.90500 (13) | 0.4262 (2) | 0.34587 (9) | 0.0442 (4) | |
| H21A | 0.9374 | 0.3535 | 0.3875 | 0.066* | |
| H21B | 0.9201 | 0.3810 | 0.2985 | 0.066* | |
| H21C | 0.8266 | 0.4318 | 0.3435 | 0.066* | |
| C22 | 1.19345 (16) | 0.9696 (3) | 0.52047 (11) | 0.0690 (6) | |
| H22A | 1.2440 | 0.9640 | 0.5686 | 0.103* | |
| H22B | 1.1358 | 1.0496 | 0.5244 | 0.103* | |
| H22C | 1.2322 | 1.0058 | 0.4810 | 0.103* | |
| N1 | 0.84386 (10) | 0.69119 (16) | 0.24129 (7) | 0.0412 (3) | |
| H1A | 0.8302 | 0.5852 | 0.2308 | 0.049* | |
| O1 | 0.67637 (9) | 0.57676 (13) | 0.14827 (6) | 0.0477 (3) | |
| O2 | 1.14758 (10) | 0.80923 (17) | 0.50230 (6) | 0.0612 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0331 (7) | 0.0372 (8) | 0.0294 (7) | −0.0008 (6) | 0.0026 (6) | −0.0002 (6) |
| C2 | 0.0323 (7) | 0.0345 (8) | 0.0264 (7) | 0.0029 (6) | 0.0024 (6) | −0.0041 (6) |
| C3 | 0.0388 (8) | 0.0420 (8) | 0.0318 (8) | −0.0004 (7) | 0.0030 (6) | −0.0009 (6) |
| C4 | 0.0585 (10) | 0.0449 (9) | 0.0338 (8) | 0.0049 (8) | 0.0114 (7) | 0.0045 (7) |
| C5 | 0.0558 (10) | 0.0458 (9) | 0.0256 (7) | 0.0141 (8) | −0.0022 (7) | −0.0025 (6) |
| C6 | 0.0435 (9) | 0.0450 (9) | 0.0326 (8) | 0.0067 (7) | −0.0062 (7) | −0.0089 (7) |
| C7 | 0.0386 (8) | 0.0360 (8) | 0.0318 (7) | 0.0014 (6) | 0.0013 (6) | −0.0056 (6) |
| C8 | 0.0314 (7) | 0.0377 (8) | 0.0299 (7) | −0.0001 (6) | −0.0013 (6) | 0.0027 (6) |
| C9 | 0.0279 (7) | 0.0321 (7) | 0.0252 (7) | −0.0033 (6) | −0.0055 (5) | 0.0023 (5) |
| C10 | 0.0399 (8) | 0.0347 (8) | 0.0331 (7) | −0.0082 (6) | −0.0024 (6) | 0.0067 (6) |
| C11 | 0.0503 (9) | 0.0265 (7) | 0.0418 (8) | −0.0024 (7) | −0.0121 (7) | 0.0012 (6) |
| C12 | 0.0401 (8) | 0.0371 (8) | 0.0362 (8) | 0.0056 (7) | −0.0076 (7) | −0.0060 (6) |
| C13 | 0.0381 (8) | 0.0399 (8) | 0.0337 (7) | −0.0025 (7) | 0.0023 (6) | −0.0008 (6) |
| C14 | 0.0373 (8) | 0.0288 (7) | 0.0314 (7) | −0.0032 (6) | −0.0006 (6) | 0.0029 (6) |
| C15 | 0.0260 (7) | 0.0444 (8) | 0.0281 (7) | 0.0042 (6) | 0.0033 (5) | 0.0017 (6) |
| C16 | 0.0279 (7) | 0.0453 (9) | 0.0315 (7) | 0.0023 (6) | 0.0084 (6) | 0.0050 (6) |
| C17 | 0.0302 (7) | 0.0542 (10) | 0.0333 (8) | 0.0001 (7) | 0.0047 (6) | 0.0151 (7) |
| C18 | 0.0297 (8) | 0.0616 (10) | 0.0295 (7) | −0.0071 (7) | −0.0016 (6) | 0.0101 (7) |
| C19 | 0.0351 (8) | 0.0499 (9) | 0.0356 (8) | −0.0089 (7) | 0.0011 (6) | 0.0052 (7) |
| C20 | 0.0337 (8) | 0.0445 (9) | 0.0323 (7) | 0.0018 (7) | 0.0007 (6) | 0.0082 (6) |
| C21 | 0.0437 (9) | 0.0476 (9) | 0.0407 (8) | −0.0011 (7) | 0.0062 (7) | 0.0079 (7) |
| C22 | 0.0614 (12) | 0.0892 (15) | 0.0463 (10) | −0.0308 (11) | −0.0170 (9) | 0.0086 (10) |
| N1 | 0.0441 (7) | 0.0359 (7) | 0.0371 (7) | 0.0081 (6) | −0.0094 (6) | −0.0035 (5) |
| O1 | 0.0572 (7) | 0.0424 (6) | 0.0380 (6) | −0.0124 (5) | −0.0057 (5) | 0.0068 (5) |
| O2 | 0.0565 (7) | 0.0759 (9) | 0.0411 (6) | −0.0243 (7) | −0.0179 (6) | 0.0195 (6) |
Geometric parameters (Å, °)
| C1—O1 | 1.2226 (19) | C12—H12A | 0.9500 |
| C1—C2 | 1.494 (2) | C13—C14 | 1.390 (2) |
| C1—C8 | 1.544 (2) | C13—H13A | 0.9500 |
| C2—C3 | 1.393 (2) | C14—H14A | 0.9500 |
| C2—C7 | 1.402 (2) | C15—C20 | 1.394 (2) |
| C3—C4 | 1.393 (2) | C15—N1 | 1.399 (2) |
| C3—H3A | 0.9500 | C15—C16 | 1.417 (2) |
| C4—C5 | 1.385 (2) | C16—C17 | 1.390 (2) |
| C4—H4A | 0.9500 | C16—C21 | 1.509 (2) |
| C5—C6 | 1.380 (2) | C17—C18 | 1.391 (2) |
| C5—H5A | 0.9500 | C17—H17A | 0.9500 |
| C6—C7 | 1.386 (2) | C18—O2 | 1.383 (2) |
| C6—H6A | 0.9500 | C18—C19 | 1.385 (2) |
| C7—H7A | 0.9500 | C19—C20 | 1.399 (2) |
| C8—N1 | 1.4443 (19) | C19—H19A | 0.9500 |
| C8—C9 | 1.535 (2) | C20—H20A | 0.9500 |
| C8—H8A | 1.0000 | C21—H21A | 0.9800 |
| C9—C10 | 1.392 (2) | C21—H21B | 0.9800 |
| C9—C14 | 1.395 (2) | C21—H21C | 0.9800 |
| C10—C11 | 1.393 (2) | C22—O2 | 1.422 (2) |
| C10—H10A | 0.9500 | C22—H22A | 0.9800 |
| C11—C12 | 1.389 (2) | C22—H22B | 0.9800 |
| C11—H11A | 0.9500 | C22—H22C | 0.9800 |
| C12—C13 | 1.382 (2) | N1—H1A | 0.8800 |
| O1—C1—C2 | 119.92 (13) | C12—C13—H13A | 120.0 |
| O1—C1—C8 | 119.27 (13) | C14—C13—H13A | 120.0 |
| C2—C1—C8 | 120.77 (13) | C13—C14—C9 | 120.87 (14) |
| C3—C2—C7 | 119.18 (14) | C13—C14—H14A | 119.6 |
| C3—C2—C1 | 123.38 (13) | C9—C14—H14A | 119.6 |
| C7—C2—C1 | 117.39 (13) | C20—C15—N1 | 122.87 (13) |
| C2—C3—C4 | 120.09 (15) | C20—C15—C16 | 119.00 (14) |
| C2—C3—H3A | 120.0 | N1—C15—C16 | 118.12 (14) |
| C4—C3—H3A | 120.0 | C17—C16—C15 | 118.33 (15) |
| C5—C4—C3 | 119.99 (16) | C17—C16—C21 | 120.70 (13) |
| C5—C4—H4A | 120.0 | C15—C16—C21 | 120.97 (14) |
| C3—C4—H4A | 120.0 | C16—C17—C18 | 122.38 (14) |
| C6—C5—C4 | 120.41 (15) | C16—C17—H17A | 118.8 |
| C6—C5—H5A | 119.8 | C18—C17—H17A | 118.8 |
| C4—C5—H5A | 119.8 | O2—C18—C19 | 125.58 (15) |
| C5—C6—C7 | 120.02 (15) | O2—C18—C17 | 115.06 (13) |
| C5—C6—H6A | 120.0 | C19—C18—C17 | 119.35 (14) |
| C7—C6—H6A | 120.0 | C18—C19—C20 | 119.34 (15) |
| C6—C7—C2 | 120.30 (15) | C18—C19—H19A | 120.3 |
| C6—C7—H7A | 119.9 | C20—C19—H19A | 120.3 |
| C2—C7—H7A | 119.9 | C15—C20—C19 | 121.61 (14) |
| N1—C8—C9 | 114.91 (13) | C15—C20—H20A | 119.2 |
| N1—C8—C1 | 106.44 (13) | C19—C20—H20A | 119.2 |
| C9—C8—C1 | 106.20 (12) | C16—C21—H21A | 109.5 |
| N1—C8—H8A | 109.7 | C16—C21—H21B | 109.5 |
| C9—C8—H8A | 109.7 | H21A—C21—H21B | 109.5 |
| C1—C8—H8A | 109.7 | C16—C21—H21C | 109.5 |
| C10—C9—C14 | 118.68 (13) | H21A—C21—H21C | 109.5 |
| C10—C9—C8 | 121.58 (13) | H21B—C21—H21C | 109.5 |
| C14—C9—C8 | 119.71 (13) | O2—C22—H22A | 109.5 |
| C9—C10—C11 | 120.40 (15) | O2—C22—H22B | 109.5 |
| C9—C10—H10A | 119.8 | H22A—C22—H22B | 109.5 |
| C11—C10—H10A | 119.8 | O2—C22—H22C | 109.5 |
| C12—C11—C10 | 120.28 (15) | H22A—C22—H22C | 109.5 |
| C12—C11—H11A | 119.9 | H22B—C22—H22C | 109.5 |
| C10—C11—H11A | 119.9 | C15—N1—C8 | 124.59 (13) |
| C13—C12—C11 | 119.71 (14) | C15—N1—H1A | 117.7 |
| C13—C12—H12A | 120.1 | C8—N1—H1A | 117.7 |
| C11—C12—H12A | 120.1 | C18—O2—C22 | 117.49 (13) |
| C12—C13—C14 | 120.05 (15) | ||
| O1—C1—C2—C3 | −160.36 (14) | C11—C12—C13—C14 | 0.1 (2) |
| C8—C1—C2—C3 | 22.1 (2) | C12—C13—C14—C9 | −0.6 (2) |
| O1—C1—C2—C7 | 17.0 (2) | C10—C9—C14—C13 | 0.8 (2) |
| C8—C1—C2—C7 | −160.60 (12) | C8—C9—C14—C13 | −177.33 (12) |
| C7—C2—C3—C4 | −1.2 (2) | C20—C15—C16—C17 | 0.0 (2) |
| C1—C2—C3—C4 | 176.15 (14) | N1—C15—C16—C17 | 178.73 (13) |
| C2—C3—C4—C5 | 0.9 (2) | C20—C15—C16—C21 | 179.37 (13) |
| C3—C4—C5—C6 | 0.3 (2) | N1—C15—C16—C21 | −1.9 (2) |
| C4—C5—C6—C7 | −1.3 (2) | C15—C16—C17—C18 | 0.4 (2) |
| C5—C6—C7—C2 | 1.0 (2) | C21—C16—C17—C18 | −178.96 (14) |
| C3—C2—C7—C6 | 0.2 (2) | C16—C17—C18—O2 | 178.75 (13) |
| C1—C2—C7—C6 | −177.25 (13) | C16—C17—C18—C19 | −0.8 (2) |
| O1—C1—C8—N1 | 21.30 (18) | O2—C18—C19—C20 | −178.70 (14) |
| C2—C1—C8—N1 | −161.10 (12) | C17—C18—C19—C20 | 0.8 (2) |
| O1—C1—C8—C9 | −101.58 (16) | N1—C15—C20—C19 | −178.65 (14) |
| C2—C1—C8—C9 | 76.02 (16) | C16—C15—C20—C19 | 0.0 (2) |
| N1—C8—C9—C10 | 135.07 (14) | C18—C19—C20—C15 | −0.4 (2) |
| C1—C8—C9—C10 | −107.56 (15) | C20—C15—N1—C8 | −14.1 (2) |
| N1—C8—C9—C14 | −46.83 (17) | C16—C15—N1—C8 | 167.24 (13) |
| C1—C8—C9—C14 | 70.54 (15) | C9—C8—N1—C15 | −57.18 (19) |
| C14—C9—C10—C11 | −0.5 (2) | C1—C8—N1—C15 | −174.41 (13) |
| C8—C9—C10—C11 | 177.64 (12) | C19—C18—O2—C22 | 1.2 (2) |
| C9—C10—C11—C12 | −0.1 (2) | C17—C18—O2—C22 | −178.33 (16) |
| C10—C11—C12—C13 | 0.3 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O1 | 0.88 | 2.22 | 2.609 (3) | 107 |
| C11—H11A···O1i | 0.95 | 2.48 | 3.352 (4) | 153 |
Symmetry codes: (i) x, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG5030).
References
- Abdulla, R. F., Boyd, D. B., Jones, N. D. & Swartzendruber, J. K. (1985). J. Org. Chem. 50, 3502–3505.
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Au, O. & Tafeenko, V. (1986). Rev. Cubana Quim. 2, 65–74.
- Au, O. & Tafeenko, V. (1987). Rev. Cubana Quim. 3, 79–86.
- Batsanov, A. S., Goeta, A. E., Howard, J. A. K., Soto, B. & Au-Alvarez, O. (2006). Acta Cryst. C62, o304–o306. [DOI] [PubMed]
- Bruker (2008). APEX2, SADABS and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruno, I. J., Cole, J. C., Edgington, P. R., Kessler, M., Macrae, C. F., McCabe, P., Pearson, J. & Taylor, R. (2002). Acta Cryst. B58, 389–397. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811015960/hg5030sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811015960/hg5030Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811015960/hg5030Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





