Abstract
Background
The accessory gene regulator (agr) is a quorum sensing cluster of genes which control colonization and virulence in Staphylococcus aureus. We evaluated agr function in community- (CA) and healthcare-associated (HA) MRSA, to compare the pharmacodynamics and bactericidal activity of vancomycin against agr functional and dysfunctional HA-MRSA and CA-MRSA.
Methods
40 clinical isolates of MRSA from the Canadian Nosocomial Infection Surveillance Program were evaluated for delta-haemolysin production, as a surrogate marker of agr function. Time kill experiments were performed for vancomycin at 0 to 64 times the MIC against an initial inoculum of 106 and 108 cfu/ml of agr functional and dysfunctional CA-MRSA and HA-MRSA and these data were fit to a hill-type pharmacodynamic model.
Results
15% isolates were agr dysfunctional, which was higher among HA-MRSA (26.3%) versus CA-MRSA (4.76%). Against a low initial inoculum of 106 cfu/ml of CA-MRSA, vancomycin pharmacodynamics were similar among agr functional and dysfunctional strains. However, against a high initial inoculum of 108 cfu/ml, killing activity was notably attenuated against agr dysfunctional CA-MRSA (USA400) and HA-MRSA (USA100). CA-MRSA displayed a 20.0 fold decrease in the maximal reduction in bacterial counts (Emax) which was 3.71 log10 CFU/ml for agr functional vs. 2.41 log10 CFU/ml for agr dysfunctional MRSA (p = 0.0007).
Conclusions
Dysfunction in agr was less common among CA-MRSA vs. HA-MRSA. agr dysfunction demonstrated an impact on vancomycin bactericidal activity and pharmacodynamics against a high initial inoculum of CA-MRSA and HA-MRSA, which may have implications for optimal antimicrobial therapy against persistent, difficult to treat MRSA infections.
Background
The accessory gene regulator (agr) is a quorum sensing cluster of genes which orchestrate the expression of cell-secreted and virulence factors, and several metabolic pathways in Staphylococcus aureus in a growth dependant fashion [1,2]. It has been hypothesized that S. aureus which exhibit agr dysfunction may possess an intrinsic survival advantage: these strains have demonstrated vancomycin tolerance and a proclivity to develop heterogeneous resistance under vancomycin selective pressure[3,4]. However, agr dysfunction has primarily been evaluated in strains of healthcare-associated (HA) MRSA where a down-regulated agr locus has been associated with prolonged and persistent bactaeremia [5,6].
Worldwide, the incidence of community-associated (CA) MRSA is rapidly increasing with the emergence of highly virulent strains in Canada [7-9]. While a large proportion of HA-MRSA display dysfunctional in the agr loci, the prevalence of agr dysfunction among CA-MRSA is relatively low from 3.5 to 9% [10,11]. Although this may account for the enhanced virulence of CA-MRSA as compared with HA-MRSA, whether dysfunction in agr would hamper vancomycin bactericidal activity among Canadian CA-MRSA has not been fully elucidated. Therefore, the objectives of this current study were to compare vancomycin pharmacodynamics of agr dysfunctional versus functional CA-MRSA and HA-MRSA clinical isolates from the Canadian Nosocomial Infection Surveillance Program at low and high initial inoculum.
Methods
Bacterial Isolates
40 clinical methicillin-resistant S. aureus (MRSA) isolates were selected for analysis obtained from the Canadian Nosocomial Infection Surveillance Program (CNISP), as shown in Table 1. Isolates were previously subtyped by using pulsed-field gel electrophoresis (PFGE) as previously described by Simor et.al. [8]. A community-associated isolate was defined as one which had a USA300/Canadian-MRSA-10 or USA400/Canadian-MRSA-7 PFGE profile, according to the nomenclature established by Simor et. al. and McDougal et. al. [8,12]. Isolates with other PFGE profiles were considered to be healthcare-associated. All isolates were evaluated for agr dysfunction using delta-haemolysin production as a surrogate marker of agr function using RN4220, as previously described [4]. Four strains subsequently were selected including two CA-MRSA (CA-MRSA 26, agr functional, USA300/Canadian-MRSA-10 and CA-MRSA 20, agr dysfunctional, USA400/Candian-MRSA-7) and two HA-MRSA (HA-MRSA 7, agr functional, USA100/Canadian-MRSA-2 and HA-MRSA 9, agr dysfunctional, USA100/Canadian-MRSA-2) for vancomycin time killing experiments. The PFGE profile of all isolates were blinded to the investigators performing time killing experiments (R.M. and B.T.) until final analyses were completed.
Table 1.
Bacterial isolates utilized in this study
| Community or Healthcare Associated | PFGE Profile (USA) | PFGE Profile (CANADA) |
|---|---|---|
| CA-MRSA (n = 21)* | USA300 (n = 15) | CMRSA-10 (n = 15) |
| USA400 (n = 6) | CMRSA-7 (n = 6) | |
| HA-MRSA (n = 19)** | USA100 (n = 8) | CMRSA-2 (n = 8) |
| USA500 (n = 3) | CMRSA-5 (n = 3) | |
| USA600 (n = 6) | CMRSA-1 (n = 6) | |
| Other | No USA Correlate (n = 2) | CMRSA-9 (n = 2) |
*No CA-MRSA were agr dysfunctional
**6 HA-MRSA were agr dysfunctional,belonging to USA 100 (n = 1), USA 400 (n = 1), and USA 500 (n = 2), and USA 600(n = 2) PFGE profile. The remaining strains were classified as agr functional
Antimicrobial, Media, and MIC Determination
Vancomycin (obtained from Sigma Chemical Company, St. Louis, Missouri) stock solutions were freshly prepared prior to each experimental run for all experiments. Minimum inhibitory concentrations were determined for each isolate in duplicate by microdilution techniques according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). Brain Heart Infusion broth (Difco, Detroit, MI) was utilized for time-kill experiments. Quantification of colony counts were determined using Tryptic Soy Agar II plates with 5% sheep Blood (BD, Franklin Lakes, NJ ).
Time Kill Experiments
Selective time-kill experiments were performed in duplicate for vancomycin against agr functional and dysfunctional CA-MRSA and HA-MRSA using methodology as previous described[13]. Briefly, fresh bacterial colonies from an overnight growth were added to normal saline and adjusted spectrophotometrically to provide a standard suspension. This suspension was diluted with BHI and antibiotic stock solutions to achieve a starting inoculum of approximately 106 or 108 CFU/ml. Vancomycin concentrations of 0, 0.25, 1, 4, 16, and 64 times the MIC were tested against all isolates. Each 10-ml culture was incubated in a water bath at 35°C with constant shaking, and 0.1-ml samples were withdrawn for the determination of bacterial counts at 0, 4,8 and 24 h. Colony counts were determined by plating 50 μl of each diluted sample onto BHI agar (Becton Dickinson, Franklin Lakes, NJ) with an automated spiral dispenser (WASP; Don Whitley Scientific Limited, West Yorkshire, England) and incubating the plates for 24 h at 35°C to confirm colony counts.
Pharmacokinetic and Pharmacodynamic Analyses
For each regimen tested, pharmcodynamic analysis was performed as previously described[13] using the Log Ratio Change comparing the changes in cfu/ml from 0 (CFU0 h) versus 24 h (CFU24 h) and calculated as shown in equation 1.
| (1) |
Using non-linear regression a four-parameter concentration-effect Hill-type model was fit to the effect parameter Systat (Version 11, Richmond, VA) using:
| (2) |
Equation 2, a sigmoidal maximal effect concentration effect relationship, was utilized to characterize the relationship between the decrease in bacterial counts from 0 to 24 h as characterized by the Log Ratio Change. The dependent variable (E, Effect) was the Log Ratio Change measuring the reduction in bacterial counts for each regimen comparing 0 h to 24 h, E0 is the measured effect at zero drug concentration which is an assessment of bacterial growth with no vancomycin, Emax is the maximal effect which describes the maximal reduction of bacterial counts in log10 CFU/ml , C:MIC is concentration of vancomycin divided by its respective MIC, EC50 is the effective concentration of vancomycin for which there is 50% maximal effect, H is the slope constant also described as the Hill or sigmoidicity constant. Statistical comparisons among each model fitted parameter were performed using the t-test (Systat 11, Richmond, VA), for which a p value of <0.05 was considered significant.
Results
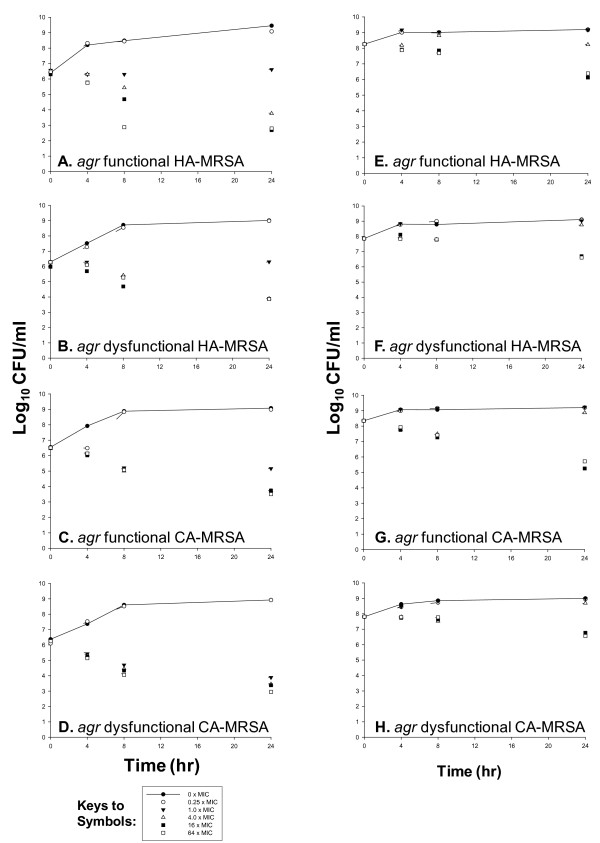
Vancomycin MICs against HA-MRSA 7, 9, were 1.0, 2.0 mg/L and for CA-MRSA 26 and 20 were 1.0, and 1.0 mg/L. 15% (6 of 40) of isolates were dysfunctional in agr which was higher among HA-MRSA (26.3%, 5 of 19) vs. CA-MRSA (4.76%, 1 of 21, USA 400/Canadian MRSA 7). The activity of vancomycin against a low (106 cfu/ml) and high (108 cfu/ml) initial inoculum of bacterial strains is depicted in Figure 1. agr functional strains (CA-MRSA 26) displayed greater killing versus agr dysfunctional MRSA (CA-MRSA 20). Maximal bacterial reductions at 64 times the MIC for CA-MRSA 20 and CA-MRSA 26 were 3.30 and 3.01 log10 cfu/ml at 24 h, respectively. However, against 106 cfu/ml inoculum differences in vancomycin killing of HA-MRSA with respect to agr function were noted with less activity versus agr dysfunctional strains. Interestingly, against a high initial inoculum of MRSA, vancomycin did not achieve bactericidal activity against any isolate. Most notably, against 108 cfu/ml, the killing activity of vancomycin against agr functional versus dysfunctional strains consistently differed for both CA-MRSA and HA-MRSA strains. Greater vancomycin killing activity occurred against agr functional (HA-MRSA 7, CA-MRSA 26) versus dysfunctional (HA-MRSA 9, CA-MRSA 20) strains. Reductions in bacterial counts at 24 h were greater against HA-MRSA 7 versus HA-MRSA 9 with 1.87 versus 1.24 log10 CFU/ml reduction at 24 h. Against CA-MRSA 20, killing approached the bactericidal threshold with a 2.63 log10 cfu/ml reduction as compared with CA-MRSA 26 where a decrease of only 1.25 log10 cfu/ml at 24 h was demonstrated.
Figure 1.
Time kill experiments evaluating the bactericidal activity of vancomycin versus agr functional HA-MRSA 7 (A,E), agr dysfunctional HA-MRSA 9 (B,F), agr functional CA-MRSA 26 (C,G), and agr dysfunctional CA-MRSA 20 (D,H) at low and high initial inoculum (106, 108 log10 cfu/ml).
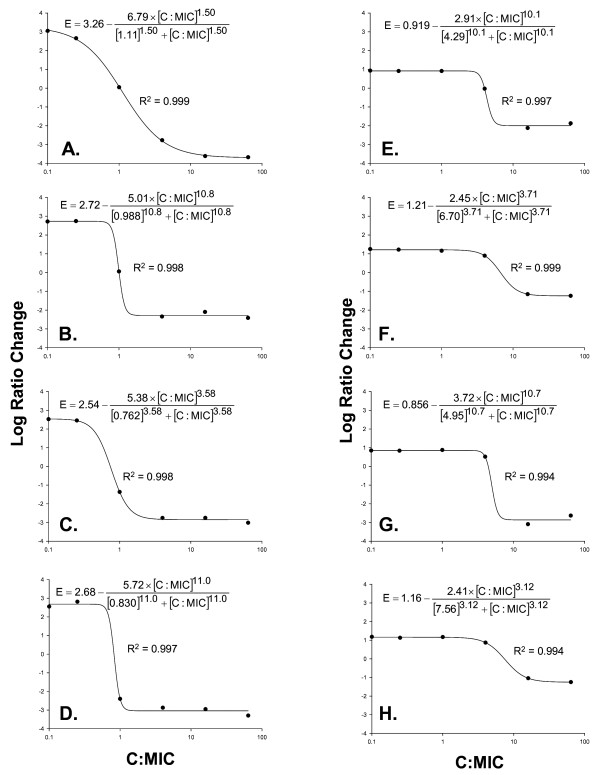
Analysis of pharmacodynamics (PD) revealed excellent model fits of the data to the Hill model. R2 for was >0.99 for all MRSA isolates at both initial inoculum. The concentration effect profile is depicted in Figure 2 with maximal likelihood fitted parameters from equation 2. Reduction in bacterial counts with vancomycin occurred in a concentration-dependent manner, as seen with the steep sigmoidicity constant (H) and low EC50 against most isolates. All pharmacodynamic parameter estimates consistently differed for both HA-MRSA and CA-MRSA when comparing agr functional versus dysfunctional isolates at the higher starting inoculum (108). There were significant differences in regards to vancomycin activity in reducing bacterial counts when comparing agr function and dysfunctional strains among HA-MRSA and CA-MRSA. HA-MRSA isolates displayed a 2.89 fold decrease in the maximal reduction in bacterial counts (Emax) which was 2.91 log10 CFU/ml for agr functional vs. 2.45 log10 CFU/ml for agr dysfunctional MRSA log10 CFU/ml (p = 0.0163).
Figure 2.
Pharmacodynamic relationship evaluating vancomycin concentration to minimum inhibitory concentration ratio (C:MIC) and change in log10 CFU/ml at 24 hours (Log Ratio Change) among agr functional HA-MRSA 7 (A,E), agr dysfunctional HA-MRSA 9 (B,F), agr functional CA-MRSA 26 (C,G), and agr dysfunctional CA-MRSA 20 (D,H) at low and high initial inoculum (106, 108 log10 cfu/ml). R2: Coefficient of Determination. Data are reported as maximum likelihood model fitted parameter estimates from equation 2 for each strain.
CA-MRSA displayed an even greater trend with a 20.0 fold decrease in the maximal reduction in bacterial counts (Emax) which was 3.71 log10 CFU/ml for agr functional vs. 2.41 log10 CFU/ml for agr dysfunctional MRSA (p = 0.0007). The sigmoidicity constant (H) was nearly three times higher and EC50 values were consistently lower in agr functional versus dysfunctional isolates, but did not achieve significance. Model fitted for each isolate parameter estimates from Equation 2 in Figure 2.
Discussion
The epidemiology of Staphylococcus aureus is rapidly changing. Highly virulent strains of CA-MRSA have emerged worldwide and have been associated with fatal infections in a plethora of recent outbreaks including severe necrotizing pneumonia and fasciitis and in both children and healthy adults [8,9,14]. A number of virulence determinants have recently been characterized in CA-MRSA including alpha-type phenol soluble modules (PSMs), the arginine catabolic mobile element (ACME), the panton- valentine leucocidin cytotoxin and a variety of virulence factors not common in HA-MRSA[15-17]. Interestingly, many of these virulence determinants are regulated via the two component agr system, the master regulator of S. aureus, controlling growth, colonization and virulence[1,17,18]. However, although agr dysfunction in S. aureus may potentially decrease virulence, the loss of agr function may increase the proclivity to develop vancomycin tolerance and resistance, although this has not been previously evaluated in strains of CA-MRSA. Therefore, we sought to determine if dysfunctional agr strains in either HA-MRSA and CA-MRSA would impact vancocmycin pharmacodynamics.
In the current investigation, agr dysfunction resulted in attenuation of vancomycin killing activity with decreases in maximal effect and lack of bactericidal activity against both HA-MRSA and CA-MRSA. We also determined that strains which exhibited agr dysfunction were far less common among CA-MRSA vs. HA-MRSA, with only one out of 18 isolates demonstrating this phenomena. Taken together, these findings may have potential implications for the use of vancomycin for the treatment of CA-MRSA infections. First, although reports of persistent bacteraemia, vancomycin treatment failure and heterogeneous resistance have been primarily in HA-MRSA, the findings of the present study highlight similarities of CA-MRSA and HA-MRSA when exposed to vancomycin [5,6]. Similar to previous reports in HA-MRSA, limited bacterial killing was evident by vancomcyin against CA-MRSA, especially in the context of agr dysfunction. Second, with reports of CA-MRSA as an emerging pathogen in infections that are sequestered or deep-seated, such as endocarditis, the potential for attenuation of bactericidal activity should be considered when vancomycin is utilized against a high initial inoculum of CA-MRSA [19]. Therefore, the inoculum effect displayed by vancomycin displays may be a potential explanation for the lack of bactericidal activity shown in this study. This is especially important to consider since eradication of S. aureus from sites of infection that are at high bacterial density or in a biofilm matrix at may be particularly difficult for vancomycin due to its pharmacodynamics properties, including slow bactericidal activity and the heterogeneous nature of vancomycin resistance. Finally, as the antimicrobial susceptibility profile of CA-MRSA is commonly greater than HA-MRSA and with initial reports of heterogeneous vancomycin-intermediate resistance in CA-MRSA emerging[20], the treatment of agr dysfunctional CA-MRSA may favor the use of combination therapy or alternative agents rather than glycopeptide monotherapy.
Potential limitations of this study include the limited number of strains including only two CA-MRSA and two HA-MRSA isolates which were utilized to evaluate vancomycin efficacy. We also acknowledge the lack of inclusion of a USA300 strain in time kill experiments because there were no agr dysfunctional CA-MRSA strains among the 40 isolates which we screened. Interestingly, these data corroborate our previous work which also shows an exceptionally low prevalence (2.5%) of agr dysfunction in community isolates of MRSA. Therefore, further in vitro and in vivo studies are necessary to confirm these findings in additional CA-MRSA before these results are applied to clinical practice.
Conclusions
This study is the first to demonstrate that vancomycin activity was impaired against a high inoculum of both community and healthcare associated strains of MRSA that were dysfunctional in agr. Additionally this study characterized the function on agr among MRSA strains which were genetically defined according to community and hospital original and determined that agr dysfunction were far less common among CA-MRSA as compared with HA-MRSA (11). These result are important to consider particularly in the treatment of infections of high bacterial density due to MRSA when vancomycin is utilized against a high initial inoculum of CA-MRSA
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BTT was responsible for overall study design, participated in the experimental work, conducted an extensive literature review, and wrote the manuscript. RCM carried out the experimental work and participated in study design. LDD participated in the study design and contributed to writing of the manuscript. MJM contributed ideas and characterization of strains. AES characterized bacterial isolates, and provided his experience in bacterial genetics, contributed to study design, was involved in the writing of the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Brian T Tsuji, Email: btsuji@buffalo.edu.
Robert D MacLean, Email: bomaclean@gmail.com.
Linda D Dresser, Email: Linda.Dresser@nygh.on.ca.
Martin J McGavin, Email: martin.mcgavin@sri.utoronto.ca.
Andrew E Simor, Email: Andrew.Simor@sunnybrook.ca.
Acknowledgements
We would like to acknowledge the Canadian Nosocomial Infection Surveillance Program (CNISP) as the source of the Canadian MRSA strains used in this study. We would like to thank Pamela Kelchlin for technical assistance. These results were presented in part at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy and 46th Infectious Diseases Society of America Meeting, Washington DC, October 25 to 28th, 2008. This study was funded by the University at Buffalo, Buffalo, NY and Sunnybrook Health Sciences Centre, Toronto, ON.
References
- Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- Wright JS, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci USA. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoulas G, Eliopoulos GM, Moellering RC Jr, Novick RP, Venkataraman L, Wennersten C, DeGirolami PC, Schwaber MJ, Gold HS. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J Infect Dis. 2003;187:929–938. doi: 10.1086/368128. [DOI] [PubMed] [Google Scholar]
- Sakoulas G, Eliopoulos GM, Moellering RC Jr, Wennersten C, Venkataraman L, Novick RP, Gold HS. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother. 2002;46:1492–1502. doi: 10.1128/AAC.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoulas G, Gold HS, Cohen RA, Venkataraman L, Moellering RC, Eliopoulos GM. Effects of prolonged vancomycin administration on methicillin-resistant Staphylococcus aureus (MRSA) in a patient with recurrent bacteraemia. J Antimicrob Chemother. 2006;57:699–704. doi: 10.1093/jac/dkl030. [DOI] [PubMed] [Google Scholar]
- Fowler VG Jr, Sakoulas G, McIntyre LM, Meka VG, Arbeit RD, Cabell CH, Stryjewski ME, Eliopoulos GM, Reller LB, Corey GR. et al. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis. 2004;190:1140–1149. doi: 10.1086/423145. [DOI] [PubMed] [Google Scholar]
- Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy ME, Etienne J. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simor AE, Ofner-Agostini M, Bryce E, McGeer A, Paton S, Mulvey MR. Laboratory characterization of methicillin-resistant Staphylococcus aureus in Canadian hospitals: results of 5 years of National Surveillance, 1995-1999. J Infect Dis. 2002;186:652–660. doi: 10.1086/342292. [DOI] [PubMed] [Google Scholar]
- Miller LG, Perdreau-Remington F, Rieg G, Mehdi S, Perlroth J, Bayer AS, Tang AW, Phung TO, Spellberg B. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med. 2005;352:1445–1453. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- Shopsin B, Drlica-Wagner A, Mathema B, Adhikari RP, Kreiswirth BN, Novick RP. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J Infect Dis. 2008;198:1171–1174. doi: 10.1086/592051. [DOI] [PubMed] [Google Scholar]
- Tsuji BT, Rybak MJ, Cheung CM, Amjad M, Kaatz GW. Community- and health care-associated methicillin-resistant Staphylococcus aureus: a comparison of molecular epidemiology and antimicrobial activities of various agents. Diagn Microbiol Infect Dis. 2007;58:41–47. doi: 10.1016/j.diagmicrobio.2006.10.021. [DOI] [PubMed] [Google Scholar]
- McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji BT, von Eiff C, Kelchlin PA, Forrest A, Smith PF. Attenuated vancomycin bactericidal activity against Staphylococcus aureus hemB mutants expressing the small-colony-variant phenotype. Antimicrob Agents Chemother. 2008;52:1533–1537. doi: 10.1128/AAC.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira-Rey M, Couzon F, Boisset S, Brown EL, Bes M, Benito Y, Barbu EM, Vazquez V, Hook M, Etienne J. et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–1133. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T. et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF. et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A. et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar BC, Prendergast BD, Moore JE. Community-associated MRSA (CA-MRSA): an emerging pathogen in infective endocarditis. J Antimicrob Chemother. 2008;61:1–7. doi: 10.1093/jac/dkm441. [DOI] [PubMed] [Google Scholar]
- Hageman JC, Patel J, Franklin P, Miscavish K, McDougal L, Lonsway D, Khan FN. Occurrence of a USA300 vancomycin-intermediate Staphylococcus aureus. Diagn Microbiol Infect Dis. 2008;62:440–442. doi: 10.1016/j.diagmicrobio.2008.08.005. [DOI] [PubMed] [Google Scholar]