Abstract
Background
The genome of Helicobacter pylori, an oncogenic bacterium in the human stomach, rapidly evolves and shows wide geographical divergence. The high incidence of stomach cancer in East Asia might be related to bacterial genotype. We used newly developed comparative methods to follow the evolution of East Asian H. pylori genomes using 20 complete genome sequences from Japanese, Korean, Amerind, European, and West African strains.
Results
A phylogenetic tree of concatenated well-defined core genes supported divergence of the East Asian lineage (hspEAsia; Japanese and Korean) from the European lineage ancestor, and then from the Amerind lineage ancestor. Phylogenetic profiling revealed a large difference in the repertoire of outer membrane proteins (including oipA, hopMN, babABC, sabAB and vacA-2) through gene loss, gain, and mutation. All known functions associated with molybdenum, a rare element essential to nearly all organisms that catalyzes two-electron-transfer oxidation-reduction reactions, appeared to be inactivated. Two pathways linking acetyl~CoA and acetate appeared intact in some Japanese strains. Phylogenetic analysis revealed greater divergence between the East Asian (hspEAsia) and the European (hpEurope) genomes in proteins in host interaction, specifically virulence factors (tipα), outer membrane proteins, and lipopolysaccharide synthesis (human Lewis antigen mimicry) enzymes. Divergence was also seen in proteins in electron transfer and translation fidelity (miaA, tilS), a DNA recombinase/exonuclease that recognizes genome identity (addA), and DNA/RNA hybrid nucleases (rnhAB). Positively selected amino acid changes between hspEAsia and hpEurope were mapped to products of cagA, vacA, homC (outer membrane protein), sotB (sugar transport), and a translation fidelity factor (miaA). Large divergence was seen in genes related to antibiotics: frxA (metronidazole resistance), def (peptide deformylase, drug target), and ftsA (actin-like, drug target).
Conclusions
These results demonstrate dramatic genome evolution within a species, especially in likely host interaction genes. The East Asian strains appear to differ greatly from the European strains in electron transfer and redox reactions. These findings also suggest a model of adaptive evolution through proteome diversification and selection through modulation of translational fidelity. The results define H. pylori East Asian lineages and provide essential information for understanding their pathogenesis and designing drugs and therapies that target them.
Background
Genome sequence comparison within a species can reveal genome evolution processes in detail and provide insights for basic and applied research. For bacteria, this approach has been quite powerful in revealing horizontal gene transfer, gene decay, and genome rearrangements underlying adaptation, such as evolution of virulence [1]. Comparison of many complete genome sequences is feasible through innovations in DNA sequencing.
Helicobacter pylori was the first species for which two complete genome sequences were available [2]. This species of ε-proteobacteria causes gastritis, gastric (stomach) ulcer, and duodenal ulcer, and is associated with gastric cancer and mucosa-associated lymphoid tissue (MALT) lymphoma [3,4]. Animal models show a causal link between H. pylori and gastric cancer [5,6]. Recent clinical work in Japan suggests that H. pylori eradication reduces the risk of new gastric carcinomas in patients with a history of the disease [7].
H. pylori shows a high mutation rate and an even higher rate of homologous recombination [8]. Phylogenetic analysis based on several genes revealed geographical differentiation since H. pylori left Africa together with Homo sapiens [9]. The analysis indicated that the East Asian type (hpEastAsia) is classified into at least three subtypes: East Asian (hspEAsia), Pacific (hspMaori) and native American (hspAmerind) [9,10]. The East Asia subtype (hspEAsia) may be related to the high incidence of gastric cancer in East Asia [4].
H. pylori CagA is considered to be a major virulence factor associated with gastric cancer. CagA is delivered into gastric epithelial cells and undergoes phosphorylation by host kinases. Membrane-localized CagA mimics mammalian scaffold proteins, perturbs signaling pathways and promotes transformation. CagA is noted for structural diversity in its C-terminal region, which interacts with host cell proteins. It is classified into Western and East Asian types, with higher activities associated with the latter [11]. The East Asian CagA-positive H. pylori infection is more closely associated with gastric cancer [12]. Geographical differences have also been noted for other genes [13-17].
To fully characterize these bacteria (hspEAsia subtype of H. pylori) and to study underlying intraspecific (within-species) evolutionary processes in detail at the genome sequence level, we determined the genome sequence of four Japanese strains and compared them to available complete H. pylori genome sequences. The sequences of the Japanese strains and two Korean strains were different in gene content from the European and West African genomes and from the Amerind genome. Unexpectedly, divergence was seen in genes related to electron transfer and translation fidelity, as well as virulence and host interaction.
Results
The complete genome sequences of four H. pylori strains (F57, F32, F30 and F16) isolated from different individuals in Fukui, Japan were determined. We compared 20 complete genomes of H. pylori (the 4 new genomes and 16 genomes in the public domain; Table 1), focusing on their gene contents.
Table 1.
Comparison of hspEAsia to other genomes
| Strain | Disease | Population | Length | % GC | CDS | Core | cagA(c) | vacA(d) | homAB | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| subpopulation | (bp)(a,b) | content | genes | |||||||
| F57 | Gastric cancer | hpEastAsia hspEAsia | 1609006 | 38.7 | 1521 | 1402 | ABD | s1a-m1-i1 | -/B | This work |
| F32 | Gastric cancer | hpEastAsia hspEAsia | 1578824, 2637 | 38.9 | 1492 | 1385 | ABD | s1a-m1-i1 | -/E(e) | This work |
| F30 | Duodenal ulcer | hpEastAsia hspEAsia | 1570564, 9129 | 38.8 | 1485 | 1385 | ABD | s1a-m1-i1 | -/B | This work |
| F16 | Gastritis | hpEastAsia hspEAsia | 1575399 | 38.9 | 1500 | 1402 | ABD | s1a-m1-i1 | -/B | This work |
| 51 | Duodenal ulcer | hpEastAsia hspEAsia | 1589954 | 38.8 | 1509 | 1424 | ABD | s1a-m1-i1 | -/B | |
| 52 | ? | hpEastAsia hspEAsia | 1568826 | 38.9 | 1496 | 1383 | (A/B)(D/B)D | (s1a)-m1-i1 (f) | -/B | |
| Shi470 | Gastritis | hpEastAsia hspAmerind | 1608548 | 38.9 | 1517 | 1401 | AB(D/C),CC(g) | s1b-m1-i1 | -/B | [21] |
| v225d | Gastritis | hpEastAsia hspAmerind | 1588278, 7326 | 39.0 | 1506 | 1377 | AB(C/D)(C/D), (tr) (g,h) | s1a-m1-i1 | -/B | [22] |
| Cuz20 | ? | hpEastAsia hspAmerind | 1635449 | 38.9 | 1527 | 1364 | AB(D/C)×5(tr) (h) | s1a-m2-i2 | -/A | |
| Sat464 | ? | hpEastAsia hspAmerind | 1629557, 8712 | 38.9 | 1465 | 1376 | AB(D/C) | s1b-m1-i1 | -/B | |
| PeCan4 | Gastric cancer | hpEastAsia hspAmerind? | 1560342, 7228 | 39.1 | 1525 | 1388 | A(B/A)BC | s1a-m1-i1 | -/B | |
| 26695 | Gastritis | hpEurope | 1667867 | 38.9 | 1575 | 1411 | ABC | s1a-m1-i1 | A/- | [28] |
| HPAG1 | Gastritis | hpEurope | 1596366, 9370 | 39.1 | 1492 | 1394 | A(B/A)C | s1b-m1-i1 | B/- | [30] |
| G27 | ? | hpEurope | 1652982, 10031 | 38.9 | 1560 | 1400 | ABCC | s1b-m1-i1 | B/- | [56] |
| P12 | Duodenal ulcer | hpEurope | 1673813, 10225 | 38.8 | 1593 | 1396 | ABCC | s1a-m1-i1 | A/- | [49] |
| B38 | MALT lymphoma | hpEurope | 1576758 | 39.2 | 1493 | 1388 | - | s2-m1-i2 | A/- | [51] |
| B8(i) | Gastric ulcer(i) | hpEurope | 1673997, 6032 | 38.8 | 1578 | 1385 | ABC | s1a-m2-i2 (j) | A/A | [57] |
| SJM180 | Gastritis | hpEurope? | 1658051 | 38.9 | 1515 | 1381 | ABC | s1b-m1-i1 | B/B | |
| J99 | Duodenal ulcer | hpAfrica1 hspWAfrica | 1643831 | 39.2 | 1502 | 1383 | (A/B)C | s1b-m1-i1 | A/B | [2] |
| 908(k) | Duodenal ulcer | hpAfrica1 hspWAfrica | 1549666 | 39.3 | 1503 | 1393 | ABC | -s1b-(-)-i1 (j,k,l) | -/-(k) | [139] |
a) The first number is the length of the chromosome and the second number (when present) is that of the plasmid.
b) Accession numbers are as follows: F57 [DDBJ:AP011945.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011945.1], F32 [DDBJ:AP011943.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011943.1, DDBJ:AP011944.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011944.1], F30 [DDBJ:AP011941.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011941.1, DDBJ: AP011942.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011942.1], F16 [DDBJ:AP011940.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011940.1], 51 [GenBank:CP000012.1], 52 [GenBank:CP001680.1], Shi470 [GenBank:NC_010698.2], v225d [GenBank:CP001582.1, GenBank:CP001583.1], Cuz20 [GenBank:CP002076.1], Sat464 [GenBank:CP002071.1, GenBank:CP002072.1], PeCan4 [GenBank:NC_014555.1, GenBank:NC_014556.1], 26695 [GenBank:NC_000915.1], HPAG1 [GenBank:NC_008086.1, GenBank:NC_008087.1], G27 [GenBank:NC_011333.1, GenBank:NC_011334.1], P12 [GenBank:NC_011498.1, GenBank:NC_011499.1], B38 [GenBank:NC_012973.1], B8 [GenBank:NC_014256.1, GenBank:NC_014257.1], SJM180 [GenBank:NC_014560.1], J99 [GenBank:NC_000921.1], 908 [GenBank:CP002184.1]. Draft sequence of the East Asian strain 98-10 [140]. 98-10, [GenBank:NZ_ABSX01000001.1] - [GenBank:NZ_ABSX01000051.1].
c) Letters in parentheses are the hybrid EPIYA segment. For example, (A/B) is a hybrid of EPIYA-A and EPIYA-B segments [21,22,141].
e) Designated as homE as it was very different from homA or homB.
f) "s" region locates outside of the ORF.
g) A second cagA gene between cagM and cagP.
h) (tr), truncation.
i) Mongolian gerbil-adapted, originally from gastric ulcer.
j) vacA gene is split.
k) According to a reference [139], the sequence might not represent a complete genome, although it is deposited as a complete circular genome in GenBank.
l) "m" region was not available because of a deletion in the center of the ORF.
Japanese/Korean core genomes diverged from the European and then the Amerind
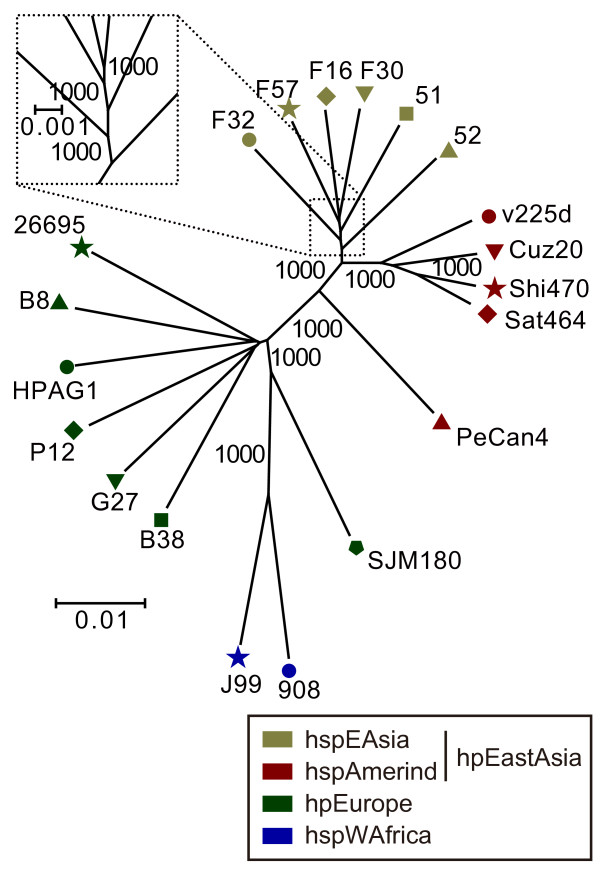
A phylogenetic tree was constructed from concatenated seven genes atpA, efp, mutY, ppa, trpC, ureI and yphC, which were used for multi-locus sequence typing (MLST) [18] and phylogenetic analyses [19,20]) (Additional file 1 (= Figure S1)). The tree showed that the 6 East Asian strains, the 4 Japanese strains (F57, F32, F30 and F16) and the 2 Korean strains (strain 51 and strain 52), are close to the known subpopulation hspEAsia of hpEastAsia, whereas 4 strains (Shi470 [21], v225d [22], Sat464 and Cuz20) are close to another subpopulation of hpEastAsia, hspAmerind. Strains 26695, HPAG1, G27, P12, B38, B8 and SJM180 were assigned to hpEurope. Strains J99 and 908 were assigned to hspWAfrica of hpAfrica1. PeCan4 was tentatively assigned to hspAmerind although it appears to be separate from the above 4 hspAmerind strains and somewhat closer to other subgroups (a subgroup of hpEurope, hspMaori and a group of "unclassified Asia" in the HpyMLST database [18]).
We deduced the common core genome structure of these 20 genomes based on the conservation of gene order using CoreAligner [23] (Table 1). CoreAligner determines the set of core genes among the related genomes not by universal conservation of genes but by conservation of neighborhood relationships between orthologous gene pairs allowing some exceptions. As a result, CoreAligner identified different numbers of core genes among strains (1364-1424), which reflect deletion, duplication and split of the core genes in the individual strains.
For phylogenetic analysis among the strains, we further extracted 1079 well-defined core orthologous groups (OGs) as those that were universally conserved, non-domain-separated, and with one-to-one correspondence (see Methods). The concatenated sequence of all well-defined core OGs resulted in a well-resolved phylogenetic tree (Figure 1). The tree was composed of two clusters, one containing the Japanese, Korean and Amerind strains and the other containing the European and West African strains. The tree strongly supported a model in which the Japanese/Korean strains (hspEAsia) and the Amerind strains (hspAmerind) diverged from their common ancestor, which in turn diverged from the ancestor shared by the European strains (hpEurope) long before. This conclusion is robust, as shown by the high bootstrap values of the internal nodes, primarily because the tree is composed of a large quantity of sequence information with approximately 1400 genes. The Japanese and Korean strains were not separated into two clusters. PeCan4 appeared diverged from the other four hspAmerind strains as expected from the result of the phylogenetic analysis based on the 7 genes described above. SJM180 appeared diverged from the other hpEurope strains in the well-defined core gene-based tree.
Figure 1.
Phylogenetic tree of 20 H. pylori strains based on their well-defined core genes. Well-defined core OGs were used for neighbor-joining method (see Methods). Numbers indicate bootstrap values. Scale bar indicates substitutions per nucleic acid residue (change/nucleotide site). The assignment of population/subpopulation was based on a phylogenetic tree constructed from the concatenated alignment of fragments of seven genes used in the H. pylori MLST database (atpA, efp, mutY, ppa, trpC, ureI and yphC) [18]. Classification of population/subpopulation was as described [10,19].
Phylogenetic profiling to identify gene contents of hspEAsia
To thoroughly characterize the gene contents specific to the Japanese/Korean (hspEAsia) strains, we conducted phylogenetic profile analysis using the DomClust program [24]. This analysis determines the presence or absence of a domain, rather than a gene, and allows detection of split genes, partially deleted genes and partially duplicated genes (detailed in Methods). Their features will be explained in the next five sections.
Differences in outer membrane proteins and related proteins in the number of loci of gene families and in alleles at each locus
One of the emerging features of the East Asian (hspEAsia) strains is the change in the number of loci of some of the outer membrane protein (OMP) families. We detected five OMP genes (gene families; oipA, hopMN, sabAB, babABC and vacA-2) with the number of loci different between the hspEAsia and hpEurope strains (Table 2). In all but one gene family, the difference in the number of locus was the result of gene decay in the East Asian (hspEAsia) strains.
Table 2.
Characteristic gene contents of East Asian (hspEAsia) H. pylori
| Population type | Strain | Locus of outer membrane proteins | Periplasmic endonuclease nucG(d) | Molybdenum- related function | ||||
|---|---|---|---|---|---|---|---|---|
| oipA/oipA-2 | hopM/hopN | babA/babB/babC(a) | sabA/sabB(b) | vacA-2(c) | ||||
| hspEAsia | F57 | A/A(e) | +/- | A/B/- | A/- | x | x | - |
| F32 | A/x | +/- | A/B(tr)/- | A/- | x | x | - | |
| F30 | A/A | +/- | A/B/- | A/- | x | x | - | |
| F16 | A/A | +/- | A/B/- | A/- | x | x | - | |
| 51 | A/A | +/- | A/B/- | A/- | + | x | - | |
| 52 | A/A | +/- | A/B/- | A/A | x | x | - | |
| hspAmerind | Shi470 | A/x | +/- | A/B/- | A/- | + | + | + |
| v225d | A/x | +/- | A/B/- | A/- | + | + | + | |
| Cuz20 | A/x | +/- | A/B/- | A/- | + | + | + | |
| Sat464 | A/x | +/- | A/B/- | A/- | + | + | + | |
| PeCan4 | A/A | +/- | A/B/- | A/B | + | + | + | |
| hpEurope | 26695 | A/- | +/+ | B/A/C | A/A | + | x | + |
| HPAG1 | A/- | x/+ | A/C/B | A/B | + | + | + | |
| G27 | A/- | +/x | C/B/A | A/B | + | + | + | |
| P12 | A/- | +/+ | A/B/B(tr) | B/B | + | + | +?(f) | |
| B38 | A/- | +/+ | A/A(tr)/- | A/- | + | + | + | |
| B8 | A/- | +/+ | A/A/- | A/Q(g) | + | + | + | |
| SJM180 | A/- | +/+ | B/C/A | A/B | + | + | + | |
| hspWAfrica | J99 | A/- | +/+ | A/B(tr)/- | A/B | + | + | + |
| 908(h) | A/- | -/-(h) | A(tr)/B(tr)/-(h) | -/-(h) | + | + | + | |
+, present; x, disrupted (nucleotide sequence partly remained); -, absent. See Additional file 2 (= Table S1) for a detailed list.
a) babA locus corresponds to HP0896; babB locus, HP1243; babC locus, HP0317.
b) sabA locus corresponds to jhp0662; sabB locus, jhp0659.
c) Paralog of vacA (HP0289), but not vacA itself (HP0887). Another paralog vacA-4 (HP0922) is in Table 6.
d) HP1382.
e)/, different loci.
f) One of 12 molybdenum-related genes was truncated.
g) hopQ gene. Two hopQ copies exist, one at sabB locus and the other, as in other strains, at the hopQ locus.
h) From the description of the reference [139], the sequence might not represent a complete genome, although it is deposited as a complete circular genome in GenBank. Hence, care should be taken in interpreting the results.
Relevant information about each family from draft sequence of the Japanese strain 98-10 (NZ_ABSX01000001.1- NZ_ABSX01000051.1) [143] are as follows: oipA/oipA-2, with at least one copy, although the exact copy number cannot be determined because of a short contig encoded only the oipA gene but not the flanking region; hopM locus, +? (partial sequence at an end of the contig); hopN locus, not applicable because it was at an end of contigs (hopN fragment is deposited but the sequence was partial at both ends of the contig, preventing locus assignment); babA/babB/babC, A?/?/? (babA at babA locus but partial at an end of the contig; babB and babC loci, not applicable because they were at ends of contigs; babB sequence was partial at both ends of the contig, preventing locus assignment); sabA/sabB, +/-; vacA-2, x; nucG split as in the other hspEAsia strains; Molybdenum-related function, x.
The notable exception was oipA, for which a secondary locus was found in hspEAsia (6/6 strains) and hspAmerind (5/5), but not in hpEurope (0/7) or hspWAfrica (0/2). This increase of the secondary locus can be explained by a novel DNA duplication mechanism associated with inversion [25]. The two hopMN loci in hpEurope (7/7 strains) and hspWAfrica (1/2) were reduced to one locus in the hspEAsia (6/6) and hspAmerind (5/5). This loss was likely caused by the same duplication mechanism [25].
For the babABC family, the babC locus [26] was empty in all the hpEastAsia strains (6/6 hspEAsia and 5/5 hspAmerind) as well as from all the hspWAfrica strains (2/2) and two hpEurope strains (B38 and B8). This is in contrast to the presence of three loci in the other (5/7) European strains (Table 2).
The strain J99 carried a sabA gene (jhp0662) at the sabA locus and a sabB gene (jhp0659) at the sabB locus [27]. All the hpEurope strains but the strain B38 (6/7) and this hspWAfrica strain (J99) had these two loci, whereas all the hpEastAsia strains but the strains 52 and PeCan4 (5/6 hspEAsia and 4/5 hspAmerind) lacked sabB locus (Table 2). These hpEastAsia strains all carried a sabA gene at the sabA locus. Genes of hpEurope differed among strains. Three strains (HPAG1, G27 and SJM180) carried a sabA gene at the sabA locus and a sabB gene at the sabB locus, as J99. The strain 26695 carried a sabA gene at both the sabA and sabB loci, whereas the strain P12 carried a sabB gene at both the loci. The strain B8 carried a sabA gene at the sabA locus and a hopQ gene at the sabB locus, along with another hopQ gene at the hopQ locus.
Some of these genes (oipA, babA and babB) and homAB genes were previously reported to diverge between the East Asian and Western strains [13,14,17]. Difference in the number of copies of homAB genes between East Asian and Western strains was reported [17].
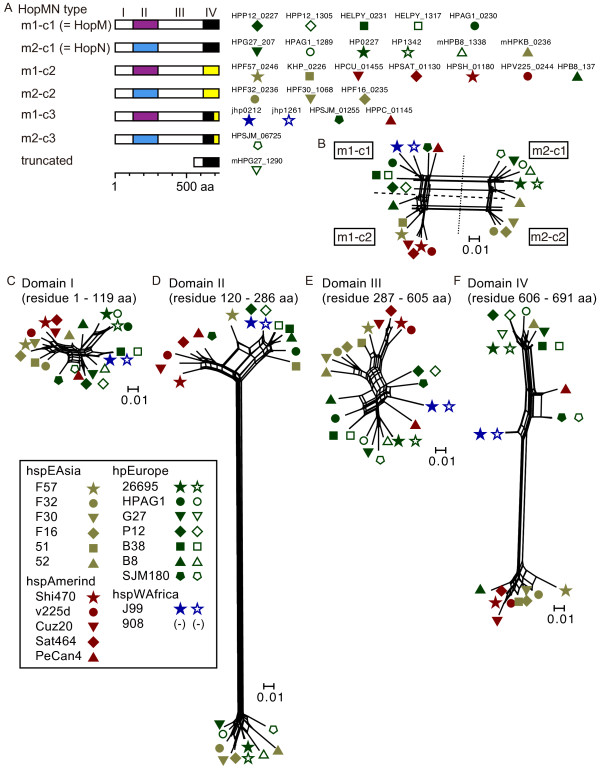
For hopMN, two gene types (hopM and hopN) have been recognized [26,27]. Phylogenetic network analysis revealed two variable regions within the hopMN family (region II and IV; Figure 2). Combining the two types of two variable regions defined four main gene types, of which two corresponded to hopM and hopN. The two types in region II were designated m1 and m2 (m for mid). The types in region IV were designated c1 and c2 (c for C-terminus); c3 was another variant type in region IV, composed of parts of c1 and c2. In this designation, previous hopM and hopN genes correspond to hopMNm1-c1 and hopMNm2-c1, respectively. All hpEastAsia strains except the strains 52 and PeCan4 (9/11) carry sequence type c2 at region IV. The c3 variant is observed in J99, PeCan4 and SJM180 (Figure 2A and 2F).
Figure 2.
East Asia-specific sequence at the C-terminus of the putative product of hopMN. (A) Four types of hopMN genes. Type c3 of m1-c3 and m2-c3 is composed of parts of c1 and c2. The c1-m1 and c2-m1 types correspond to hopM and hopN, respectively. (B) Phylogenetic network of whole region of proteins. Types m1-c3 and m2-c3 cannot be clearly distinguished from m1-c1 and m2-c1 in this figure. (C)-(F) Phylogenetic networks for the four domains. Scale bar indicates substitutions per amino acid residue (change/amino-acid site). Positions are for HP0227 of strain 26695.
Three vacA paralogs and vacA itself were found in 26695 [28]. Those paralogs share the auto-transporter domain at the C-terminus with vacA [28]. A large deletion in vacA-2 (HP0289) (approximately 2400 amino acids) was found in all the hspEAsia strains except the strain 51 (5/6) (Table 2 and Additional file 2 (= Table S1)).
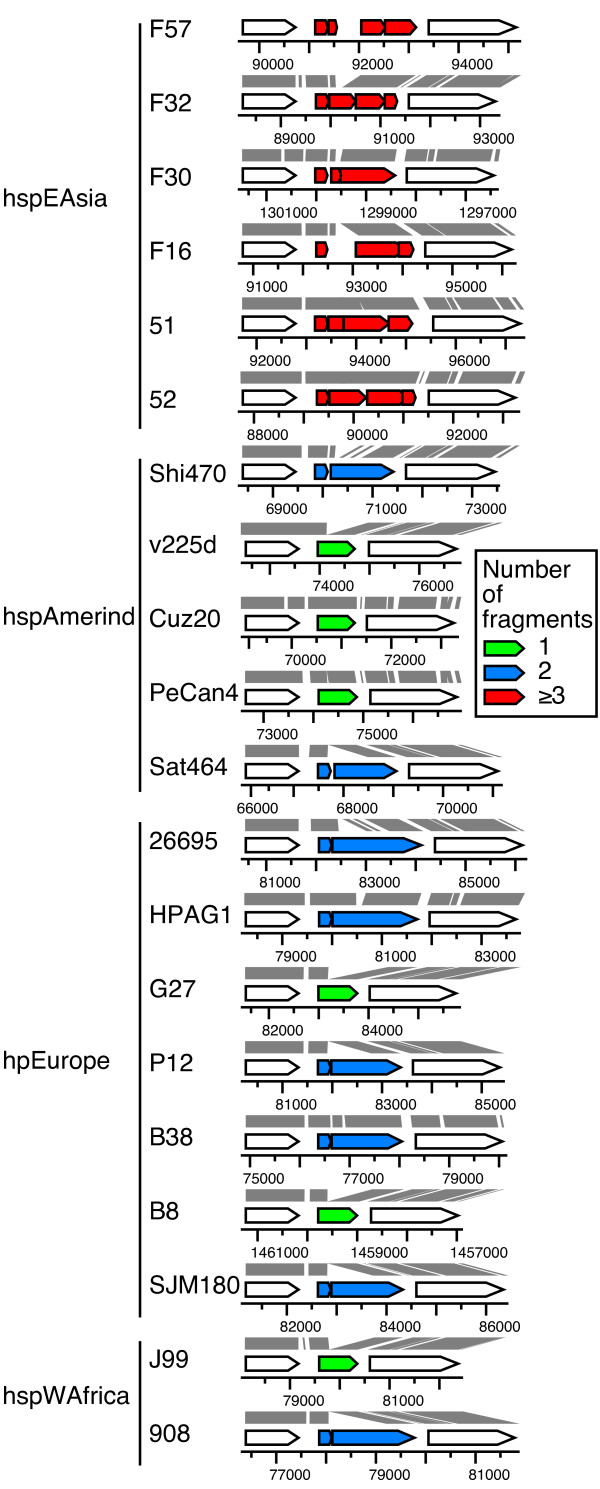
It was described earlier that horA OMP locus in 26695 is composed of two open reading frames (ORFs) (HP0078/HP0079) whereas that in J99 is composed of one ORF (jhp0073) [27]. The horA locus in all the hspEAsia strains shows apparent gene decay by fragmentation through various mutations (Figure 3). Whether the genes in the other strains are functional is not known.
Figure 3.
Fragmentation of horA OMP gene through various mutations in the hspEAsia strains. Genes homologous to horA in J99 (jhp0073) are classified by the number of ORFs. Numbers indicate coordinates on the genome sequence. Nucleotide similarity between each pair of strains is indicated by gray parallelogram. The state in strain 98-10 is: two ORFs.
A putative periplasmic endonuclease gene (nucG, HP1382) was split in all the hspEAsia strains examined (Table 2 and Additional file 2 (= Table S1)). Detailed analysis revealed that the split was mediated by recombination between short similar sequences [25].
Massive decay of molybdenum-related genes for two-electron reduction-oxidation reactions
Unexpectedly, our profiling suggested that functions related to molybdenum (Mo) were lost specifically in the hspEAsia strains (Table 3 and Additional file 2 (= Table S1)). The trace element Mo is essential for nearly all organisms [29]. After transport into the cell as molybdate, it is incorporated into metal cofactors for specific enzymes (molybdo-enzymes) that catalyze reduction-oxidation (redox) reactions mediated by two-electron transfer.
Table 3.
Decay of molybdenum-related genes
| Type | hspEAsia | hspAmerind | hpEurope | hspWAfrica | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | F57 | F32 | F30 | F16 | 51 | 52 | (a) | (b) | P12 | (c) |
| Molybdenum (MoO42-) transport | ||||||||||
| modA | x | x | x | + | + | x | + | + | + | + |
| modB | x | + | + | + | x | x | + | + | + | + |
| modC | x | x | x | x | x | + | + | + | + | + |
| Molybdenum cofactor synthesis | ||||||||||
| moaA | x | x | x | x | + | x | + | + | + | + |
| moaC | x | + | + | + | + | + | + | + | + | + |
| moaE | x | + | + | + | + | + | + | + | + | + |
| moaD | + | x | + | + | + | + | + | + | x | + |
| moeB | + | + | + | + | + | + | + | + | + | + |
| mogA | x | + | x | x | x | + | + | + | + | + |
| moeA | x | x | x | x | x | x | + | + | + | + |
| mobA | + | + | + | + | + | x | + | + | + | + |
| Molybdenum cofactor-containing enzyme | ||||||||||
| bisC | x | x | x | x | x | x | + | + | + | + |
+, present; x, disrupted (nucleotide sequence remained).
a) Strains Shi470, v225d, Cuz20, Sat464 and PeCan4.
b) Strains 26695, HPAG1, G27, B38, B8 and SJM180.
c) Strains J99 and 908
The states in strain 98-10 are: x for modA, modB, mobA, moaA, moeB and bisC; + for modC, moaD, moaE, mogA, moaC and moeA.
In the 20 H. pylori genomes, the only gene for molybdo-enzymes identified was bisC. At least one gene in each of the three Mo-related functions, Mo transport, Mo cofactor synthesis and a Mo-containing enzyme, decayed in all hspEAsia strains (Table 3 and Figure 4). Detailed analysis of nucleotide sequences revealed a mutation in 10 of 12 Mo-related genes in some of the hspEAsia strains (Table 3 and Additional file 3 (= Table S2)). The occurrence of apparently independent multiple mutations (Additional file 3 (= Table S2)) suggests some selection against use of Mo in the hspEAsia strains. All other strains but P12 possessed all intact genes. The strain P12 had a truncation of moaD (Additional file 3 (= Table S2)). Tungsten sometimes substitutes for Mo, but genes for known tungstate/molybdate binding proteins (TupA and WtpA) were not found in the H. pylori genomes.
Figure 4.
Decay of Mo-related genes in the hspEAsia strains. Mo-related genes are indicated by color. Homologs are indicated by the same color. See Additional file 3 (= Table S2) for nucleotide sequences.
The sequences in the four Japanese strains were confirmed by polymerase chain reaction (PCR) with the primers listed in the Additional file 4 (= Table S3).
The Mo-related genes were in a list of "chronic gastritis-associated" genes [30], primarily because they are absent from three Amerind strains from the Athabaskan people [31]. The 5 Amerind strains analyzed in the present study are different from the three Amerind strains in this respect. This difference could reflect the later migration of the Athabaskans to the Americas [32].
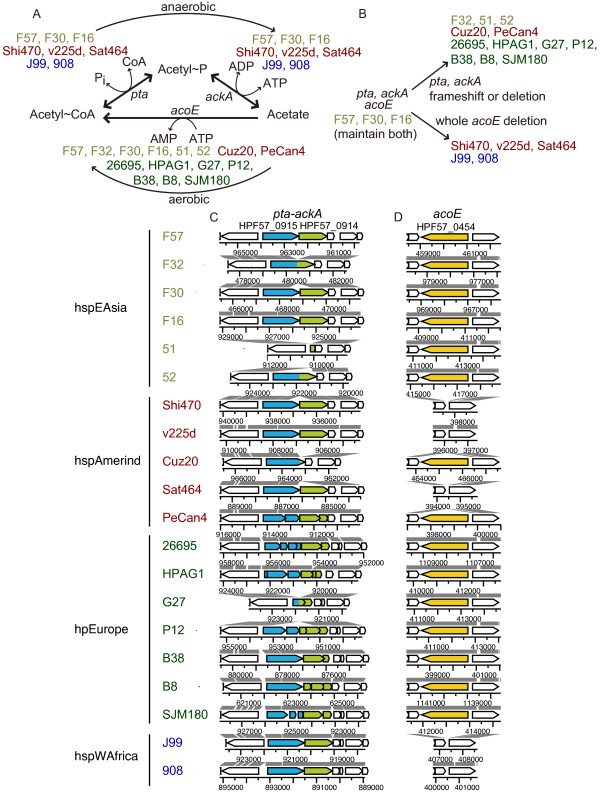
Two pathways between acetyl~CoA and acetate in some Japanese strains
Our profiling revealed an important change at the center of energy and carbon metabolism related to acetyl~CoA. Two pathways connect acetyl~CoA and acetate (Figure 5A). In anaerobic fermentation, acetyl~CoA is converted into acetate by phosphoacetyl transferase (pta product) and acetyl kinase (ackA product) with generation of ATP (anaerobic pta-ackA pathway) [33]. The intermediate acetyl~P, a high-energy form of phosphate, likely serves as a global signal. Although these reactions are reversible, assimilation of acetate may be irreversibly mediated by acetyl~CoA synthetase (acoE product) by the generation of acetyl~CoA, which enters the TCA cycle to generate energy under aerobic conditions (aerobic acoE pathway).
Figure 5.
Variation in genes connecting acetyl-CoA and acetate. (A) Functional states of three genes in two pathways inferred for 20 strains. (B) Reconstruction of pathway evolution. (C) Genome comparison for the pta-ackA region. (D) Genome comparison for the acoE region. Homologs are indicated by the same color in (C) and (D). The states in strain 98-10 are: pta+ ackA+/acoE+ as F57.
It has been suggested that strain 26695 (hpEurope) carries a mutation in pta for the former pathway whereas strain J99 (hspWAfrica) lacks acoE for the latter [28,34]. All European strains in this study (7/7) had at least one inactivated pta and ackA gene through a variety of mutations (Figure 5C). Two of five Amerind strains, PeCan4 and Cuz20, also had a mutated pta and ackA, whereas the other 3/5 Amerind, 2/2 African, and 3/6 hspEAsia strains had a pta and ackA intact but had a deletion of acoE. Exceptions to such apparent incompatibility between the two pathways were found for 3/4 of the Japanese strains (F16, F30 and F57), which had intact genes for both pathways (Figure 5BCD). The sequences in the four Japanese strains were confirmed (see Methods and Additional file 4 (= Table S3)).
A gene for an amino acid utilization
An ortholog of jhp0585 in J99 is absent from 26695 [2]. An ortholog is present in the six other hpEurope strains and both hspWAfrica strains, but absent from all hpEastAsia strains (hspEAsia and hspAmerind) (Additional file 2 (= Table S1)). It encodes a homolog of 3-hydroxy-isobutyrate dehydrogenase and the related beta-hydroxyacid dehydrogenase (COG2084). The 3-hydroxy-isobutyrate dehydrogenase degrades the branched-chain amino acid valine. H. pylori requires branched amino acids for growth. It is not known what the substrates or products of reactions catalyzed by this gene product are, or the biological relevance of its distribution.
Gene contents unique to other groups
Phylogenetic profiling involving four groups (6 hspEAsia, 5 hspAmerind, 7 hpEurope, and 2 hspWAfrica strains) (Additional file 2 (= Table S1), second sheet) revealed the following group-specific genes:
(i) tas (HP1193) for aldo-ketoreductase was present in all hpEurope strains except one (HPAG1) and one hspWAfrica strain (J99), but was absent from all hpEastAsia strains (hspEAsia and hspAmerind). Aldo-keto reductases (AKRs) constitute a large protein superfamily of mainly NAD(P)-dependent oxidoreductases involved in carbonyl metabolism [35]. This gene is fragmented in H. acinonychis strain Sheeba [36].
(ii) homB encoding an outer membrane protein was present in all but two (B8 and SJM180) hpEurope strains (5/7) but absent from the others. This result is in agreement with an earlier study [17].
(iii) trl was detected in all hpEastAsia (hspEAsia and hspAmerind) strains and 2/7 hpEurope strains (26695 and HPAG1). It is present between tRNA(Gly) and tRNA(Leu), and co-transcribed with tRNA(Gly) [37]. It is found in roughly half the clinical isolates in Ireland [37]. Its homologs are present at two loci in 26695 [38].
(iv) A part of xseA for Exonuclease VII large subunit was duplicated in all the hspAmerind strains but the strain PeCan4. Escherichia coli exonuclease VII degrades single-stranded DNA and contributes to DNA damage repair and methyl-directed DNA mismatch repair to avoid mutagenesis [39-41]. This part of xseA was present in the neighbor of 3 other genes in these hspAmerind strains. These 4 genes may form a genomic island.
(v) IS606 transposase gene was present in all hspAmerind and hspWAfrica strains, and one hpEurope (26695) strain, but was absent from the others.
(vi) Most of fecA-2 gene, a fecA paralog, was deleted in the hspAmerind strains. The fecA gene, for Iron (III) dicitrate transport protein, is important under aerobic conditions [42]. There are several links between iron metabolism and oxidative stress defense in H. pylori [43].
(vii) The hopZ OMP gene was split in the hspAmerind strains. The hopZ gene is involved in adhesion [44].
(viii) The hopQ OMP gene decayed in the hpEastAsia strains (hspEAsia and hspAmerind). This observation agrees with an earlier work [45].
(ix) H. pylori can ferment pyruvate to ethanol via an alcohol dehydrogenase [46]. Duplication of the alcohol dehydrogenase gene as in J99 (jhp1429) [2] was seen only in the two hspWAfrica strains (J99 and 908).
Prophage-related genomic islands and other mobile elements
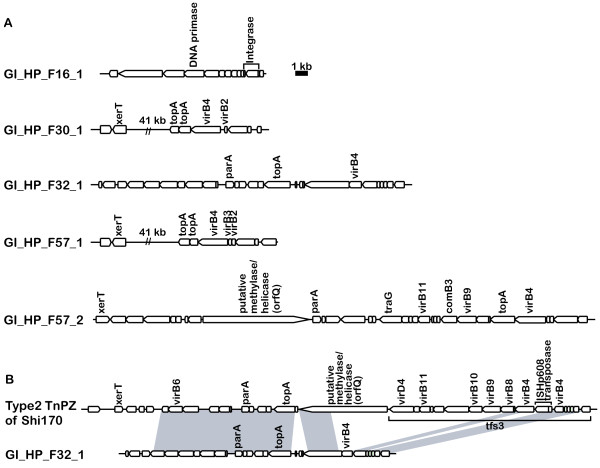
Except for the cag pathogenicity island (cagPAI), five genomic islands (GIs) were identified in the genomes of the four Japanese strains (Table 4, Figure 6 and Figure 7). In F32, the cagPAI was flanked by a 44-bp direct repeat, which extended the 22-bp sequence found in the other strains (Table 4). This length of sequence identity would allow homologous recombination [47] leading to the excision of cagPAI flanked by the repeat.
Table 4.
Genomic islands in the four Japanese H. pylori strains
| Strain | GI number | Type | Length | Start-end | Flanking repeat (bp) | Secretion system | Left gene (annotation) | Right gene (annotation) |
|---|---|---|---|---|---|---|---|---|
| F16 | GI_HP_F16_1 | prophage-like | 12245 | 471964 - 484208 (HPF16_0465 - HPF16_0478) | N/D(a) | N/D(a) | HPF16_0464 (IS605 transposase (tnpB)) | HPF16_0479 (typeIIR) |
| GI_HP_F16_2 | cagPAI | 36761 | 871413 - 834651 (HPF16_0834 - HPF16_0810) | 22(b) | Type IV | HPF16_0835 (hypothetical protein) | HPF16_0809 (glutamate racemase) | |
| F30 | GI_HP_F30_1 (left) | type 1b TnPZ partial | 7246 | 1280406 - 1287651 (HPF30_1205 - HPF30_1211) | N/D(a) | tfs3b partial | HPF30_1204 (outer membrane protein horB) | HPF30_1212 (rpoD) |
| GI_HP_F30_1 (right) | type 1b TnPZ partial | 1655 | 1237267 - 1238921 (HPF30_1166 - HPF30_1167) | N/D(a) | N/D(a) | HPF30_1165 (hypothetical protein) | HPF30_1168 (5'-methylthioadenosine/S -adenosylhomocysteine nucleosidase) | |
| GI_HP_F30_2 | cagPAI | 37153 | 867993 - 830839 (HPF30_0803 - HPF30_0778) | 22(c) | Type IV | HPF30_0804 (hypothetical protein) | HPF30_0777 (glutamate racemase) | |
| F32 | GI_HP_F32_1 | type 2 TnPZ partial | 24283 | 1058236 - 1082518 (HPF32_0988 - HPF32_1014) | N/D(a) | tfs3 partial | HPF32_0987 (hypothetical protein) | HPF32_1015 (hypothetical protein) |
| GI_HP_F32_2 | cagPAI | 36609 | 534488 - 571096 (HPF32_0500 - HPF32_0524) | 44(d) | Type IV | HPF32_0499 (hypothetical protein) | HPF32_0525 (glutamate racemase) | |
| F57 | GI_HP_F57_1 (left) | type 1b TnPZ partial | 7246 | 103791 - 111036 (HPF57_0102 - HPF57_0109) | N/D(a) | tfs3b partial | HPF57_0101 (RNA polymerase sigma factor RpoD) | HPF57_0110 (hypothetical protein) |
| GI_HP_F57_1 (right) | type 1b TnPZ partial | 1625 | 152699 - 154323 (HPF57_0147 - HPF57_0148) | N/D(a) | N/D(a) | HPF57_0146 (5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase) | HPF57_0149 (hypothetical protein) | |
| GI_HP_F57_2 | type 1 TnPZ | 38991 | 284353 - 323343 (HPF57_0279 - HPF57_0311) | 8(e) | tfs3b partial | HPF57_0278 (typeIIM) | HPF57_0312 (type II DNA modification enzyme ) | |
| GI_HP_F57_3 | cagPAI | 36797 | 562215 - 599011 (HPF57_0550 - HPF57_0575) | 22(c) | Type IV | HPF57_0549 (hypothetical protein) | HPF57_0576 (glutamate racemase) | |
a) N/D, not detected
b) TTATAATTTGAGCCATTATTTA
c) TTTCAATTTGAGCCATTCTTTA
d) TTATAATTTGAGCCATTCTTTAGCTTGTTTTTCTAGCCAAACCA
e) ACATTCTT
Figure 6.
GIs inserted into restriction-modification systems. (A) Insertion of a prophage-like GI (GI_HP_F16_1) into a restriction-modification system. (B) Insertion of a GI into a modification gene. (See Table 4 for detail).
Figure 7.
GIs detected in Japanese H. pylori strains. (A) GIs. (B) Decay of type 2 TnPZ in F32 strain inferred from comparison to the Shi170 strain. The sequence of Type 2 TnPZ of Shi170 is deposited under the accession number [GenBank:EU807988] [48] (Table 4).
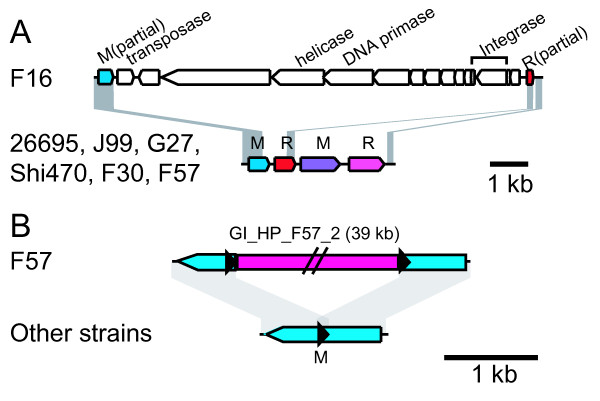
A GI found in strain F16 lacked similarity to known GIs of H. pylori whereas the other four GIs were homologous to transposable elements TnPZs, as recently reported [48,49]. The GI in F16 appears to be a remnant of a prophage inserted into a restriction-modification system (Figure 6A). It is homologous to the 5'-half of the Hac II prophage found in H. acinonychis Sheeba. The F16 GI appeared to have lost its 3'-half, presumably through deletion mediated by the inserted IS605 copy. The GI included putative phage integrase genes (HPF16_0475 and HPF16_0476) that suggest the mobility of this region, and a DNA primase gene (HPF16_0468). The gene (HPF16_0469) next to the DNA primase gene had weak sequence similarity to a putative phage helicase gene (ORF35 of bacteriophage phi3626, e-value 5e-5 by TBLASTN against phage nucleotide database), which can be assumed to be the primase-helicase system found in several bacteriophages such as T3, T4, T7 and P4 [50]. Recently, a partial Hac II prophage region was reported for another H. pylori strain [51].
The other four GIs in the other three strains had sequence similarity to TnPZs [48]. One GI in F57 was entirely homologous to the type 1 TnPZ inserted into the coding region for a DNA methyltransferase with 8-bp target duplication (5' ACATTCTT) (Figure 6B). The GI in F32 appeared to have been deleted by a type 2 TnPZ (Figure 7B). Among the Korean strains, a Type 2 TnPZ was observed only in strain 51.
The plasmid in F32 (pHPF32) was similar in sequence to known theta replication plasmids with a RepB family (Rep_3 superfamily) replication protein and R3 iterons [52-54].
The plasmid in F30 (pHPF30) was similar to a group of previously characterized H. pylori plasmids such as pHel4 in H. pylori [52,55]. This carries genes for microcin (7-aa peptide; MKLSYRN), MccB (microcin C7 biosynthesis protein), MccC (microcin C7 secretion protein), MobBCD (for plasmid mobilization), a replication initiator protein, and two relaxases. When compared to other related plasmids, a substitution in mobB and a deletion covering several small ORFs were seen. Homologous plasmids are found in G27 (pHPG27 [56]), P12 (HPP12 [49]), and v225d [22]. HPAG1 [30], B8 [57], PeCan4 and Sat464 carry a similar plasmid without the MccBC genes.
Insertion sequences (ISs) were searched for in the Japanese strains using GIB-IS [58]. An apparently intact known IS was detected in two strains: IS607 in F16; IS605 in F32.
Divergence of genes between the East Asian (hspEAsia) and the European (hpEurope) strains
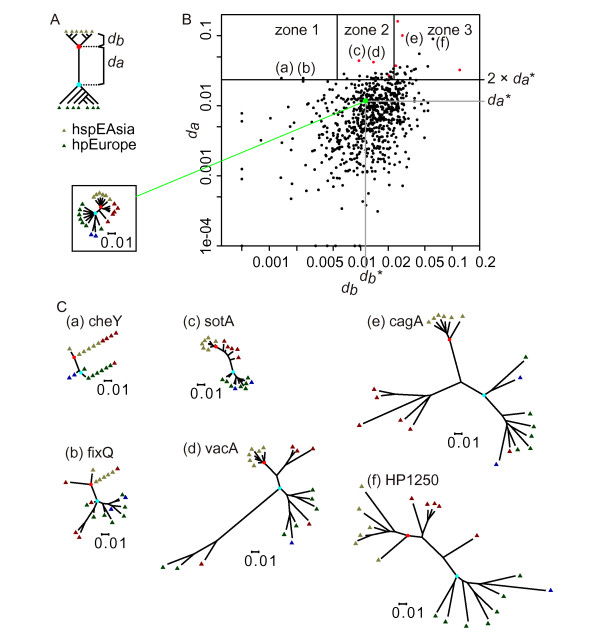
We systematically examined the amino acid-based phylogenetic trees of the orthologous genes (gene families) common to the six hspEAsia genomes and the seven hpEurope genomes. Trees of 687 OGs were selected with genes of the hspEAsia strains forming a sub tree with no genes of the hpEurope strains and vice versa. Each of the orthologs was plotted according to two distance parameters: da for the hspEAsia-hpEurope divergence and db for intra-hspEAsia divergence (Figure 8A). An hspEAsia-hpEurope divergence greater than twice that of the well-defined core tree (da*) was seen in 47 gene families (Table 5 and 6; genes of those orthologs in each strain are listed in Additional file 5 (= Table S4)). These genes were further divided by the intra-hspEAsia divergence (db) into zone 1 (lowest divergence), zone 2 (average divergence) and zone 3 (highest divergence) (Figure 8B). Six typical trees are depicted in Figure 8C. The cagA tree (e) (zone 3) has large da and db values and a low db/da value, primarily because of the divergence in a C-terminal region of the ORF. This region, including sequences known as EPIYA (Gln-Pro-Ile-Tyr-Ala) motif, is involved in host interaction [22,59]. The tree here is consistent with previous results [22].
Figure 8.
Genes diverged between East Asian and European strains. (A) Diagram of phylogenetic tree-based analysis. Black dots, last common ancestors of Eastern and Western strains. da, length of the branch separating the two; db, average branch length of the Eastern strains. (B) Plot of gene trees based on the two distance values. Large green dot, well-defined core tree; da*, da for the well-defined core tree; db*, db for the well-defined core tree; inset box, well-defined core tree; zone 1, db < 0.00550; zone 2, 0.00550 ≤ db ≤ 0.0231; zone 3, db > 0.0231; red dot, genes with positive selection for amino acid change and with da > 2 × da*, that is, da >0.02324; (a), cheY; (b), fixQ; (c), sotA; (d), vacA; (e), cagA; (f), HP1250. N = 692 genes. (C) Representative trees with high divergence between hspEAsia and hpEurope strains. Lowest common ancestor (LCA) of hspEAsia (red) and hpEurope (cyan).
Table 5.
Selected genes diverged between East Asian (hspEAsia) and European (hpEurope) H. pylori
| Function | Genes (classified by divergence within hspEAsia) | ||
|---|---|---|---|
| Conserved(a) | Average(b) | Diverged(c) | |
| Known virulence genes | vacA, tipα | cagA, hcpD | |
| Outer membrane proteins | oipA/oipA-2, vacA, vacA-4 | hpaA-2, homC, hopJ/hopK, horI | |
| Lipopolysaccharide synthesis (Lewis antigen mimicry) | agt | futA/futB | |
| Transport | secG, sotB, comH, cvpA | yajC | |
| Motility and chemotaxis | cheY | maf, fliT | fliK |
| Redox | fixQ | hypD, frxA, pgl, nuoF | fixS, hydE |
| Nuclease | rnhA | addA, rnhB, hsdR | |
| Protein synthesis | def, prmA, tilS | miaA | |
| Antibiotic-related | def, frxA, ftsA | ||
Table 6.
Genes diverged between East Asian and European H. pylori
| Gene | Description | Representative of the gene family(a,b,c) | Distance da(d) | Distance db(e) | db zone | Reference |
|---|---|---|---|---|---|---|
| hpaA-2 | HpaA paralog | HP0492(f) | 0.1608 | 0.0253 | 3 | [68] |
| cagA | Cag pathogenicity island protein | HP0547(f) | 0.1009 | 0.0285 | 3 | [11] |
| Bacterial SH3 domain | HP1250 | 0.0901 | 0.0615 | 3 | ||
| futA, futB | α-(1,3)-fucosyltransferase | HP0379, HP0651 | 0.0553 | 0.0436 | 3 | [15] |
| sotB | Sugar efflux transporter | HP1185 | 0.0441 | 0.0095 | 2 | |
| vacA | Vacuolating cytotoxin A | HP0887 | 0.0420 | 0.0137 | 2 | [67] |
| miaA | tRNA delta(2)-isopentenylpyrophosphate transferase | mHP1415 | 0.0373 | 0.0241 | 3 | [64,144] |
| Hypothetical protein | HPAG1_0619 | 0.0366 | 0.0540 | 3 | ||
| hcpD | Cysteine-rich protein, SLR (Sel1-like repeat) protein | HP0160 | 0.0363 | 0.0323 | 3 | [16] |
| yajC | Preprotein translocase subunit YajC | HP1551 | 0.0353 | 0.0268 | 3 | [81] |
| agt | β-1,3-N-acetyl-glucosaminyl transferase | HP1105 | 0.0338 | 0.0228 | 2 | |
| rnhB | Ribonuclease HII | mHP1323(f) | 0.0337 | 0.0398 | 3 | [103,104] |
| fliK | Flagellar hook length control | HP0906 | 0.0328 | 0.0382 | 3 | [85] |
| homC | Putative outer membrane protein | HP0373 | 0.0325 | 0.1207 | 3 | |
| hopJ,hopK | Outer membrane protein | HP0477, HP0923 | 0.0313 | 0.0357 | 3 | [27] |
| frxA | NAD(P)H-flavin oxidoreductase | HP0642 | 0.0306 | 0.0212 | 2 | [120] |
| secG | Preprotein translocase subunit SecG | mHP1255 | 0.0300 | 0.0226 | 2 | [80] |
| Hypothetical protein | HP0384 | 0.0296 | 0.0302 | 3 | ||
| tipα | Tumor necrosis factor alpha-inducing protein | HP0596 | 0.0293 | 0.0145 | 2 | [66] |
| hydE | Membrane-bound, nickel containing, hydrogen uptake hydrogenase | HP0635 | 0.0288 | 0.0252 | 3 | [92] |
| tilS | tRNA(Ile) lysidine synthase | HP0728 | 0.0286 | 0.0193 | 2 | [96,97] |
| comH | Periplasmic competence protein | HP1527 | 0.0285 | 0.0194 | 2 | [82] |
| def | Peptide deformylase | HP0793 | 0.0285 | 0.0065 | 2 | [98] |
| vacA-4 | Putative vacuolating cytotoxin-like protein | HP0922 | 0.0284 | 0.0222 | 2 | |
| hypD | Hydrogenase expression/formation protein | HP0898 | 0.0284 | 0.0169 | 2 | [91,145,146] |
| addA | Helicase | HP1553 | 0.0283 | 0.0308 | 3 | [100] |
| hsdR | Type I restriction enzyme, R protein | mHP1402 | 0.0282 | 0.0245 | 3 | |
| Hypothetical protein | mHP0174 | 0.0268 | 0.0203 | 2 | ||
| oipA,oipA-2 | Outer membrane protein OipA | HP0638 | 0.0267 | 0.0097 | 2 | [70] |
| prmA | Ribosomal protein L11 methyltransferase | HP1068 | 0.0261 | 0.0118 | 2 | [99] |
| maf | Maf family (motility accessory family of flagellin-associated proteins) homolog | HP0465 | 0.0259 | 0.0214 | 2 | [86] |
| Hypothetical protein | HP0097 | 0.0257 | 0.0207 | 2 | ||
| Hypothetical protein | HP1143 | 0.0254 | 0.0146 | 2 | ||
| cvpA | Membrane protein required for colicin V production and secretion | mHP0181 | 0.0252 | 0.0169 | 2 | [83] |
| pgl | 6-phosphogluconolactonase | HP1102 | 0.0250 | 0.0130 | 2 | |
| horI | Outer membrane protein Horl | HP1113 | 0.0248 | 0.0348 | 3 | |
| fixQ | cbb3-type cytochrome c oxidase subunit Q | mHP0146 | 0.0248 | 0.0023 | 1 | |
| Hypothetical protein | HP0150 | 0.0248 | 0.0154 | 2 | ||
| cheY | Chemotaxis effector | HP1067 | 0.0248 | 0.0014 | 1 | [84] |
| fliT | Flagellar chaperone | HP0754 | 0.0245 | 0.0138 | 2 | [84] |
| ftsA | Cell division protein | HP0978 | 0.0244 | 0.0071 | 2 | [105,106] |
| rnhA | Ribonuclease H | HP0661 | 0.0243 | 0.0217 | 2 | [103,104] |
| ilvE | Branched-chain amino acid aminotransferase | HP1468 | 0.0239 | 0.0136 | 2 | |
| fixS | Cation transport subunit for cbb3-type oxidase | HP1163 | 0.0237 | 0.0250 | 3 | [87] |
| nuoF | NADH-ubiquinone oxidoreductase chain F | HP1265 | 0.0236 | 0.0202 | 2 | |
| Putative thiol:disulfide interchange protein | HP0861 | 0.0234 | 0.0185 | 2 | ||
| Hypothetical protein | HP0806 | 0.0233 | 0.0233 | 3 |
(a) m, different assignment of start codon from the RefSeq entry in the GenBank database
(b) All paralogous genes in each orthologous group are counted.
(c) Assignments to gene families are in Additional file 5 (= Table S4).
(d) Distance between the last common ancestor of hspEAsia and the last common ancestor of hpEurope.
(e) Average of distances between the last common ancestor of hspEAsia and each hspEAsia strain.
(f) A homolog in the draft genome sequence of another East Asian strain 98-10 has been reported to be diverged from four Western strains [143]. The other genes listed as diverged in 98-10 [143], HP0806, HP0061, HP1524, HP0519 and HP1322, did not meet the criteria of this study. HP0806 was below the da threshold; for the others, the hspEAsia genes did not form a separate sub tree from hpEurope.
This tree-based analysis effectively extracted known pathogenesis-related genes (Table 5 and Table 6) as discussed below. The list also included several genes related to antibiotics. Amino acid alignments (Additional file 6) located the divergent sites. The distribution pattern of these sequences suggests a possible relationship between structure and function as detailed below for each protein. The divergence could be related to differential activity and adaptation.
The variable da for an orthologous group is expected to be sensitive to the presence of a member with an exceptional phylogeny. The strain B8, assigned to hpEurope in this work (Additional file 1 (= Figure S1)), has been adapted to a mongolian gerbil [57]. The strain SJM180, also assigned to hpEurope based on the tree of seven MLST genes (Additional file 1 (= Figure S1)), clustered with hspWAfrica strains rather than with hpEurope strains in the tree of the well-defined core genes (Figure 1). To examine robustness of the above classification into diverged genes, the same analysis was conducted using the 6 hspEAsia strains and 5 hpEurope strains excluding B8 and SJM180 (Additional file 7 (= Table S5)). These two analyses used all the 20 strains, because we expected inclusion of the hspAmerind and hspWAfrica strains may provide better classification of the sub trees. In addition to these two analyses, analysis with the 6 hspEAsia and 7 hpEurope strains or with the 6 hspEAsia and 5 hpEurope strains was carried out, which allowed assignment of a bootstrap value to the branch separating the hspEAsia and hpEurope strains. Comparison of these 4 analyses is summarized in Additional file 7 (= Table S5). The four sets of results agreed rather well, especially for those genes with larger da value: 34 among the 47 genes in Table 6 were extracted in all the 4 analyses. The bootstrap value supported the separation of hspEAsia and hpEurope well in most cases, with the bootstrap value ≥ 900 in 41 among the 47 genes.
Positively-selected amino-acid changes between the East Asian (hspEAsia) and European (hpEurope) strains
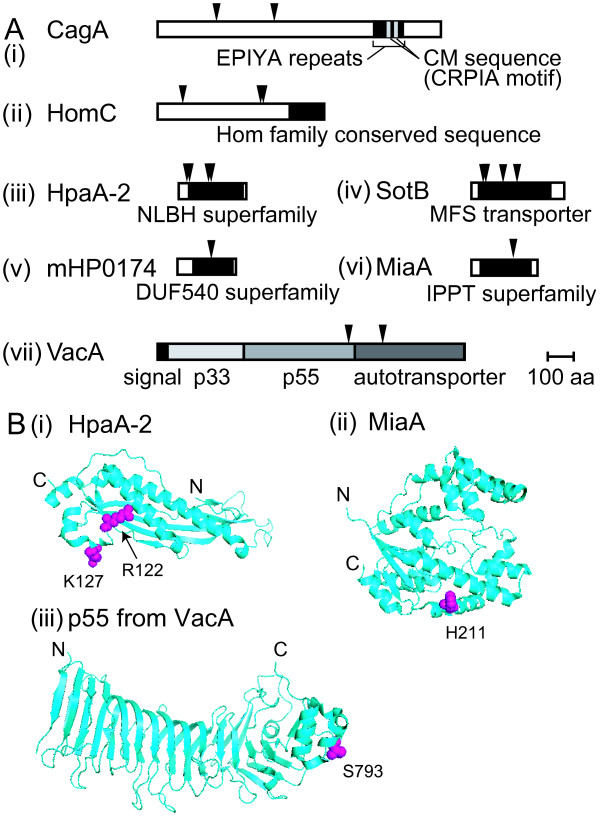
Divergence could be adaptive or neutral. We searched for sites where the hspEAsia-hpEurope changes in amino acids were positively selected [60] and found that 7 of 47 genes passed the likelihood test (Table 7; red dots in Figure 8B). These selected sites were mapped on the coding sequences (Figure 9A). For CagA, several sites were found outside the area of EPIYA segments.
Table 7.
Genes with positively selected amino-acid changes between the East Asian and the European H. pylori
| Locus tag | Gene | Description | p-value(a) | Positively selected sites (b,c) |
|---|---|---|---|---|
| HP0547 | cagA | Cag pathogenicity island protein | < 1E-21 | V238R (0.994) |
| A482Q (0.953) | ||||
| HP0373 | homC | Putative outer membrane protein | < 1E-14 | E110N (0.978) |
| K428H (0.986) | ||||
| T437D (0.979) | ||||
| HP0492 | hpaA-2 | Hpa paralog | < 1E-5 | S34V (0.970) |
| A46Q (0.993) | ||||
| R122F (0.967) | ||||
| K127S (0.962) | ||||
| HP1185 | sotB | Sugar efflux transporter protein | 0.00005 | T50S (0.956) |
| A57L (0.990) | ||||
| N134G (0.983) | ||||
| W186Y (0.980) | ||||
| mHP0174 | Hypothetical protein | 0.0007 | F144W (0.952) | |
| mHP1415 | miaA | General tRNA delta(2)-isopentenylpyrophosphate transferase | 0.0002 | H174A (0.992) |
| HP0887 | vacA | Vacuolating cytotoxin A | 0.002(d) | S793A (0.964) (d) N931A (0.960) (d) |
Figure 9.
Genes with positively selected amino acid changes between East Asian and non-East Asian strains. (A) Position of the positively selected amino-acid residues in ORF (triangles). In (i), EPIYA segments and CM sequences [138] are marked. (B) Position of positively selected amino acids in the three-dimensional structure. (i) HpaA-2 [PDB:2I9I]. (ii) E. coli MiaA [PDB:3FOZ] [61] with the residue corresponding to H174 of H. pylori MiaA. (iii) p55 fragment of VacA [PDB:2QV3] [61] (Table 7).
Three-dimensional structure was available for mapping some of the selected sites for three of these genes (Figure 9B). The three-dimensional structure of part of VacA, the p55 fragment, is determined [61]. S793A mapped on the surface of the p55 at its C-terminal region (Figure 9B). Deletion of the p55 region reduces VacA binding to cells [62], so S793A might affect cell binding of the hspEAsia and hpEurope strains. Two selected residues of HpaA-2 were mapped (Figure 9B). The residue (H211) corresponding to the selected residue H174 of H. pylori MiaA mapped to the alpha helix 10 of E. coli MiaA [63,64] (Figure 9B).
Diverged genes and possible biological significance
We explored the possible biological significance of the observed divergence in genes in Table 6 using gene and protein properties, as summarized in Table 5.
Known virulence genes
Four genes in Table 6, cagA, vacA, hcpD and tipα, are virulence genes.
CagA is introduced in the Background section and discussed above in the section "Divergence of genes between the East Asian (hspEAsia) and the European (hpEurope) strains". VacA is another important virulence protein [65]. The hcpD (HP0160) is a member of the Hcp (H. pylori cysteine-rich protein) family, which contains repeat motifs characteristic to the eukaryotic Sel1 regulatory proteins, is secreted and interacts with the host immune systems [16]. Geographical divergence and positive selection for amino acid changes in this family, including HcpD, are reported [16]. HP0596 encodes tumor necrosis factor alpha-inducing protein (Tipα), a DNA-binding protein [66]. This enters the gastric cells and induces TNF-alpha, an essential cytokine for tumor promotion.
The cagA gene is discussed above in the section "Divergence of genes between the East Asian (hspEAsia) and the European (hpEurope) strains". The vacA gene showed a qualitatively similar pattern of intra-hspEAsia divergence and overall divergence as cagA (Figure 8C (d)). The overall tree pattern was consistent with previous studies (for review, see [67]). Intra-hspEAsia divergence was large for hcpD. Positively-selected residues of cagA and vacA are described above.
Outer membrane proteins
Nine genes in Table 6 are outer membrane protein genes (Table 5).
The vacA gene is discussed above. vacA-4 is a vacA paralog. The hpaA-2 is of unknown function [68], but is a paralog of hpaA [27] which is essential for adhesion [69]. The homA/B genes are homologs of homC and known to have diverse copy number and genomic localization in Western and East Asian strains (Table 1) [17]. OipA (also known as HopH) induces IL-8 from host cells [70]. Geographical divergence of oipA has been reported [14].
The hpaA-2 showed a very large hspEAsia-hpEurope divergence (the largest da value; Figure 8B and Table 6).
Intra-hspEAsia divergence was intermediate for oipA/oipA-2 (Table 6).
The da value (hspEAsia-hpEurope divergence) of homC (0.0325) was larger than the threshold distance (Table 6). Moreover, the homC genes of all hpEastAsia and hpAfrica1 strains but the strain 52 were greatly diverged from those of the hpEurope strains and the strain 52: distance 0.1387 for this separation was comparable to the largest da values for hpaA-2 and cagA. Diverged residues were clustered in a specific region. Positively selected amino-acid changes of the putative homC product were identified (Table 7 and Figure 9).
The hopJ and hopK genes (HP0477 and HP0923) were similar within each strain but different between strains [26,27]. This earlier observation, seen for 26695, J99 and HPAG1, was confirmed with the other genomes except for 908 and B8. This similarity of hopJ and hopK genes in one strain is likely to be caused by concerted evolution by homologous interaction, possibly with selection.
The babA and alpA genes were not included in the 687 OGs that showed complete separation between genes of the six hspEAsia strains and those of the seven hpEurope strains on the phylogenetic tree. BabA binds to Lewis b antigens [71,72]. Geographic variation of BabA has been reported [13]. AlpAB proteins are necessary for specific adherence to human gastric tissue [73]. In the East Asian strains but not the Western strains, AlpA activates NF-κB-related pro-inflammatory signaling pathways [74].
The reason that the babA is not in Table 6 was mainly because babA genes of the hpEurope strains B8 and SJM180 grouped together with the hspEAsia strains (Additional file 7 (= Table S5)). The alpA in the hpEurope strain SJM180 grouped with the hspEAsia strains (Additional file 7 (= Table S5)).
Lipopolysaccharide synthesis and Lewis antigen mimicry
Three genes in Table 6, futA, futB and HP1105 (designated here as agt), are related to lipopolysaccharide (LPS) synthesis and Lewis antigen mimicry.
The lipopolysaccharides of H. pylori are important for host interaction. H. pylori can express Lewis and related antigens in the O-chains of its surface lipopolysaccharide that mimic the hosts. O-chains are commonly composed of internal Lewis X units with terminal Lewis X or Lewis Y units or, in some strains, with additional units of Lewis a, Lewis b, Lewis c, sialyl-Lewis X and H-1 antigens, as well as blood groups A and B, producing a mosaic of antigenic units [75]. The activity and specificity of the fucosyltransferases may vary between the two paralogs in one strain, as well as between the orthologs in different strains [76]. Mechanism of these changes is phase variation involving simple repeats and longer repeats [77,78]. Such diversity could be adaptive and related to differences in pathogenicity [79].
The two fucosyltransferase genes (futA = HP0379, futB = HP0651) showed large hpEurope-hspEAsia divergence (the 4th largest da value), as reported earlier [15]. Intra-hspEAsia divergence was large for them (in zone 3). HP1105 (agt) was β-1,3-N-acetyl-glucosaminyl transferase gene for LPS synthesis. Another transfereaseα-1,6-glucosyltransferase gene (HP0159 = rfaJ-1) was in the list of 6 hspEAsia - 5 hpEurope comparison (Additional file 7 (= Table S5)).
Transport
Four genes in Table 6, sotB, secG, yajC, comH and cvpA, are related to motility and chemotaxis.
The sotB gene was similar to genes for sugar efflux transporters and multi-drug resistance transporters (COG2814, TIGR00880). SecG forms the machinery for protein translocation across the cytoplasmic membrane [80]. YajC is a member of the preprotein translocase machinery, SecDF-YajC. SecDF-YajC inhibits disulfide bond formation between two SecG molecules [81]. ComH is essential for natural transformation [82]. Its putative N-terminal secretion signal suggests that it is either anchored in the cytoplasmic membrane or exported to the periplasm [82]. The cvpA gene of E. coli is suggested to encode a membrane protein required for colicin V production/secretion [83].
The secG homolog, mHP1255, showed divergence focused around residues 150-160. The nucleotide sequence AAAGAGAAG encoding Lys-Glu-Asn was present once in hpEurope and hspWAfrica strains whereas repeated 2 to 4 times in tandem in all hpEastAsia strains (4 in F16, 3 in Sat464, and 2 in the others).
Positively-selected amino-acid changes of the putative sotB product were identified (Table 7). Of these, W186Y lay at the end of a transmembrane helical region away from the substrate tranlocation pores.
Motility and chemotaxis
Four genes in Table 6, fliT, fliK, maf and cheY, are related to motility and chemotaxis.
The fliT product is a flagellar chaperone [84], whereas the fliK product controls the hook length of flagella [85]. The maf gene encodes a member of motility accessory family of flagellin-associated proteins implicated in flagellar assembly [86]. The cheY gene (HP1067) encodes a response regulator of a two-component signal transduction system regulating chemotaxis [84]. CheY does not act as a transcriptional activator. Instead, when activated, it interacts directly with the flagellar motor-switch complex, causing a clockwise rotation of the flagella that results in cell tumbling.
Intra-hspEAsia divergence was very small for cheY (Table 6 and Figure 8C (a)). It would be interesting to see whether this divergence is related to differences in chemotaxis.
Electron transfer
Seven genes in Table 6, fixQ, fixS, frxA, hypD, hydE, pgl and nuoF, are related to electron transfer.
Aerobic respiration in H. pylori has been analyzed experimentally and by genome sequences. A cb-type cytochrome c oxidase is the sole terminal oxidase present in H. pylori [87]. FixQ (= CcoQ) is a component of the oxidase. The fixS gene likely encodes the cation transport subunit of the oxidase [34]. It has been proposed that FixS plays a role in the uptake and metabolism of copper required for oxidase assembly [87]. Aerobic respiration results in production of toxic superoxide at this terminal oxidase, which is involved in bacterial death [88]. The frxA gene, NAD(P)H-flavin oxidoreductase, is involved in redox of flavins, which are important electron transfer mediators [89]. Reduced flavins reduce ferric complexes or iron proteins with low redox potential. FrxA is one of the enzymes that make H. pylori sensitive to metronidazole [90]. H. pylori is capable of hydrogen oxidation [87]. HypD is involved in maturation of the [NiFe] H2-uptake hydrogenase, and catalyzes insertion and cyanation of the iron center [91]. The hydE gene is also necessary for the hydrogenase activity [92]. The pgl gene (HP1102) encodes a 6-phosphogluconolactonase, which catalyzes the second step of the phosphopentose pathway. This phase of the phosphopentose pathway generates reducing power in the form of NADPH and is important in other organisms in defense against reactive oxygen species and oxidative stress response [93,94].
Intra-hspEAsia divergence was very small for fixQ (Figure 8C (b), Table 5 and Table 6).
Translation
Four genes in Table 6, miaA, tilS, def, and prmA, are important for translation.
MiaA and TilS affects translation fidelity [95-97]. MiaA isopentenyl-tRNA transferase modifies the tRNAs that read codons starting with U to minimize peptidyl-tRNA slippage in translation. TilS, the tRNA(Ile2) lysidine synthetase, modifies cytidine to lysidine (2-lysyl-cytidine) at the first anticodon of tRNA(Ile2), thereby switching tRNA(Ile2) from a methionine-specific to an isoleucine-specific tRNA. Def removes a formyl group from the N-terminus of a nascent polypeptide and is a potential drug target [98]. PrmA is a trimethyltransferase that methyates multiple residues in the N-terminal domain of ribosomal protein L11, a universally conserved component of the large ribosomal subunit [99].
There was evidence that divergence in miaA was adaptive (Table 7), and the relevant amino acid residue was mapped on the structure (Figure 9B ii), as described above. Intra-hspEAsia divergence was not large for def (located in zone 2), whereas large for miaA (in zone 3).
Nucleases
Four genes in Table 6, addA, rnhA, rnhB and hsdR, are nucleases.
AddA (AdnA, PcrA) is a RecB-like helicase that promotes DNA recombination repair and survival during colonization [100]. Upon encounter with a DNA double-strand break, E. coli RecBCD enzyme degrades non-self DNA, but repairs self DNA marked by a genomic identification sequence through RecA-mediated homologous recombination. The identification sequence varies among bacterial groups [101] and can be altered by a mutation in RecBCD [102].
The rnhA and rnhB genes encode RNase HI and RNase HII, which hydrolyze RNA hybridized to DNA. Their biological role remains unclear, although they affect DNA replication, repair and transcription [103,104].
An AT-rich region of the addA gene linking the helicase domain and the nuclease domain showed an interesting divergence: the sequence AAAGAAAG(T/C)AAA encoding Lys-Glu-Ser-Lys was repeated in tandem 2 to 8 times in the hspWAfrica and hpEurope strains but was absent or present only once in the hspEAsia strains. The hspAmerind strains have a single copy (4 strains) or two copies (1 strain).
Cell division
Gene ftsA encodes an actin-like, membrane-associated protein that interacts with the tubulin-like FtsZ protein, helps it assemble into the Z ring, anchors it to the cytoplasmic membrane, and recruits other proteins for cell division [105]. It is a potential drug target [106].
Amino acid
The ilvE gene (HP1468) encodes a branched-chain amino acid aminotransferase that generates glutamic acid from branched-chain amino acids (valine, leucine, isoleucine) that are essential to H. pylori. We do not know whether its divergence is related to loss of jhp0585, encoding a branched-amino-acid dehydrogenase, in all hpEastAsia strains (see above), or whether it is related to a possible geographical divergence in the amino acid content of food.
Discussion
We closely compared complete genome sequences through phylogenetic profiling, phylogenetic tree construction, and nucleotide sequence analysis. The results distinguished decaying from intact genes and revealed drastic evolutionary changes within the H. pylori species. Our results clearly define the H. pylori East Asian lineage as distinct at the genome level from the African, European or Amerind lineages (Table 2). The East Asian lineage consists of Japanese and Korean genomes and corresponds to hspEAsia in the phylogenetic tree of the concatenated seven genes used for multi-locus sequence typing. The hspEAsia and hspAmerind lineages form a phylogenetic group hpEastAsia. The outstanding differences are in proteins related to: (i) host-interaction; (ii) electron transfer and redox metabolism; and (iii) translation fidelity.
Host-interaction proteins
Many of the virulence factors show wide divergence between hspEAsia and hpEurope, most likely because of co-evolution with the host. We anticipate that the list of well-diverged genes (Table 6) is enriched for host-interaction and potential virulence genes. We detected positively-selected amino-acid changes in two virulence factors: cagA and vacA (Table 7).
Many OMP families showed loss of one of their resident loci (hopMN, babABC, sabAB), whereas one family (oipA) showed duplication of its locus. Some OMP genes showed internal deletions (vacA-2) or interallelic homologous recombination (hopMN). A group-specific repertoire was seen for other OMP genes (homB, hopZ and hopQ), for other criteria. We also found substantial hspEAsia-hpEurope divergence in many OMPs (Table 5). The OMPs play important roles in host interaction such as adhesion to the host cells and induction of immune responses [26]. For example, OipA induces IL-8 from host cells [70]. Systematic decay of OMP genes occurred during adaptation of H. pylori to a new host of large felines, generating the new species of H. acinonychis [36]. Hence, the above OMP changes might reflect selection and/or fine regulation in host interaction, and more specifically, may help avoid the host immune system. At least two OMPs show evidence for positive selection (Table 7). We do not yet know whether these OMP changes are related to immune response or adhesin activity.
Lewis antigen mimicry is important for gastric colonization and adhesion. The mimicry affects innate immune recognition, inflammatory response, and T-cell polarization. Long-term infection by H. pylori might induce autoreactive anti-Lewis antigen antibodies [107]. Divergence in transferase genes for LPS biosynthesis may have resulted from co-evolution with the host immune system and could be related to changes in Lewis antigens in human populations. For example, the Le(a+b+) phenotype is almost absent in Caucasian persons whereas it occurs with a higher frequency in the Asian population [108]. This might be related to differences in pathogenicity and adaptation [109].
Changes in transporter genes, the loss of a putative amino acid utilization gene, divergence in a branched chain amino acid metabolism gene, differences in acetate metabolism genes, and divergence in motility and chemotaxis genes could also be related to host interaction, because these are related to the stomach environment. An interesting question is if these changes are related to variation in human diets.
Electron transfer
Several key electron transfer components were diverged between hspEAsia and hpEurope. The multiple and drastic changes in redox metabolism were unexpected. The systematic decay of all Mo-related genes through mutations in all (6/6) hspEAsia strains was the most striking. We do not know whether our findings reflect the biased environmental occurrence of Mo or the dietary habits of human populations. The richest sources of Mo include legumes, cereal grains (and baked products), leafy vegetables, milk, beans, liver, and kidney, whereas fruits, stem and root vegetables, and muscle meats are poor Mo sources [110].
The BisC homolog, the only molybdoenzyme found in the H. pylori genome, is similar to a number of periplasmic reductases for alternative oxidants such as dimethylsulfoxide or trimethylamine N-oxide [87]. Western strains of H. pylori might be able to use N- and/or S-oxide as an electron acceptor in energy metabolism in addition to oxygen and fumarate. One hypothesis about decay of the Mo-related genes is that this anaerobic electron transport system became maladaptive in the East Asian lineage. One possibility is the radical reaction mediated by MoaA in molybdopterin synthesis is dangerous in the presence of oxygen. This could explain the observed changes in oxidative phosphorylation and acetate metabolism.
A candidate for the BisC substrate is an oxidized form of methionine, free or within a protein. Methionine is sensitive to oxidation, which converts it to a racemic mixture of methionine-S-sulfoxide (Met-S-SO) and methionine-R-sulfoxide (Met-R-SO) [111]. The reductive repair of oxidized methionine residues performed by methionine sulfoxide reductase is important in many pathogenic bacteria in general, and specifically for H. pylori to maintain persistent stomach colonization [112,113]. H. pylori methionine sulfoxide reductase (Msr, HP0224 product) is induced under oxidative stress control and can repair methionine-R-sulfoxide but not the S isomer, even though it is a fusion of an R-specific and an S-specific enzyme [114]. BisC from other bacteria can reduce and repair the S but not the R form [111].
If the sole function of BisC is to repair methionine-S-sulfoxide, another means to repair methionine-S-sulfoxide may have appeared in the East Asian H. pylori, for example by higher expression of Msr. In this case, BisC may have been inactivated because Mo-related reactions were no longer necessary. The substitution by a DNA element downstream of the msr gene in the hspEAsia strains (5/6, all but strain 52) could be involved in the hypothesized methionine-S-sulfoxide repair activity of its product.
Another possibility is decrease of oxidative stress generating methionine-S-sulfoxide in the East Asian H. pylori. Oxidative stress is induced by acid exposure, and msr is among the oxidative stress genes induced by acid [115]. H. pylori infection has different effects on acid secretion in Europe and Asia [116]. In Europe, antral-predominant gastritis with increased acid secretion is frequent, whereas in Asia, pan-gastritis and subsequent atrophic gastritis with decreased acid secretion are common. The decrease in acid experienced by East Asian H. pylori lineages may have decreased their methionine-S-sulfoxide and made its repair by BisC unnecessary.
Downregulation of some of the Mo-related genes in a European strain under acidic conditions may be related to their decay [30]. Downregulation may occur to avoid the possible toxic effects of Mo metabolism under conditions of acid adaptation.
Taken together, our results led us to predict that the East Asian H. pylori strains are different from the European strains in electron transfer reactions and responses to oxygen and acid. Possibly related to this alteration in redox is the presence of the two acetate-related pathways in 3 out of 4 Japanese strains. These are expected to be able to switch from acetate fermentation to acetate utilization under aerobic conditions, as seen for E. coli [117]. The European strains, some of the hspAmerind strains, and the other hspEAsia strains may be regarded as mutants that lack the pta-ackA pathway and the supposedly important acetyl~P signal. Global effects of these defects on chemotaxis, nitrogen and phosphate assimilation, osmo-regulation, flagellar biogenesis, biofilm development, and pathogenicity are expected, based on the various phenotypes of E. coli strains defective in these genes [33].
Translation fidelity
Translational proteins also diverged between hpEurope and hspEAsia strains. MiaA (tRNA delta(2)-isopentenylpyrophosphate transferase) and TilS (tRNA lysidine synthetase) affect accuracy in elongation. The amino-acid change in MiaA turned out to be adaptive (Table 7). TilS affects translation efficiency at various stages. Ambiguity in translation is proposed to be important in the evolution of novel proteins by generating phenotypic and genetic diversity in the proteome for selection [118]. This role of ambiguity is similar to the evolutionary role of genome-wide modulation of mutation rates by genes such as mutS [119].
Implications for medicine
East Asian (Japanese/Korean) H. pylori appear to be quite different from European H. pylori. Our results provide a solid starting point for understanding the biology, host interaction, and pathogenesis of the East Asian H. pylori, which in most previous works were inferred from a European strain. Divergences included virulence, cell surface-related, and drug target genes. These results will affect our strategy in developing effective therapies and drugs. Questions raised by our findings include whether East Asian VacA (Figure 9B) interacts with host cells in the same way as European VacA.
The diverged gene frxA is associated with resistance to antibiotics metronidazole [120], which is frequently used in H. pylori eradication. The divergence in the frxA could affect resistance to this group of drugs in various ways. More generally, if redox metabolism differs between hspEAsia and hpEurope strains, the same drug might produce different effects, depending on intra-bacterial redox reactions.
The diverged genes included two potential drug targets (def and ftsA), so drugs that target these proteins may have different effects in East Asian and European strains. We do not know, for example, whether anti-H. pylori drugs designed from structure of European Def [98] will be as effective against East Asian H. pylori.
Remaining questions
Clearly, many studies are needed to answer these and other questions raised by the genomics results presented here. Phylogenetic analysis in the present study used OGs where genes of hspEAsia were clustered separately from those of hpEurope. Some genes do not share this topology, as suggested above for acoE deletion and hopMN recombination. We plan to study the distortion in the tree. We focused on differences between a limited numbers of strains from each group. However, there are variations within East Asian strains (Table 5). Further experimental examination of the divergence within hspEAsia, and between hspEAsia and the other strains are necessary to understand their divergence in detail. Such examination might reveal complexity in evolution and will be the subject of a separate study. The mechanisms underlying the variation, such as mutations and rearrangements, will be a subject of a separate study [25].
Conclusions
Taking advantage of the extreme genome plasticity of H. pylori, we demonstrated how drastically a genome can change during evolution within a species. Our results revealed drastic changes in proteins for host interaction and electron transfer and suggested their importance in adaptive evolution. These results define the H. pylori East Asian and Western lineages at the genome level, enhance our understanding of their host interaction, and contribute to the design of effective drugs and therapies. The approach of fine comparative analysis of closely-related multiple genomes may reveal subtle but important evolutionary changes in other populations.
Methods
H. pylori culture
Four strains were isolated from patients with diffuse type gastric cancer, intestinal type gastric cancer, duodenal ulcer, and gastritis (F57 [121], F32, F30 and F16 [122]). The ABO blood groups of the hosts were: F57, B; F32, A; F30, O; F16, B. Studies were performed according to the principles of the Declaration of Helsinki, and consent obtained from each individual after a full description of the nature and protocol of the study.
Gastric biopsy specimens from each patient were inoculated onto a trypticase soy agar (TSA)-II/5% sheep blood plate and cultured under microaerobic conditions (O2, 5%; CO2, 15%; N2, 80%) at 37°C for 5 days. A single colony was picked from each primary culture plate, inoculated onto a fresh TSA-II plate, and cultured under the conditions described above. A few colonies were picked from each plate and transferred into 20 ml of Brucella broth liquid culture medium containing 10% fetal calf serum, and cultured for 3 days under the conditions as described above. A part of the liquid culture sample was stored at -80°C in 0.01 M phosphate-buffered saline (PBS) containing 20% glycerol. DNA from each H. pylori isolate was extracted from the culture pellet by the protease/phenol-chloroform method, suspended in 300 μl of TE buffer (10 mM Tris HCl, 1 mM EDTA) and stored at 4°C for PCR analysis and nucleotide sequencing.
Genome sequencing
The genome sequences of H. pylori strains F16, F30, F32 and F57 were determined by a whole-genome shotgun strategy. We constructed small-insert (2 kb) and large-insert (10 kb) plasmid libraries from genomic DNA, and sequenced both ends of the clones to obtain 26,112 (F16 and F57), 30,720 (F30) and 33,792 (F32) sequences using ABI 3730xl sequencers (Applied Biosystems), with coverage of 10.0 (F16)-, 11.5 (F30)-, 12.7 (F32)- and 10.0 (F57)-fold. Sequence reads were assembled with the Phred-Phrap-Consed program, and gaps were closed by direct sequencing of clones that spanned the gaps or with PCR products amplified using oligonucleotide primers designed against the ends of neighboring contigs. The overall accuracy of the finished sequence was estimated to have an error rate of less than 1 per 10,000 bases (Phrap score of ≥40). Sequences of the molybdenum-related genes and the genes in the acetate pathway of the four Japanese strains were verified by resequencing PCR fragments directly amplified from genomic DNA (primers are in Additional file 4 (= Table S3)). The genome sequences of other strains were obtained from National Center for Biotechnology Information (NCBI) [123]. Accession numbers are in Table 1.
Gene finding and annotation
We used the same protocol to identify genes in the four new strains and 16 other complete genomes (Table 1; gene assignment differences are in Additional file 8 (= Table 6)).
Protein-coding genes were identified by integrating predictions from programs GeneMarkS [124] and GLIMMER3 [125]. All ORFs longer than 10 amino acids were searched using BLASTP [126] against two databases, one composed of genes of 6 H. pylori genomes in RefSeq database at NCBI ("close" database), and the other composed of genes of 300 complete prokaryote genomes (one genome per one genus) available at the end of 2008, except for those in the Helicobacter genus ("distant" database). When the predicted start position differed in GeneMarkS and GLIMMER3, assignments were made by consensus of hits, with consensus against the "distant" database taking priority over the "close" one. The consensus start position among bidirectional best hits with 50% or more amino acid sequence identity for each matched region for each genome pair was determined by majority rule. Overlap of genes was resolved by comparing the results from four prediction programs. Genes encoding fewer than 100 amino acids and predicted only by Glimmer3 were dropped except for the microcin gene.
tRNA genes were detected using tRNAscan-SE [127]. rRNA genes were identified based on sequence conservation. Putative replication origins were predicted by GC-skew (window size 500 bp, window shift 250 bp).
Core genome analysis
The common core structure conserved among 20 H. pylori genomes was identified based on conservation of gene order among orthologs using the CoreAligner program [23] implemented in the RECOG system. Briefly, CoreAligner identifies the genomic core of the input genomes by taking the longest path of the neighborhood graph that consists of conserved neighborhood gene pairs, which are defined as pairs of OGs that are within a neighborhood of 20 genes in at least half of the genomes. For this analysis, we used as input a set of OGs generated by the DomClust program [24] (see "Phylogenetic profile analysis" section below for details about identification of OGs by DomClust). Absence of a gene in some genomes (at least half of the genomes) in each OGs among the core is allowed. In addition, as identified OGs are at the domain level, if a counterpart of a gene in one genome is split in another genome, different number of genes can participate in the OGs in different genomes. Thus, the number of core genes in each genome can vary. Still, the numbers of core genes varied less (1364-1424; SD = 13.5) than the total number of genes among the strains (1465-1593; SD = 33.9) (Table 1). Among those core OGs, 1079 OGs were universally conserved (conserved in the all genomes), non-domain-separated, with one-to-one correspondence, and designated "well-defined core OGs". Those 1079 OGs were used for phylogenetic analysis (Figure 1). Nucleotide sequences of genes in well-defined core OGs were aligned by the Mafft program [128], from which conserved blocks were extracted by the Gblocks program [129].
Phylogenetic profile analysis
Phylogenetic profiling was carried out using the set of OGs generated by DomClust [24]. We identified OGs with East Asian-specific features as those whose phylogenetic profiles were highly correlated to the template pattern (taking 1 for hspEAsia and 0 for hpEurope). The DomClust clustering program can identify OGs at the domain level, and was used to identify genes truncated in particular strains. Clustering was performed based on PAM (point accepted mutation) distance rather than score to ensure proper evaluation of evolutionary distances, even if one gene was truncated; in the latter case, scores may underestimate evolutionary relatedness. To clarify differences in gene-splitting patterns among strains, we did not use DomClust options to suppress domain splitting.
To identify genes with characteristic patterns of hspEAsia strains, we constructed a phylogenetic profile for each OG as a vector of examined property values (e.g., number of domains or number of duplications). For surveying patterns of gene splitting and deletion, a phylogenetic profile was constructed for each OG using the number of domains for each gene that resulted from the clustering. For surveying patterns of gene duplication, a phylogenetic profile was constructed using the number of duplicated genes (in-paralogs). To find OGs with a characteristic hspEAsia pattern, equality of the medians among different populations was tested by Kruskal-Wallis test. Tests between East Asian and European strains used the six hspEAsia strains and the seven hpEurope strains. Tests among four subpopulations used six hspEAsia, five hspAmerind, seven hpEurope, and two hspWAfrica strains.
Phylogenetic network analysis of the hopM/N family was carried out using NeighborNet [130] implemented on SpritsTree [131].
Analyses of molybdenum-related genes
H. pylori protein sequences were searched against the CDD conserved protein domain database, by RPS-BLAST [132]. Protein families extracted from the search results for Mo-cofactor synthesis or binding domain were: PF03404 (Mo-co_dimer), PF03205 (MobB), PF02738 (Ald_Xan_dh_C2), PF01568 (Molydop_binding), PF02730 (AFOR_N), PF02597 (ThiS), PF03454 (MoeA_C), PF06463 (Mob_synth_C), PF03453 (MoeA_N), PF01315 (Ald_Xan_dh_C), PF01493 (GXGXG), PF02579 (Nitro_FeMo-Co, PF01967 (MoaC), PF03459 (TOBE), PF02391 (MoaE), PF00384 (Molybdopterin), PF04879 (Molybdop_Fe4S4), PF02665 (Nitrate_red_gam), PF00174 (Oxidored_molyb), PF00994 (MoCF_biosynth), PF03473 (MOSC), PF02625 (XdhC_CoxI), PF01314 (AFOR_C), PF01547 (SBP_bac_1) (pfam name in parentheses). Homologs of two molybdoproteins [133] that were not detected in the above protein families were absent in the H. pylori genomes.
bisC was the only molybdoenzyme gene in the 20 H. pylori genomes with detected domains PF01568 (Molydop_binding) and PF00384 (Molybdopterin). A multidomain TIGR00509 (bisC_fam) was also detected in bisC.
Analyses of horizontally transferred regions
GIs were detected by searching for regions that fulfilled the conditions of: (i) longer than 5 kb; (ii) continuous ORFs not perfectly conserved in all 20 H. pylori strains; and (iii) whole regions assumed as extrinsic by Alien Hunter [134]. Counterparts of detected GIs in Amerind strains were previously reported as TnPZ [48,49].
Genes with a large distance between East Asian and European strains
OGs diverged between six hspEAsia and seven hpEurope strains were screened based on two values related to their phylogenetic tree. The da value was the distance between the last common ancestral (LCA) node of hspEAsia and the LCA node of hpEurope. The db value was the average distance of hspEAsia from its LCA node. OGs with hspEAsia-diverged genes were screened by introducing the following conditions (with hspAmerind omitted): (i) OGs in which all the hspEAsia genes of the OG formed a sub tree without any hpEurope genes in the phylogenetic tree; (ii) OGs universally conserved (not less than 12 of the 13 genomes; not less than 10 among 11 genomes for comparison of 6 hspEAsia and 5 hpEurope strains in Additional file 7 (= Table S5)); (iii) genes with no domain fusion/fission event among the 13 genomes (within ± 20% of the mean length of the OG, measured in amino acid residues); (iv) da value greater than twice the da value of the concatenated well-defined core tree (of amino-acid sequences) (denoted as da*; with the resulting cutoff of da > 0.02324; 1079 OGs; see "core genome analysis" section above). Among 1248 OGs that satisfied the criteria (ii) and (iii), 692 OGs satisfied the criteria (i), that is, complete separation of genes of hspEAsia from those of hpEurope. The db* ± sd values in logarithmic scale, corresponding to 0.00550 and 0.0231 (db* = 0.01128) in the original scale, were used as threshold values for the three zones (N = 687; five OGs with db = 0 were excluded from 692 OGs satisfying the above criteria (i)-(iii)).
Amino acid sequences of the genes were aligned by the einsi command of the MAFFT program [128], from which a neighbor-joining tree was constructed by the ClustalW program [135].
A branch-site likelihood ratio test of positive selection was carried out using PAML [60] based on the multiple alignment by the einsi command of MAFFT [128]. Only residues aligned at the same site by the einsi command and by PRANK (with codon option) [136] were considered. Positively-selected residues were mapped on the p55 structure of VacA using PyMol).
Statistics
The equality of means for phylogenetic profiling between East Asian and European strains was tested by Kruskal-Wallis one-way analysis of variance by ranks, a non-parametric method for testing equality of population medians among groups. The tests were conducted using the R statistics package [137].
Accession Numbers
The accession numbers of the H. pylori genome sequences reported in this paper are: F16 [DDBJ:AP011940.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011940.1 ], F30 [DDBJ:AP011941.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011941.1, DDBJ:AP011942.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011942.1], F32 [DDBJ:AP011943.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011943.1, DDBJ:AP011944.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query= AP011944.1] and F57 [DDBJ:AP011945.1 http://getentry.ddbj.nig.ac.jp/cgi-bin/get_entry2.pl?database=ver_ddbj&query=AP011945.1].
List of Abbreviations
bp: base pair(s); hpEastAsia: hpEurope and hpAfrica1, populations of H. pylori; hspEAsia and hspAmerind: sub-populations of hpEastAsia; hspWAfrica: a sub-population of hpAfrica1; GI: genomic island; Mo: molybdenum; OG: orthologous group; OMP: outer membrane protein; ORF: open reading frame; redox: reduction-oxidation.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MK and YF contributed to informatics analysis and wrote the manuscript. YF carried out experimental verification of sequences of molybdenum-related genes and acetate pathway related genes. KY, TT, and IU contributed to informatics analysis. NH and NT contributed to genome DNA preparation. KO and MH contributed to sequencing and assembly. MY and TA provided the strains. I.K. contributed to design, analysis and writing. All the authors discussed the results and commented on the manuscript. All the authors read and approved the final manuscript.
Author information
Current position of MK: Institute of Biogeosciences, Japan Agency for Marine-Earth Science and Technology, Yokosuka, Kanagawa, 237-0061, Japan
Supplementary Material
Phylogenetic tree of H. pylori based on MLST genes
Genes characterizing East Asian strains: domain-based analysis.
Mutations in molybdenum-related genes of H. pylori.
Primers for sequence validation.
Distance values of 692 genes with complete separation of hspEAsia and hpEurope.
Multiple sequence alignments of diverged genes.
Examination of robustness of extraction of diverged genes.
Differences in gene assignment.
Contributor Information
Mikihiko Kawai, Email: kawaim@jamstec.go.jp.
Yoshikazu Furuta, Email: yfuruta@ims.u-tokyo.ac.jp.
Koji Yahara, Email: yahara@world.email.ne.jp.
Takeshi Tsuru, Email: tsuru@ims.u-tokyo.ac.jp.
Kenshiro Oshima, Email: oshima@cb.k.u-tokyo.ac.jp.
Naofumi Handa, Email: nhanda@ims.u-tokyo.ac.jp.
Noriko Takahashi, Email: renekt@ims.u-tokyo.ac.jp.
Masaru Yoshida, Email: myoshida@med.kobe-u.ac.jp.
Takeshi Azuma, Email: azumat@med.kobe-u.ac.jp.
Masahira Hattori, Email: hattori@k.u-tokyo.ac.jp.
Ikuo Uchiyama, Email: uchiyama@nibb.ac.jp.
Ichizo Kobayashi, Email: ikobaya@ims.u-tokyo.ac.jp.
Acknowledgements
YF, TT, NH, NT and IK are grateful to Hitomi Mimuro and Chihiro Sasakawa for introduction to H. pylori experiments. This work was supported by the Institute for Bioinformatics Research and Development, the Japan Science and Technology Agency. I.U. was supported by a Grant-in-Aid for Scientific Research (20310125) from the Japan Society for the Promotion of Science. N. H. was supported by grants from Ministry of Education, Culture, Sports, Science and Technology-Japan (MEXT), by Takeda Foundation, by Sumitomo Foundation, by Kato Memorial Bioscience Foundation and by Naito Foundation. I.K. was supported by the global COE (Center of Excellence) project of "Genome Information Big Bang" from MEXT, by the Suzuken Memorial Foundation, and by the Urakami Food and Food Culture Foundation. M.H. was supported by Grants-in-Aid for Scientific Research on Priority Areas "Comprehensive Genomics" from MEXT.
References
- Fitzgerald JR, Musser JM. Evolutionary genomics of pathogenic bacteria. Trends Microbiol. 2001;9:547–553. doi: 10.1016/S0966-842X(01)02228-4. [DOI] [PubMed] [Google Scholar]
- Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- Mobley HLT, Mendz GL, Hazell SL. Helicobacter pylori: physiology and genetics. Amer Society for Microbiology; 2001. [PubMed] [Google Scholar]
- Yamaoka Y. Helicobacter pylori: molecular genetics and cellular biology. Caister Academic Pr; 2008. [Google Scholar]
- Honda S, Fujioka T, Tokieda M, Satoh R, Nishizono A, Nasu M. Development of Helicobacter pylori-induced gastric carcinoma in Mongolian gerbils. Cancer Res. 1998;58:4255–4259. [PubMed] [Google Scholar]
- Watanabe T, Tada M, Nagai H, Sasaki S, Nakao M. Helicobacter pylori infection induces gastric cancer in mongolian gerbils. Gastroenterology. 1998;115:642–648. doi: 10.1016/S0016-5085(98)70143-X. [DOI] [PubMed] [Google Scholar]
- Fukase K, Kato M, Kikuchi S, Inoue K, Uemura N, Okamoto S, Terao S, Amagai K, Hayashi S, Asaka M. Effect of eradication of Helicobacter pylori on incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: an open-label, randomised controlled trial. Lancet. 2008;372:392–397. doi: 10.1016/S0140-6736(08)61159-9. [DOI] [PubMed] [Google Scholar]
- Kraft C, Suerbaum S. Mutation and recombination in Helicobacter pylori: mechanisms and role in generating strain diversity. Int J Med Microbiol. 2005;295:299–305. doi: 10.1016/j.ijmm.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Megraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Moodley Y, Linz B, Yamaoka Y, Windsor HM, Breurec S, Wu JY, Maady A, Bernhoft S, Thiberge JM, Phuanukoonnon S, Jobb G, Siba P, Graham DY, Marshall BJ, Achtman M. The peopling of the Pacific from a bacterial perspective. Science. 2009;323:527–530. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA. 2002;99:14428–14433. doi: 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satomi S, Yamakawa A, Matsunaga S, Masaki R, Inagaki T, Okuda T, Suto H, Ito Y, Yamazaki Y, Kuriyama M, Keida Y, Kutsumi H, Azuma T. Relationship between the diversity of the cagA gene of Helicobacter pylori and gastric cancer in Okinawa, Japan. J Gastroenterol. 2006;41:668–673. doi: 10.1007/s00535-006-1838-6. [DOI] [PubMed] [Google Scholar]
- Pride DT, Meinersmann RJ, Blaser MJ. Allelic Variation within Helicobacter pylori babA and babB. Infect Immun. 2001;69:1160–1171. doi: 10.1128/IAI.69.2.1160-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose C, Perez-Perez GI, Dominguez-Bello MG, Pride DT, Bravi CM, Blaser MJ. East Asian genotypes of Helicobacter pylori strains in Amerindians provide evidence for its ancient human carriage. Proc Natl Acad Sci USA. 2002;99:15107–15111. doi: 10.1073/pnas.242574599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaün L, Saunders NJ. Population-associated differences between the phase variable LPS biosynthetic genes of Helicobacter pylori. BMC Microbiol. 2006;6:79. doi: 10.1186/1471-2180-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Perez JC, Mittl PRE, Lee HK, Dailide G, Tan S, Ito Y, Secka O, Dailidiene D, Putty K. Helicobacter pylori evolution: lineage-specific adaptations in homologs of eukaryotic Sel1-like genes. PLoS Comput Biol. 2007;3:e151. doi: 10.1371/journal.pcbi.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleastro M, Cordeiro R, Menard A, Yamaoka Y, Queiroz D, Megraud F, Monteiro L. Allelic diversity and phylogeny of homB, a novel co-virulence marker of Helicobacter pylori. BMC Microbiol. 2009;9:248. doi: 10.1186/1471-2180-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- H. pylori MLST database. http://pubmlst.org/helicobacter/
- Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadstrom T, Suerbaum S, Achtman M. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Chan MS, Maiden MC. mlstdbNet - distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Kalia A, Gilman RH, Mendez M, Herrera P, Cabrera L, Velapatiño B, Balqui J, Paredes Puente de la Vega F, Rodriguez Ulloa CA, Cok J, Hooper CC, Dailide G, Tamma S, Berg DE. Helicobacter pylori from Peruvian amerindians: traces of human migrations in strains from remote Amazon, and genome sequence of an Amerind strain. PLoS One. 2010;5:e15076. doi: 10.1371/journal.pone.0015076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mane SP, Dominguez-Bello MG, Blaser MJ, Sobral BW, Hontecillas R, Skoneczka J, Mohapatra SK, Crasta OR, Evans C, Modise T, Shallom S, Shukla M, Varon C, Megraud F, Maldonado-Contreras AL, Williams KP, Bassaganya-Riera J. Host-interactive genes in Amerindian Helicobacter pylori diverge from their Old World homologs and mediate inflammatory responses. J Bacteriol. 2010;192:3078–3092. doi: 10.1128/JB.00063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama I. Multiple genome alignment for identifying the core structure among moderately related microbial genomes. BMC Genomics. 2008;9:515. doi: 10.1186/1471-2164-9-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama I. Hierarchical clustering algorithm for comprehensive orthologous-domain classification in multiple genomes. Nucleic Acids Res. 2006;34:647–658. doi: 10.1093/nar/gkj448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Kawai M, Yahara K, Takahashi N, Handa N, Tsuru T, Oshima K, Yoshida M, Azuma T, Hattori M, Uchiyama I, Kobayashi I. Birth and death of genes linked to chromosomal inversion. Proc Natl Acad Sci USA. 2011;108:1501–1506. doi: 10.1073/pnas.1012579108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y, Alm RA. In: Helicobacter pylori Molecular Genetics and Cellular Biology. Yamaoka Y, editor. Norfolk, UK: Caister Academic Press; 2008. Helicobacter pylori Outer Membrane Proteins; pp. 37–60. [Google Scholar]
- Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68:4155–4168. doi: 10.1128/IAI.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM. et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Schwarz G, Mendel RR, Ribbe MW. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- Oh JD, Kling-Backhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, Cordum HS, Wang C, Elliott G, Edwards J, Mardis ER, Engstrand LG, Gordon JI. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc Natl Acad Sci USA. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1:e43. doi: 10.1371/journal.pgen.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson RP. Autosomal recessive diseases among the Athabaskans of the southwestern United States: recent advances and implications for the future. Am J Med Genet A. 2009;149A:2602–2611. doi: 10.1002/ajmg.a.33052. [DOI] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig P, de Jonge BL, Alm RA, Brown ED, Uria-Nickelsen M, Noonan B, Mills SD, Tummino P, Carmel G, Guild BC, Moir DT, Vovis GF, Trust TJ. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol Mol Biol Rev. 1999;63:675–707. doi: 10.1128/mmbr.63.3.675-707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratzer R, Wilson DK, Nidetzky B. Catalytic mechanism and substrate selectivity of aldo-keto reductases: insights from structure-function studies of Candida tenuis xylose reductase. IUBMB Life. 2006;58:499–507. doi: 10.1080/15216540600818143. [DOI] [PubMed] [Google Scholar]
- Eppinger M, Baar C, Linz B, Raddatz G, Lanz C, Keller H, Morelli G, Gressmann H, Achtman M, Schuster SC. Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLoS Genet. 2006;2:e120. doi: 10.1371/journal.pgen.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundon WG, Marshall DG, Moráin CA, Smyth CJ. A novel tRNA-associated locus (trl) from Helicobacter pylori is co-transcribed with tRNA(Gly) and reveals genetic diversity. Microbiology. 1999;145(Pt 6):1289–1298. doi: 10.1099/13500872-145-6-1289. [DOI] [PubMed] [Google Scholar]
- Bocs S, Danchin A, Medigue C. Re-annotation of genome microbial coding-sequences: finding new genes and inaccurately annotated genes. BMC Bioinformatics. 2002;3:5. doi: 10.1186/1471-2105-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JW, Rabin BA, Murphy JB, Stone KL, Williams KR. Escherichia coli exonuclease VII. Cloning and sequencing of the gene encoding the large subunit (xseA) J Biol Chem. 1986;261:14929–14935. [PubMed] [Google Scholar]
- Chase JW, Richardson CC. Escherichia coli mutants deficient in exonuclease VII. J Bacteriol. 1977;129:934–947. doi: 10.1128/jb.129.2.934-947.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc Natl Acad Sci USA. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassbinder F, van Vliet AH, Gimmel V, Kusters JG, Kist M, Bereswill S. Identification of iron-regulated genes of Helicobacter pylori by a modified fur titration assay (FURTA-Hp) FEMS Microbiol Lett. 2000;184:225–229. doi: 10.1111/j.1574-6968.2000.tb09018.x. [DOI] [PubMed] [Google Scholar]
- Stoof J, Belzer C, van Vliet A. Metal Metabolism and Transport in Helicobacter pylori. Helicobacter pylori: molecular genetics and cellular biology. 2008. pp. 165–177.
- Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325–3333. doi: 10.1093/nar/27.16.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P, Lee KJ, Blaser MJ, Cover TL. Analysis of hopQ alleles in East Asian and Western strains of Helicobacter pylori. FEMS Microbiol Lett. 2005;251:37–43. doi: 10.1016/j.femsle.2005.07.023. [DOI] [PubMed] [Google Scholar]
- Chalk PA, Roberts AD, Blows WM. Metabolism of pyruvate and glucose by intact cells of Helicobacter pylori studied by 13C NMR spectroscopy. Microbiology. 1994;140(Pt 8):2085–2092. doi: 10.1099/13500872-140-8-2085. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Yamamoto K, Kobayashi I. Dependence of frequency of homologous recombination on the homology length. Genetics. 1995;140:797–809. doi: 10.1093/genetics/140.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Lee W, Subramaniam D, Anant S, Herrera P, Cabrera L, Balqui J, Barabas O, Kalia A, Gilman RH, Berg DE. Helicobacter Pylori's plasticity zones are novel transposable elements. PLoS One. 2009;4:e6859. doi: 10.1371/journal.pone.0006859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W, Windhager L, Rohrer S, Zeiller M, Karnholz A, Hoffmann R, Zimmer R, Haas R. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 2010;38:6089–6101. doi: 10.1093/nar/gkq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina TV, Gorbalenya AE, Koonin EV. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J Mol Evol. 1992;34:351–357. doi: 10.1007/BF00160243. [DOI] [PubMed] [Google Scholar]
- Thiberge JM, Boursaux-Eude C, Lehours P, Dillies MA, Creno S, Coppee JY, Rouy Z, Lajus A, Ma L, Burucoa C, Ruskone-Foumestraux A, Courillon-Mallet A, De Reuse H, Boneca IG, Lamarque D, Megraud F, Delchier JC, Medigue C, Bouchier C, Labigne A, Raymond J. From array-based hybridization of Helicobacter pylori isolates to the complete genome sequence of an isolate associated with MALT lymphoma. BMC Genomics. 2010;11:368. doi: 10.1186/1471-2164-11-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofler C, Fischer W, Hofreuter D, Haas R. Cryptic plasmids in Helicobacter pylori: putative functions in conjugative transfer and microcin production. Int J Med Microbiol. 2004;294:141–148. doi: 10.1016/j.ijmm.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Hosaka Y, Okamoto R, Irinoda K, Kaieda S, Koizumi W, Saigenji K, Inoue M. Characterization of pKU701, a 2.5-kb plasmid, in a Japanese Helicobacter pylori isolate. Plasmid. 2002;47:193–200. doi: 10.1016/S0147-619X(02)00003-3. [DOI] [PubMed] [Google Scholar]
- Song JY, Choi SH, Byun EY, Lee SG, Park YH, Park SG, Lee SK, Kim KM, Park JU, Kang HL, Baik SC, Lee WK, Cho MJ, Youn HS, Ko GH, Bae DW, Rhee KH. Characterization of a small cryptic plasmid, pHP51, from a Korean isolate of strain 51 of Helicobacter pylori. Plasmid. 2003;50:145–151. doi: 10.1016/S0147-619X(03)00059-3. [DOI] [PubMed] [Google Scholar]
- Hofreuter D, Haas R. Characterization of two cryptic Helicobacter pylori plasmids: a putative source for horizontal gene transfer and gene shuffling. J Bacteriol. 2002;184:2755–2766. doi: 10.1128/JB.184.10.2755-2766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnbacher M, Jahns T, Willrodt D, Daniel R, Haas R, Goesmann A, Kurtz S, Rieder G. Sequencing, annotation and comparative genome analysis of the gerbil-adapted Helicobacter pylori strain B8. BMC Genomics. 2010;11:335. doi: 10.1186/1471-2164-11-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIB-IS. http://gib-is.genes.nig.ac.jp/
- Nesic D, Miller MC, Quinkert ZT, Stein M, Chait BT, Stebbins CE. Helicobacter pylori CagA inhibits PAR1-MARK family kinases by mimicking host substrates. Nat Struct Mol Biol. 2010;17:130–132. doi: 10.1038/nsmb.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, Lacy DB. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc Natl Acad Sci USA. 2007;104:16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HJ, Wang WC. Expression and binding analysis of GST-VacA fusions reveals that the C-terminal approximately 100-residue segment of exotoxin is crucial for binding in HeLa cells. Biochem Biophys Res Commun. 2000;278:449–454. doi: 10.1006/bbrc.2000.3820. [DOI] [PubMed] [Google Scholar]
- Seif E, Hallberg BM. RNA-protein mutually induced fit: structure of Escherichia coli isopentenyl-tRNA transferase in complex with tRNA(Phe) J Biol Chem. 2009;284:6600–6604. doi: 10.1074/jbc.C800235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminska KH, Baraniak U, Boniecki M, Nowaczyk K, Czerwoniec A, Bujnicki JM. Structural bioinformatics analysis of enzymes involved in the biosynthesis pathway of the hypermodified nucleoside ms(2)io(6)A37 in tRNA. Proteins. 2008;70:1–18. doi: 10.1002/prot.21640. [DOI] [PubMed] [Google Scholar]
- Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- Jang JY, Yoon HJ, Yoon JY, Kim HS, Lee SJ, Kim KH, Kim dJ, Jang S, Han BG, Lee BI, Suh SW. Crystal structure of the TNF-alpha-Inducing protein (Tipalpha) from Helicobacter pylori: Insights into Its DNA-binding activity. J Mol Biol. 2009;392:191–197. doi: 10.1016/j.jmb.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Chung C, Olivares A, Torres E, Yilmaz O, Cohen H, Perez-Perez G. Diversity of VacA intermediate region among Helicobacter pylori strains from several regions of the world. J Clin Microbiol. 2010;48:690–696. doi: 10.1128/JCM.01815-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerman T, McGee D, Mobley H. Adherence and colonization. Helicobacter pylori: physiology and genetics. 2001. pp. 381–417.
- Carlsohn E, Nystrom J, Bolin I, Nilsson CL, Svennerholm AM. HpaA is essential for Helicobacter pylori colonization in mice. Infect Immun. 2006;74:920–926. doi: 10.1128/IAI.74.2.920-926.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci USA. 2000;97:7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspholm-Hurtig M, Dailide G, Lahmann M, Kalia A, Ilver D, Roche N, Vikstrom S, Sjostrom R, Linden S, Backstrom A, Lundberg C, Arnqvist A, Mahdavi J, Nilsson UJ, Velapatino B, Gilman RH, Gerhard M, Alarcon T, Lopez-Brea M, Nakazawa T, Fox JG, Correa P, Dominguez-Bello MG, Perez-Perez GI, Blaser MJ, Normark S, Carlstedt I, Oscarson S, Teneberg S, Berg DE. et al. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science. 2004;305:519–522. doi: 10.1126/science.1098801. [DOI] [PubMed] [Google Scholar]
- Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol. 1999;31:1537–1548. doi: 10.1046/j.1365-2958.1999.01300.x. [DOI] [PubMed] [Google Scholar]
- Lu H, Wu JY, Beswick EJ, Ohno T, Odenbreit S, Haas R, Reyes VE, Kita M, Graham DY, Yamaoka Y. Functional and intracellular signaling differences associated with the Helicobacter pylori AlpAB adhesin from Western and East Asian strains. J Biol Chem. 2007;282:6242–6254. doi: 10.1074/jbc.M611178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran AP, Trent MS. Helicobacter pylori: molecular genetics and cellular biology. Caister Academic Pr; 2008. Helicobacter pylori Lipopolysaccharides and Lewis Antigens; p. 7. [Google Scholar]
- Rasko DA, Wang G, Palcic MM, Taylor DE. Cloning and characterization of the alpha(1,3/4) fucosyltransferase of Helicobacter pylori. J Biol Chem. 2000;275:4988–4994. doi: 10.1074/jbc.275.7.4988. [DOI] [PubMed] [Google Scholar]
- Bergman M, Del Prete G, van Kooyk Y, Appelmelk B. Helicobacter pylori phase variation, immune modulation and gastric autoimmunity. Nat Rev Microbiol. 2006;4:151–159. doi: 10.1038/nrmicro1344. [DOI] [PubMed] [Google Scholar]
- Nilsson C, Skoglund A, Moran AP, Annuk H, Engstrand L, Normark S. Lipopolysaccharide diversity evolving in Helicobacter pylori communities through genetic modifications in fucosyltransferases. PLoS One. 2008;3:e3811. doi: 10.1371/journal.pone.0003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund A, Bäckhed HK, Nilsson C, Björkholm B, Normark S, Engstrand L. A changing gastric environment leads to adaptation of lipopolysaccharide variants in Helicobacter pylori populations during colonization. PLoS One. 2009;4:e5885. doi: 10.1371/journal.pone.0005885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Kato Y, Nishiyama K, Tokuda H. Depletion of SecDF-YajC causes a decrease in the level of SecG: implication for their functional interaction. FEBS Lett. 2003;550:114–118. doi: 10.1016/S0014-5793(03)00847-0. [DOI] [PubMed] [Google Scholar]
- Smeets LC, Bijlsma JJ, Boomkens SY, Vandenbroucke-Grauls CM, Kusters JG. comH, a novel gene essential for natural transformation of Helicobacter pylori. J Bacteriol. 2000;182:3948–3954. doi: 10.1128/JB.182.14.3948-3954.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath MJ, Mahanty HK, Kolter R. Characterization of a purF operon mutation which affects colicin V production. J Bacteriol. 1989;171:3158–3161. doi: 10.1128/jb.171.6.3158-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M, Schweinitzer T, Josenhans C. Helicobacter Flagella, Motility and Chemotaxis. Helicobacter pylori: molecular genetics and cellular biology. 2008. p. 61.
- Ryan KA, Karim N, Worku M, Penn CW, O'Toole PW. Helicobacter pylori flagellar hook-filament transition is controlled by a FliK functional homolog encoded by the gene HP0906. J Bacteriol. 2005;187:5742–5750. doi: 10.1128/JB.187.16.5742-5750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan SM. Flagellar glycosylation - a new component of the motility repertoire? Microbiology. 2006;152:1249–1262. doi: 10.1099/mic.0.28735-0. [DOI] [PubMed] [Google Scholar]
- Kelly DJ, Hughes NJ, Poole RK. Microaerobic physiology: aerobic respiration, anaerobic respiration, and carbon dioxide metabolism. Helicobacter pylori: physiology and genetics. 2001. pp. 113–124. [PubMed]
- Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelman M, Ramaswamy S, Nivière V, Fontecave M, Eklund H. Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli. Biochemistry. 1999;38:7040–7049. doi: 10.1021/bi982849m. [DOI] [PubMed] [Google Scholar]
- Kwon DH, El-Zaatari FA, Kato M, Osato MS, Reddy R, Yamaoka Y, Graham DY. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:2133–2142. doi: 10.1128/AAC.44.8.2133-2142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Matsumi R, Arai T, Atomi H, Imanaka T, Miki K. Crystal structures of [NiFe] hydrogenase maturation proteins HypC, HypD, and HypE: insights into cyanation reaction by thiol redox signaling. Mol Cell. 2007;27:29–40. doi: 10.1016/j.molcel.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Benoit S, Mehta N, Wang G, Gatlin M, Maier RJ. Requirement of hydD, hydE, hypC and hypE genes for hydrogenase activity in Helicobacter pylori. Microb Pathog. 2004;36:153–157. doi: 10.1016/j.micpath.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Hazell S, Harris A, Trend M. In: Helicobacter pylori: physiology and genetics. Mobley H, Mendz G, Hazell S, editor. ASM Press; 2001. Evasion of the toxic effects of oxygen; pp. 167–175. [PubMed] [Google Scholar]
- Giró M, Carrillo N, Krapp AR. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology. 2006;152:1119–1128. doi: 10.1099/mic.0.28612-0. [DOI] [PubMed] [Google Scholar]
- Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. Embo J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Bonnefond L, Kimura S, Suzuki T, Ishitani R, Nureki O. Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase. Nature. 2009;461:1144–1148. doi: 10.1038/nature08474. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miyauchi K. Discovery and characterization of tRNAIle lysidine synthetase (TilS) FEBS Lett. 2010;584:272–277. doi: 10.1016/j.febslet.2009.11.085. [DOI] [PubMed] [Google Scholar]
- Cai J, Han C, Hu T, Zhang J, Wu D, Wang F, Liu Y, Ding J, Chen K, Yue J, Shen X, Jiang H. Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation. Protein Sci. 2006;15:2071–2081. doi: 10.1110/ps.062238406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci H, Gregory ST, Dahlberg AE, Jogl G. Recognition of ribosomal protein L11 by the protein trimethyltransferase PrmA. Embo J. 2007;26:567–577. doi: 10.1038/sj.emboj.7601508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen SK, Fero J, Hansen LM, Cromie GA, Solnick JV, Smith GR, Salama NR. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol Microbiol. 2008;69:994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourice S, Biaudet V, El Karoui M, Ehrlich SD, Gruss A. Identification of the Chi site of Haemophilus influenzae as several sequences related to the Escherichia coli Chi site. Mol Microbiol. 1998;27:1021–1029. doi: 10.1046/j.1365-2958.1998.00749.x. [DOI] [PubMed] [Google Scholar]
- Handa N, Ohashi S, Kusano K, Kobayashi I. Chi-star, a chi-related 11-mer sequence partially active in an E. coli recC1004 strain. Genes Cells. 1997;2:525–536. doi: 10.1046/j.1365-2443.1997.1410339.x. [DOI] [PubMed] [Google Scholar]
- Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Lock RL, Harry EJ. Cell-division inhibitors: new insights for future antibiotics. Nat Rev Drug Discov. 2008;7:324–338. doi: 10.1038/nrd2510. [DOI] [PubMed] [Google Scholar]
- Moran AP. Relevance of fucosylation and Lewis antigen expression in the bacterial gastroduodenal pathogen Helicobacter pylori. Carbohydr Res. 2008;343:1952–1965. doi: 10.1016/j.carres.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Broadberry RE, Lin-Chu M. The Lewis blood group system among Chinese in Taiwan. Hum Hered. 1991;41:290–294. doi: 10.1159/000154015. [DOI] [PubMed] [Google Scholar]
- Anstee DJ. The relationship between blood groups and disease. Blood. 2010;115:4635–4643. doi: 10.1182/blood-2010-01-261859. [DOI] [PubMed] [Google Scholar]
- Rajagopalan KV. Molybdenum: an essential trace element in human nutrition. Annual review of nutrition. 1988;8:401–427. doi: 10.1146/annurev.nu.08.070188.002153. [DOI] [PubMed] [Google Scholar]
- Ezraty B, Bos J, Barras F, Aussel L. Methionine sulfoxide reduction and assimilation in Escherichia coli: new role for the biotin sulfoxide reductase BisC. J Bacteriol. 2005;187:231–237. doi: 10.1128/JB.187.1.231-237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamuri P, Maier RJ. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Molecular microbiology. 2004;53:1397–1406. doi: 10.1111/j.1365-2958.2004.04190.x. [DOI] [PubMed] [Google Scholar]
- Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006;61:847–860. doi: 10.1111/j.1365-2958.2006.05302.x. [DOI] [PubMed] [Google Scholar]
- Alamuri P, Maier RJ. Methionine sulfoxide reductase in Helicobacter pylori: interaction with methionine-rich proteins and stress-induced expression. J Bacteriol. 2006;188:5839–5850. doi: 10.1128/JB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs G, Weeks D, Melchers K, Scott D. The gastric biology of Helicobacter pylori. Helicobacter pylori: molecular genetics and cellular biology. 2008. p. 137.
- McColl KE. Helicobacter pylori and acid secretion: where are we now? Eur J Gastroenterol Hepatol. 1997;9:333–335. doi: 10.1097/00042737-199704000-00004. [DOI] [PubMed] [Google Scholar]
- El-Mansi M, Cozzone AJ, Shiloach J, Eikmanns BJ. Control of carbon flux through enzymes of central and intermediary metabolism during growth of Escherichia coli on acetate. Curr Opin Microbiol. 2006;9:173–179. doi: 10.1016/j.mib.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Moura GR, Carreto LC, Santos MA. Genetic code ambiguity: an unexpected source of proteome innovation and phenotypic diversity. Curr Opin Microbiol. 2009;12:631–637. doi: 10.1016/j.mib.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Denamur E, Lecointre G, Darlu P, Tenaillon O, Acquaviva C, Sayada C, Sunjevaric I, Rothstein R, Elion J, Taddei F, Radman M, Matic I. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell. 2000;103:711–721. doi: 10.1016/S0092-8674(00)00175-6. [DOI] [PubMed] [Google Scholar]
- Jenks PJ, Edwards DI. Metronidazole resistance in Helicobacter pylori. Int J Antimicrob Agents. 2002;19:1–7. doi: 10.1016/S0924-8579(01)00468-X. [DOI] [PubMed] [Google Scholar]
- Ito Y, Azuma T, Ito S, Suto H, Miyaji H, Yamazaki Y, Kohli Y, Kuriyama M. Full-length sequence analysis of the vacA gene from cytotoxic and noncytotoxic Helicobacter pylori. J Infect Dis. 1998;178:1391–1398. doi: 10.1086/314435. [DOI] [PubMed] [Google Scholar]
- Azuma T, Yamakawa A, Yamazaki S, Ohtani M, Ito Y, Muramatsu A, Suto H, Yamazaki Y, Keida Y, Higashi H, Hatakeyama M. Distinct diversity of the cag pathogenicity island among Helicobacter pylori strains in Japan. J Clin Microbiol. 2004;42:2508–2517. doi: 10.1128/JCM.42.6.2508-2517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov
- Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic A, Menon AL, Thorgersen MP, Scott JW, Poole FL, Jenney FE Jr, Lancaster WA, Praissman JL, Shanmukh S, Vaccaro BJ, Trauger SA, Kalisiak E, Apon JV, Siuzdak G, Yannone SM, Tainer JA, Adams MW. Microbial metalloproteomes are largely uncharacterized. Nature. 2010;466:779–782. doi: 10.1038/nature09265. [DOI] [PubMed] [Google Scholar]
- Vernikos GS, Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2010. http://www.R-project.org/
- Ren S, Higashi H, Lu H, Azuma T, Hatakeyama M. Structural basis and functional consequence of Helicobacter pylori CagA multimerization in cells. J Biol Chem. 2006;281:32344–32352. doi: 10.1074/jbc.M606172200. [DOI] [PubMed] [Google Scholar]
- Devi SH, Taylor TD, Avasthi TS, Kondo S, Suzuki Y, Megraud F, Ahmed N. Genome of Helicobacter pylori strain 908. J Bacteriol. 2010;192:6488–6489. doi: 10.1128/JB.01110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Yamaoka Y, Zhu Q, Matha I, Gao X. A comprehensive sequence and disease correlation analyses for the C-terminal region of CagA protein of Helicobacter pylori. PLoS One. 2009;4:e7736. doi: 10.1371/journal.pone.0007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn LJ, Figueiredo C, Rossau R, Jannes G, van Asbroek M, Sousa JC, Carneiro F, Quint WG. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol. 1998;36:1271–1276. doi: 10.1128/jcm.36.5.1271-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M, Atherton JC. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133:926–936. doi: 10.1053/j.gastro.2007.06.056. [DOI] [PubMed] [Google Scholar]
- McClain MS, Shaffer CL, Israel DA, Peek RM Jr, Cover TL. Genome sequence analysis of Helicobacter pylori strains associated with gastric ulceration and gastric cancer. BMC Genomics. 2009;10:3. doi: 10.1186/1471-2164-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Zhou C, Huang RH. Structure of tRNA dimethylallyltransferase: RNA modification through a channel. J Mol Biol. 2007;367:872–881. doi: 10.1016/j.jmb.2007.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch M, Albracht SP, Matzanke BF, Drapal NM, Jacobi A, Bock A. The complex between hydrogenase-maturation proteins HypC and HypD is an intermediate in the supply of cyanide to the active site iron of [NiFe]-hydrogenases. J Mol Biol. 2004;344:155–167. doi: 10.1016/j.jmb.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Blokesch M, Bock A. Properties of the [NiFe]-hydrogenase maturation protein HypD. FEBS Lett. 2006;580:4065–4068. doi: 10.1016/j.febslet.2006.06.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of H. pylori based on MLST genes
Genes characterizing East Asian strains: domain-based analysis.
Mutations in molybdenum-related genes of H. pylori.
Primers for sequence validation.
Distance values of 692 genes with complete separation of hspEAsia and hpEurope.
Multiple sequence alignments of diverged genes.
Examination of robustness of extraction of diverged genes.
Differences in gene assignment.