Abstract
Interactions of sulfhydryl reagents with introduced cysteines in the pore-forming (Kir6.2) subunits of the KATP channel were examined. 2-Aminoethyl methanethiosulfonate (MTSEA+) failed to modify Cd2+-insensitive control-Kir6.2 channels, but rapidly and irreversibly modified Kir6.2[L164C] (L164C) channels. Although a single Cd2+ ion is coordinated by L164C, four MTSEA+ “hits” can occur, each sequentially reducing the single-channel current. A dimeric fusion of control-Kir6.2 and L164C subunits generates Cd2+-insensitive channels, confirming that at least three cysteines are required for coordination, but MTSEA+ modification of the dimer occurs in two hits. L164C channels were not modified by bromotrimethyl ammoniumbimane (qBBr+), even though qBBr+ caused voltage-dependent block (as opposed to modification) that was comparable to that of MTSEA+ or 3-(triethylammonium)propyl methanethiosulfonate (MTSPTrEA+), implying that qBBr+ can also enter the inner cavity but does not modify L164C residues. The Kir channel pore structure was modeled by homology with the KcsA crystal structure. A stable conformation optimally places the four L164C side chains for coordination of a single Cd2+ ion. Modification of these cysteines by up to four MTSEA+ (or three MTSPTrEA+, or two qBBr+) does not require widening of the cavity to accommodate the derivatives within it. However, like the KcsA crystal structure, the energy-minimized model shows a narrowing at the inner entrance, and in the Kir6.2 model this narrowing excludes all ions. To allow entry of ions as large as MTSPTrEA+ or qBBr+, the entrance must widen to >8 Å, but this widening is readily accomplished by minimal M2 helix motion and side-chain rearrangement.
Keywords: inner pore, Cd2+, MTS reagents, subconductance
Until recently, the structures of ion channel pores were restricted to naive models, extrapolated from biophysical measurements of channel current and blocking characteristics (1). Doyle et al. (2) published a K+ channel (KcsA) crystal structure, confirming the general correctness of such models. All K+ channels share a similar overall structure, consisting of a tetramer of like subunits, each of which minimally contains two transmembrane α-helices. A narrow selectivity filter is formed by the conserved external P-loop between the two helices. A vestibule or cavity on the interior side is lined by the less conserved S6 or M2 helices (3–11). In inward rectifier (Kir) channels, the inner cavity structure has been suggested to be “unprecedentedly wide” on the basis of experiments (10) with the substituted cysteine accessibility method (12). Lu et al. (10) used methanethiosulfonate (MTS) reagents to modify the introduced cysteines in Kir2.1. When cysteines were introduced to one, two, three, or all four subunits at positions 172 or 176 within the inner vestibule, a graded reduction of single-channel current (i) was observed after 2-aminoethyl methanethiosulfonate (MTSEA+) modification. Up to four MTSEA+, two 3-(triethylammonium)propyl methanethiosulfonates (MTSPTrEA+), or two bromotrimethyl ammoniumbimane (qBBr+) moieties could modify the channel. These data were taken to suggest that the internal vestibule of Kir2.1 must be unprecedentedly wide, with an inner pore diameter greater than 10 Å, considerably wider than that of the KcsA crystal structure. In contrast to these conclusions, a substituted cysteine accessibility method study of Kir6.2 (the pore-forming subunit of the KATP channel), with the use of Cd2+ as the cysteine-interacting probe (11), predicted a narrow inner pore very similar to KcsA (11, 13, 14). Residue 164 in Kir6.2 is equivalent to residue 176 in Kir2.1, and in KATP channels generated by Kir6.2[L164C] mutant subunits, a single Cd2+ ion was coordinated by at least three cysteines, indicating a narrow diameter (<6 Å) at the level of L164C (11).
Thus, the picture emerging from the study of Lu et al. (10) is that the Kir inner vestibule may be unprecedentedly wide, whereas the study of Loussouarn et al. (11) concludes that the Kir inner vestibule should be narrow and that the structure should be quite similar to that of KcsA. The objective of the present study was to reconcile the two data sets. In Kir6.2[L164C] mutant channels, we now demonstrate that although a single Cd2+ ion is coordinated, four “hits” by MTSEA+ and two or three by MTSPTrEA+ can also occur. Hence the derivatives must fit in the inner vestibule. Contrary to the notion that this fit requires a vestibule wider than that of KcsA, these derivatives are all accommodated in a Kir6.2 pore modeled by homology to KcsA. Entry of these reagents from the cytoplasm into the pore requires a widening of the entrance, but this widening is readily accomplished in the model by minimal motions of M2 helices and side-chain rearrangements.
Materials and Methods
Expression of KATP Channels in COSm6 Cells.
COSm6 cells were transfected with pCMV6b-Kir6.2 (with mutations as described), pECE-SUR1, and pGreenLantern (BRL), as described (15).
Generation of a Nonreactive Background and Introduction of Cysteines.
Point mutations were prepared by overlap extension at the junctions of relevant residues by sequential PCR as described (15). The “control” Kir6.2 construct had a deletion of 36 amino acids from the C-terminal end, as well as N160D and C166S mutations (11). These mutations generate channels that are insensitive to cytoplasmic Cd2+ or MTSEA+ (11) and that can be expressed without SUR1. Cysteine substitutions were introduced on this control-Kir6.2 background. Dimeric constructs consisted of the control-Kir6.2 at the N terminus linked by six glycine residues to a second control-Kir6.2 with mutations as described.
Patch-Clamp Measurements.
Patch-clamp experiments were made at room temperature, in a chamber that allowed the solution bathing the exposed surface of the isolated patch to be changed rapidly. Data were normally filtered at 0.5–2 kHz (1–2 kHz for single channel), signals were digitized at 88 kHz (Neurocorder; Neurodata, New York) and stored on videotape, or digitized at 5 kHz and stored directly on a computer hard drive with the use of clampex software (Axon Instruments, Foster City, CA). Tape-recorded experiments were digitized into a computer with the use of axotape, fetchex, or clampex software (Axon Instruments). The standard bath (intracellular) and pipette (extracellular) solution used in these experiments (K-INT) had the following composition: 140 mM KCl, 10 mM K-Hepes, 1 mM K-EGTA (pH 7.3). In some single-channel experiments, 0.1 mM BaCl2 was added to the pipette solution to produce a low-frequency (≈1 s−1), long-lived (≈10 ms) closure, to improve resolution of the zero current level. Cadmium was added as a chloride salt and is expected to be present in solution as approximately equal parts of Cd2+ and CdCl+ ions. In Cd2+-containing solutions, K-EGTA was omitted and 20 mM KCl was replaced by KF (as a replacement Ca2+ chelator). Other additions to solutions are described in the text.
Data Analysis.
Off-line analysis was performed with the use of fetchan, pstat (Axon Instruments), and Microsoft excel programs. Wherever possible, data are presented as means ± SEM. Microsoft solver was used to fit data by least-squares algorithm.
Computational Procedures for Structural Modeling.
The sequence of the Kir6.2 fragment 78–176 was threaded over the structure of the KcsA fragment 23–119 (1BL8 from the Research Collaboratory for Structural Bioinformatics database, Rutgers University, New Brunswick, NJ) according to a clustal x multialignment of multiple Kir channels, fixing the local alignment of the M2 helix as described (11). Residue substitutions, insertions, and hydrogen additions were performed with Insight (Molecular Simulation, San Diego). Three K+ ions and one water molecule were held in the selectivity filter through the modeling process to keep the selectivity filter open. The optimization process was run with discover and the CFF91 forcefield (Molecular Simulation, San Diego): backbone loops and side-chain orientations were explored by torsion of dihedral angles according to a Monte Carlo/Metropolis algorithm, then minimized with 200 steepest descent and 1,500 conjugate gradient cycles, using no cutoff for nonbound interactions and a dielectric constant set to 1. Models were checked with procheck (16). A model of the open Kir6.2 channel was built with an unbound qBBr+ (7 × 10 × 12 Å) molecule constrained between four F168 residues at the otherwise narrowest part of the pore. The model was minimized as described above; the backbone was free to move, but the secondary structure was constrained by backbone torsional angles.
Derivatizations of L164C by MTSEA+, MTSPTrEA+, and qBBr+ were processed with biopolymer from the starting Kir6.2 model. Optimizations were performed as for the “wild type,” and added covalent groups were treated as side chains of the modified cysteines. No 4-fold symmetry was required to preserve the chain symmetry. Cd-6-metallothionein α and β domains (1DMC and 1DME from Research Collaboratory for Structural Bioinformatics) were used as templates for the tetrahedral coordination of Cd2+: all four distances between Cys Sγ and Cd2+ were set at 2.55 Å. Thermolysin and α-toxin (1LNE and 1CA1 from Research Collaboratory for Structural Bioinformatics) were used as models for the octahedral coordination of Cd2+: the four distances between Cys Sγ and Cd2+ were set at 2.55 Å as equatorial coordinations, whereas the distances with the two axial water molecule were set at 2.65 Å. During the modeling process involving Cd2+ binding, constraints were applied to keep the coordination shell intact. The surface of the inner vestibule was determined by filling the space between atoms of the protein with 1-Å hydrogen atoms and building a Connolly surface around these hydrogens by rolling a sphere of 1-Å radius on their surfaces.
Results and Discussion
One Cd2+ Is Sufficient to Inhibit the Channel by Interaction with Three or Four L164C Residues.
Expression of control-Kir6.2+SUR1 generates ATP-sensitive (11) channels that are inhibited only weakly (<15%), and rapidly reversibly, by intracellular Cd2+ (Fig. 1). Kir6.2[L164C] mutant channels (L164C) are strongly and only very slowly reversibly (τ ≈ 30 s; see Fig. 3 A and B) inhibited by 100 μM Cd2+. The open probability of L164C mutant channels is very high (≈0.9), and only a single short-lived closed time is observed [τ ≈ 0.5 ms (17)]. Accordingly, Cd2+ blocking and unblocking events are easily resolved in single-channel recordings. As shown in Fig. 2A, both inhibition by Cd2+ and recovery from inhibition occur as single hits, i.e., only full conductance transitions are observed. In addition, steady-state dose–response curves for inhibition by Cd2+ are well fit by a Hill equation, with mean values for half-maximal inhibition of 4.6 ± 1.5 μM and a Hill coefficient very close to 1 (1.04 ± 0.11). Together, these data indicate that a single Cd2+ ion is responsible for inhibition (11).
Figure 1.
Cd2+ and MTSEA+ modification of L164C residues (A) Effects of Cd2+ and MTSEA+ on representative inside-out patch currents from COSm6 cells cotransfected with SUR1 and Control-Kir6.2 (Control, Top), Kir6.2[L164C] (L164C, Middle), or control-Kir6.2-G6-Kir6.2[L164C] (control-L164C dimer, Bottom). In this and subsequent figures (except Fig. 4 C and D), inward currents are shown as upward deflections, zero current is indicated by a dashed line, and cartoons indicate the tetrameric makeup of L164C (●) and wild-type 164L (○) subunits. Cd2+ or MTSEA+ was applied as indicated. (B) Decrease in current in Cd2+, or after MTSEA+, as a fraction of control current (relative block), for each construct. Bars show mean ± SEM for n patches as indicated. (Inset) Structure of MTSEA+.
Figure 3.
MMTS and MTSEA+ modification abolishes high-affinity Cd2+ inhibition. (A) Macroscopic current recorded from an inside-out SUR1 + L164C patch. Cd2+ was applied as indicated. τrec indicates the time constant for recovery from Cd2+, obtained by fitting a single exponential. (B) τrec (mean ± SEM) for n = 4 similar experiments. (C) Representative record of single SUR1 + L164C channel in inside-out patch. In control, i (Upper, Left) was 4 pA, and the application of 100 μM Cd2+ generated a very long-lived (≫5 s) closed state (Upper, Right). MTSEA+ (1 mM) was applied until the first clear hit was observed (Lower, Left). The subsequent application of 100 μM Cd2+ caused rapidly reversible blocking events (Lower, Right). (D) Lifetime analysis of ≈30 s of recording from C (Lower, Left and Right) showed a single fast closed time of τc = 1 ms, before Cd2+, and an additional longer closed time of τc,2 = 31 ms, in the presence of Cd2+. (E) Effect of MMTS on representative inside-out patch currents from cells cotransfected with SUR1 and either Control-Kir6.2 (Control, Left) or L164C (Right). MMTS was applied as indicated. (F) Decrease in current after MMTS as a fraction of control current (relative block) for each construct. Bars show the mean ± SEM for n patches as indicated. (Inset) Structure of MMTS. (G) Effect of MMTS on Cd2+ block of representative L164C inside-out patch current. MMTS or Cd2+ was applied as indicated. (H) Decrease in current in Cd2+ as a fraction of control current (relative block), before (pre) and after (postMMTS) treatment with MMTS. Bars show the mean ± SEM for n patches as indicated.
Figure 2.
Multiple hits by MTSEA+ at residue L164C. (A) Representative single SUR1 + L164C channel in inside-out patch. Cd2+ was applied as indicated. The channel activity disappeared by a full conductance transition. After Cd2+ removal, channel conductance reappeared as a full transition. (B and C) (Upper) Representative single SUR1 + L164C (A) and SUR1 + Control-L164C dimeric (B) channels in inside-out patch, after exposure to 0.1 mM MTSEA+. (Lower) Fast time-base segments showing the change in conductance associated with each MTSEA+ hit. (D) Conductance levels observed after MTSEA+ hits (solid symbols) as a percentage of full conductance (open symbols) for individual patches of five homomeric L164C channels and three dimeric Control-L164C channels. Presumed missed hits are indicated by question marks. * indicates a L164C channel expressed without SUR1.
MTSEA+ Blocks L164C Channels in Four Hits.
As shown in Fig. 1, 1 mM MTSEA+ fails to modify control-Kir6.2 channels, but rapidly and irreversibly reduces L164C mutant channel current by ≈90% (Fig. 1), as shown for the corresponding I176C residue in Kir2.1 (10). Moreover, as with Kir2.1[I176C], modification of individual residues leads to only a partial reduction of i. After exposure of a single L164C channel to MTSEA+, up to four hits occur (Fig. 2B), each sequentially reducing i. The amplitude of each hit was variable, but a consistent picture emerges from analysis of multiple single-channel patches (Fig. 2D). First hits appear to reduce i by only ≈10% and may be missed (indicated by ?) in some recordings. Second hits can either cause a reduction to ≈70% or ≈45% of full conductance. In the former case, third and fourth hits then reduce i to ≈45% and ≈10%, respectively, of full conductance. In the latter case, only one more distinct hit is apparent, reducing i to ≈10% of full, as in Fig. 2D. In a tetramer, a second hit could theoretically occur either adjacent to or opposite the first. In the control + L164C dimer, with two diagonally opposed cysteine residues, we only observed a larger second hit that reduced conductance to ≈45% of full (see below). Therefore, we suggest that adjacent hits result in less reduction of i than opposite hits. Because all mutations were made on the control-Kir6.2 background, which included a deletion of 36 amino acids from the C terminus, functional channels can be obtained in the absence of SUR1 (18, 19). Essentially the same pattern of MTSEA+ hits is observed in the absence of SUR1 (Fig. 2D), excluding the possibility that the SUR1 subunit confers a unique pore conformation on Kir6.2 channels.
Control + L164C Dimer Has No High-Affinity Cd2+ Block but Is Inhibited by MTSEA+.
To gain further insight to the nature of the Cd2+ and MTSEA+ interactions with L164C, we constructed a dimeric fusion (control + L164C) of control-Kir6.2 and L164C subunits. As shown in Fig. 1 (Bottom), the dimer (+ SUR1) generates channels that show only rapidly reversibly and are inhibited by about 15% by 100 μM Cd2+, which is indistinguishable from the behavior of homomeric control-Kir6.2 channels (Fig. 1, Top). Unlike homomeric L164C channels, both macroscopic (Fig. 1) and single-channel (Fig. 2B) dimer currents are a little more than halved by MTSEA+ (Fig. 2C), the latter occurring in two hits, consistent with the two cysteines reacting with two MTSEA+.
MTS Modification Abolishes High Cd2+ Affinity.
The above data indicate that multiple MTSEA+ hits are possible, even though a single Cd2+ ion is coordinated in L164C channels. In macroscopic L164C patches, recovery from Cd2+ inhibition takes place with τrec ≈ 30 s (Fig. 3 A and B), indicating slow unbinding of Cd2+ from the pore. Fig. 3C shows that after a hit by MTSEA+, high-affinity Cd2+ inhibition is abolished; 100 μM Cd2+ now results only in brief closures (≈30 ms). This end of Cd2+ inhibition is resolved as a second component in the closed time histogram and indicates a ≈1,000-fold increase in the Cd2+ off-rate. MTSEA+ modification places a positive charge on the modified side chain, and it is conceivable that the effect on Cd2+ affinity is a long-range electrostatic effect rather than modification of a side chain directly involved in coordination. Accordingly, we attempted to modify L164C with the use of uncharged methyl methanethiosulfonate (MMTS). There is a small reduction of macroscopic current in L164C (Fig. 3 E and F), but MMTS did not affect i (not shown), making it impossible to judge whether or when modification had occurred. However, as shown in Fig. 3 G and H, modification with 1 mM MMTS clearly abolished high-affinity Cd2+ sensitivity. These data provide a strong indication that both MTS reagents and Cd2+ ions are interacting with the same 164C residues.
MTSPTrEA+, but Not qBBr+, Modifies Kir6.2[L164C].
Coordination of a single Cd2+ ion at L164C requires that the pore be <6-Å diameter at this point, although the data above clearly show that four MTSEA+ moieties can modify these same residues. As discussed below, modification of all four L164C residues by MTSEA+ is actually sterically feasible, even in such a narrow pore (11, 13), and requires no movement of backbone M2 helices. However, Lu et al. (10) demonstrated that considerably larger sulfhydryl agents can also modify the Kir2.1 pore at the equivalent position, so it is important to know whether the same holds true in Kir6.2. Fig. 4 (A and B) shows original records and averaged data from control-Kir6.2, L164C, and dimeric channels exposed to 5 mM MTSPTrEA+. There was no significant modification of control-Kir6.2 by this agent, but >95% modification of L164C. The dimer was also ≈75% inhibited by MTSPTrEA+. This level of inhibition is significantly less modification than what occurs in the homomeric L164C channel, indicating that at least two cysteines can be modified in the latter [two cysteines are apparently modified by MTSPTrEA+ in Kir2.1[I176C] (10)]. In contrast to the effects of MTSPTrEA+, and in contrast to the report of Lu et al. (10), there was no modification of L164C channels by 5 mM qBBr+ (Fig. 4C), even with long (>10 min) applications. This lack of modification might indicate that qBBr+ did not gain access to the channel pore. However, qBBr+ did cause a steeply voltage-dependent (Z = 1.7 ± 0.2, n = 3) block of the channel (Fig. 4 C and D), comparable to the voltage dependence of block (as opposed to modification) by MTSEA (Z = 1.8 ± 0.1, n = 7) or MTSPTrEA+ (Z = 1.7 ± 0.1, n = 3) (data not shown). Such a steep voltage dependence indicates that qBBr+ accesses the inner cavity, but the lack of modification suggests steric unfavorability.
Figure 4.
MTSPTrEA+, but not qBBr+, modifies L164C. (A) Effect of MTSPTrEA+ on representative inside-out patch currents from cells cotransfected with SUR1 and Control-Kir6.2 (Control, Top), L164C (Middle), or Control-L164C dimer (Bottom). (B) Decrease in current after MTSPTrEA+ modification as a fraction of control current (relative block), for each construct. Bars show the mean ± SEM for n patches as indicated. (Inset) Structure of MTSPTrEA+ and qBBr+. (C) Effect of qBBr+ on representative L164C inside-out patch current. The voltage was continuously ramped (4 s every 5 s, from −100 mV to +50 mV). (D) Representative current (relative to current at −100 mV)–voltage relationships obtained from ramps as indicated in C.
Modeling the Kir6.2 Pore: A Narrow Inner Cavity.
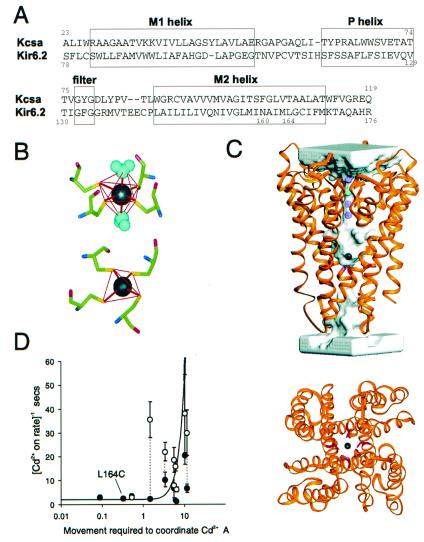
A sequence alignment of Kir6.2 fragment 78–176 with KcsA fragment 23–119 was generated (see Materials and Methods). Fourteen identities match the 98 aligned residues with two very short inserts in Kir6.2 of one (H115) and two (E140–E141) residues and a deletion of one residue (Y45) from KcsA (Fig. 5A). This match is equivalent to the alignment of Kir1.1 with KcsA proposed by Doyle et al. (2), and the hydropathy profiles of the two sequences are superimposable over the sequence length (not shown). The Kir6.2 sequence was threaded over the KcsA structure (1BL8 from the Research Collaboratory for Structural Bioinformatics database) according to the selected alignment and optimized (see Materials and Methods). The very small gaps in the loop do not significantly change the position of the loops around the common core of bundled helices (Fig. 5C). Whereas G56 and T85 are two facing residues in KcsA, this homology modeling positions the analogous C110 and C142 side chains of Kir6.2 to form a disulfide bridge. In accord with this prediction, a disulfide between the two homologous cysteine residues in Kir2.1 has recently been demonstrated experimentally (20).
Figure 5.
Cd2+ coordination by inner pore cysteines in a molecular model of Kir6.2. (A) Alignment of KcsA with Kir6.2 (B) Cd2+ coordination by four sulfhydryl groups may be tetrahedral (Lower) or octahedral, with two water molecules participating (Upper), if the sulfhydryl groups are arranged in a plane. (C) Molecular model of Kir6.2 based on homology with KcsA, including a Cd2+ ion in octahedral coordination by sulfhydryl groups in the side chains of residue 164C. (Upper) The backbone structure is shown in a ribbon representation superimposed on a transparent representation of the water-accessible pore. Three K+ ions and one water molecule are indicated in the selectivity filter, and a Cd2+ ion is coordinated by residues L164C. (Lower) Cd2+ coordinated by four L164C-Sγ sulfhydryls, viewed from beneath the membrane. (D) Apparent Cd2+ on-rate for inhibition of cysteine-substituted M2 mutants, before (solid) and after (open) treatment with phosphatidyl inositol-4-5-bisphosphate (from ref. 11), plotted against the movement required to bring the substituted sulfhydryls (Cys-Sγ) into the correct position to coordinate Cd2+. The side chains of residue L164C (indicated) are optimally positioned in the model structure for coordination of Cd2+.
To examine the likelihood of Cd2+ coordination by L164C, a homotetrameric model of Kir6.2[L164C] fragment 78–176 was built with a Cd2+ ion. The distance between two C164-Sγ is about 5 Å, too long to allow a disulfide bridge, but perfect for binding a cadmium ion (the Sγ-Cd2+ distance is about 2.55 Å). We used two template models for coordination of Cd2+: a tetrahedral organization of the four C164-Sγ coordinated to one central Cd2+ (Fig. 5B, Lower) (21) and an octahedral organization of four equatorial C164-Sγ and two added axial water molecules coordinate to one central Cd2+ (Fig. 5B, Upper) (22, 23). Tetracoordination of Cd2+ changes the backbone structure of residues 163–166 (1.2-Å rms deviation). However, hexacoordination by four cysteines and two water molecules (one above, one below, in the central axis) is very compatible with the Cd2+-free conformation and requires no backbone modification. Fig. 5C (Upper) shows a ribbon model of the Kir6.2 backbone, with the pore lining indicated by a transparent surface. The L164C side chains, near the bottom of the cavity, are pointing to the central axis and coordinate to the Cd2+ ion, as is more clearly visible in the 90° projection (Fig. 5C, Lower). The overall model structure is very similar to that of the KcsA crystal, although there is a discontinutity of the pore at the inner entrance, i.e., it is actually closed by the large F168 side chains (see below).
In our previous study the apparent on-rate for Cd2+ inhibition varied considerably between cysteine substituents, but the off-rate was fairly constant (11). At all tested residues, the on-rate was slowed (and the off-rate unaffected) by treatment with phosphatidyl inositol-4-5-bisphosphate, which stabilizes the open state of the channel (17), and Cd2+ block was abolished when the channels were closed by ATP. We thus suggested that Cd2+ enters the open channel, but that effective coordination depends on the ability of the side chains to move into close enough proximity to coordinate the Cd2+ ion. We systematically replaced each residue with cysteine in the four chains at each position of the M2 helix of the Kir6.2 model and measured, after local optimizations, the shortest distance between the corresponding four Cys Sγ atoms. The difference between these Sγ–Sγ distances and the optimal separation for Cd2+ coordination (5.1 Å) are plotted in Fig. 5D versus the estimated Cd2+ sensitivity (11). Consistent with the above hypothesis, there is a clear overall correlation: the greater the Sγ–Sγ separation, the slower the apparent on-rate.
Derivatization of Accessible Cysteines by MTSEA+, MTSPTrEA+, and qBBr+ Sulfhydryl Reagents.
Models of Kir6.2 in which the four L164C are derivatized by MTSEA+, MTSPTrEA+, and qBBr+ are shown in Fig. 6B, as well as KcsA derivatized at the equivalent residue by MTSEA+ (Fig. 6A). The internal lining of the pore, before derivatization, is displayed as a solid surface. The atoms of the labeling group bound to the four Cys-164 are displayed as colored CPK, where they protrude from this surface after derivatization and optimization. Contrary to the notion that the vestibule must be larger than that of KcsA to accommodate four MTSEA+ moieties, four Cys-164-CH2S-CH2-CH2-NH3+ (MTSEA+ derivatives) can actually fit within Kir6.2[L164C] (Fig. 6B, Left) or the equivalent KcsA[T107C] (Fig. 6A), with maximum deviation of the Cα position from each chain <0.7 Å away from the tetrafold axis through the pore. This deviation is less than the standard deviation observed for minimized underivatized structures (<0.9 Å). As shown in Fig. 6B, however, four MTSPTrEA+ (Middle) or qBBr+ (Right) derivatives protrude significantly outside the underivatized pore. Nevertheless, two or three Cys-164-CH2S-CH2-CH2-N+(CH2-CH3)3 (MTSPTrEA+ derivatives) would fit with a maximal deviation of less than 0.5 Å or1.3 Å, respectively. Furthermore, two Cys-164-qB would fit with a maximal deviation less than 0.5 Å (not shown). These results are consistent with very small motions or “breathing” (24) being necessary to accommodate the number of derivatives (4 MTSEA+, 2 or 3 MTSPTrEA+, 0–2 qBBr+) that are experimentally observed in either Kir2.1 (10) or Kir6.2 (above).
Figure 6.
Space-filling representations of sulfhydryl-reactive moieties within the Kir inner pore. (A) Transparent representations of the water-accessible pore of KcsA. Four MTSEA+ derivatives are attached to mutant T107C (equivalent to L164C in Kir6.2). Only a couple of H atoms (yellow) of the MTSEA+ derivatives appear outside of the pore cavity. (B) Transparent representations of the water-accessible pore of Kir6.2, with space-filling representations of MTS derivatives attached to L164C as indicated. Increasing portions of the derivatives appear outside of the pore cavity. (C) Transparent representation of the open Kir6.2 channel (see text).
The inner entrance of the channel is very narrow (<2-Å diameter in KcsA; Fig. 6A) or completely closed in the Kir6.2 model (Fig. 6B). In KcsA the narrowest part is lined by A111; complete closure in Kir6.2 is due to the much larger side chains of F168 at the equivalent position, arranged rather like the iris diaphragm of a camera. Given this closure, some rearrangement must occur to allow entry of ions and blockers. MTSPTrEA+ and qBBr+ are the largest sulfhydryl reagents tested in this study and in the study of Lu et al. (10). Suggesting that these reagents should provide a minimal diameter estimate for the inner pore in the “open channel,” we minimized the Kir6.2 model with a qBBr+ constrained to stay between the four F168 residues and optimized the structure. Fig. 6C shows the resulting open channel pore surface. Rearrangement of the F168 side chains initiates a slight rotation of the M2 helix, enlarging the inner entrance to a minimum 8-Å diameter, even though M2 helices are not displaced more than 0.8 Å from the axis, and the volume of the cavity is unchanged (within 10%).
Implications and Conclusions.
The primary objective of the present study was to reconcile the finding of an apparently narrow pore at residue 164 of Kir6.2 (11) with the finding that multiple large MTS reagents can be accommodated at the equivalent position in Kir2.1 (10). We now show that four MTSEA+ hits can also occur at residue 164C in Kir6.2, the effects on single-channel conductance being similar to those occurring in Kir2.1 (10). Even MTSPTrEA+ modification reduces the conductance of homomeric L164C channels almost completely but reduces the current through dimeric channels by only ≈70%, indicating that at least two moieties can simultaneously occupy the Kir6.2 pore. Although we see no evidence of qBBr+ modification of L164C channels, this reagent does cause voltage-dependent pore block, indicating that at least one qBBr+ molecule can also enter the pore but may be sterically restricted from modifying the 164C residue. Thus none of the experiments in this or previous studies indicate that the Kir6.2 and Kir2.1 pore structures should be fundamentally different from one another. Homology modeling indicates that several bulky MTS reagents can modify the 164 position and be accommodated within the cavity. Moreover, minimal backbone movements of the M2 helices can open the “neck” below the cavity sufficiently to allow these bulky derivatives to enter the cavity in the first place (Fig. 6C). Thus the overall structure of the Kir pore may be very similar to that of KcsA, and as in KcsA (25, 26), there should be some mobility of M2, to adopt the configuration necessary to coordinate a single Cd2+ ion in the central axis, but conversely to allow MTS reagents to enter the cavity. The former configuration may then correspond to the apparently closed KcsA crystal structure, whereas the latter may correspond to the open state, and possibly to the arrangement of helices predicted by the assays of Minor et al. (9). Spin–spin interactions between labeled residues in M2 of KcsA show periodic changes, indicative of rigid-body rotation and displacement during gating (26). It is tempting to suggest that the M2 motions predicted in the present study may be fundamentally the same as the gating motions of KcsA and represent the structural motions that underlie channel closing and opening in Kir channels (17).
Acknowledgments
The original Kir6.2 cDNA was provided by Dr. S. Seino (Chiba University, Chiba, Japan). This work was supported by Grant HL54171 from the National Institutes of Health (C.G.N.) and by the Washington University Diabetes Research and Training Center.
Abbreviations
- MTS
methanethiosulfonate
- MTSEA+
2-aminoethyl methanethiosulfonate
- MTSPTrEA+
3-(triethylammonium)propyl methanethiosulfonate
- qBBr+
bromotrimethyl ammoniumbimane
- MMTS
methyl methanethiosulfonate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 2.Doyle D A, Morais Cabral J, Pfuetzner R A, Kuo A, Gulbis J M, Cohen S L, Chait B T, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 3.MacKinnon R, Yellen G. Science. 1990;250:276–279. doi: 10.1126/science.2218530. [DOI] [PubMed] [Google Scholar]
- 4.Hartmann H A, Kirsch G E, Drewe J A, Taglialatela M, Joho R H, Brown A M. Science. 1991;251:942–944. doi: 10.1126/science.2000495. [DOI] [PubMed] [Google Scholar]
- 5.Yool A J, Schwarz T L. Nature (London) 1991;349:700–704. doi: 10.1038/349700a0. [DOI] [PubMed] [Google Scholar]
- 6.Heginbotham L, Abramson T, MacKinnon R. Science. 1992;258:1152–1155. doi: 10.1126/science.1279807. [DOI] [PubMed] [Google Scholar]
- 7.Stanfield P R, Davies N W, Shelton P A, Sutcliffe M J, Khan I A, Brammar W J, Conley E C. J Physiol (London) 1994;478:1–6. doi: 10.1113/jphysiol.1994.sp020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nichols C G, Lopatin A N. Annu Rev Physiol. 1997;59:171–191. doi: 10.1146/annurev.physiol.59.1.171. [DOI] [PubMed] [Google Scholar]
- 9.Minor D L, Jr, Masseling S J, Jan Y N, Jan L Y. Cell. 1999;96:879–891. doi: 10.1016/s0092-8674(00)80597-8. [DOI] [PubMed] [Google Scholar]
- 10.Lu T, Nguyen B, Zhang X, Yang J. Neuron. 1999;22:571–580. doi: 10.1016/s0896-6273(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 11.Loussouarn G, Makhina E N, Rose T, Nichols C G. J Biol Chem. 2000;275:1137–1144. doi: 10.1074/jbc.275.2.1137. [DOI] [PubMed] [Google Scholar]
- 12.Akabas M H, Kaufmann C, Archdeacon P, Karlin A. Neuron. 1994;13:919–927. doi: 10.1016/0896-6273(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 13.Capener C E, Shrivastava I H, Ranatunga K M, Forrest L R, Smith G R, Sansom M S. Biophys J. 2000;78:2929–2942. doi: 10.1016/S0006-3495(00)76833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson G A, Leyland M L, Ashmole I, Sutcliffe M J, Stanfield P R. J Physiol (London) 2000;526:231–240. doi: 10.1111/j.1469-7793.2000.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shyng S L, Nichols C G. Science. 1998;282:1138–1141. doi: 10.1126/science.282.5391.1138. [DOI] [PubMed] [Google Scholar]
- 16.Laskowski R A, Rullmannn J A, MacArthur M W, Kaptein R, Thornton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 17.Enkvetchakul D, Loussouarn G, Makhina E, Shyng S L, Nichols C G. Biophys J. 2000;78:2334–2348. doi: 10.1016/S0006-3495(00)76779-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker S J, Gribble F M, Zhao C, Trapp S, Ashcroft F M. Nature (London) 1997;387:179–183. doi: 10.1038/387179a0. [DOI] [PubMed] [Google Scholar]
- 19.Zerangue N, Schwappach B, Jan Y N, Jan L Y. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 20.Cho H C, Tsushima R G, Nguyen T T, Guy H R, Backx P H. Biochemistry. 2000;39:4649–4657. doi: 10.1021/bi992469g. [DOI] [PubMed] [Google Scholar]
- 21.Narula S S, Brouwer M, Hua Y, Armitage I M. Biochemistry. 1995;34:620–631. doi: 10.1021/bi00002a029. [DOI] [PubMed] [Google Scholar]
- 22.Holland D R, Hausrath A C, Juers D, Matthews B W. Protein Sci. 1995;4:1955–1965. doi: 10.1002/pro.5560041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naylor C E, Eaton J T, Howells A, Justin N, Moss D S, Titball R W, Basak A K. Nat Struct Biol. 1998;5:738–746. doi: 10.1038/1447. [DOI] [PubMed] [Google Scholar]
- 24.Ishima R, Torchia D A. Nat Struct Biol. 2000;7:740–743. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]
- 25.Perozo E, Cortes D M, Cuello L G. Nat Struct Biol. 1998;5:459–469. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 26.Perozo E, Cortes D M, Cuello L G. Science. 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]