SUMMARY
Skeletal muscle atrophy is a common and debilitating condition that lacks a pharmacologic therapy. To develop a potential therapy, we identified 63 mRNAs that were regulated by fasting in both human and mouse muscle, and 29 mRNAs that were regulated by both fasting and spinal cord injury in human muscle. We used these two unbiased mRNA expression signatures of muscle atrophy to query the Connectivity Map, which singled out ursolic acid as a compound whose signature was opposite to those of atrophy-inducing stresses. A natural compound enriched in apples, ursolic acid reduced muscle atrophy and stimulated muscle hypertrophy in mice. It did so by enhancing skeletal muscle insulin/IGF-I signaling, and inhibiting atrophy-associated skeletal muscle mRNA expression. Importantly, ursolic acid’s effects on muscle were accompanied by reductions in adiposity, fasting blood glucose and plasma cholesterol and triglycerides. These findings identify a potential therapy for muscle atrophy and perhaps other metabolic diseases.
INTRODUCTION
Skeletal muscle atrophy is characteristic of starvation and a common consequence of aging. It also complicates a wide range of severe human illnesses, including diabetes, cancer, chronic renal failure, congestive heart failure, chronic respiratory disease, acute critical illness, chronic infections such as HIV/AIDS, spinal cord injury, muscle denervation, and many other medical and surgical conditions that limit muscle use. However, we currently lack medical therapies to prevent or reverse skeletal muscle atrophy in humans. Sequelae of muscle atrophy (including weakness, falls, fractures, opportunistic respiratory infections, and loss of independence) are thus commonplace in hospital wards and extended care facilities.
Previous studies demonstrated that skeletal muscle atrophy is driven by conserved changes in skeletal muscle gene expression (Bodine et al., 2001a; Sandri et al., 2004). We therefore hypothesized that pharmacologic compounds with opposite effects on gene expression might inhibit skeletal muscle atrophy. To test this, we first determined an mRNA expression signature of one atrophy-inducing stress (fasting) in human and mouse skeletal muscle. We then used these unbiased data in conjunction with the Connectivity Map (Lamb et al., 2006) to identify candidate small molecule inhibitors of muscle atrophy. This approach identified a natural compound that may have applications in the treatment of human skeletal muscle atrophy.
RESULTS
Effects of Fasting on Skeletal Muscle mRNA Expression in Humans
Prolonged fasting induces muscle atrophy, but its effects on global mRNA expression in human skeletal muscle are not known. To determine this, we studied seven healthy adult humans (3 male and 4 female) with ages ranging from 25 to 69 years (mean = 46 years). The mean body mass index of these subjects (± SEM) was 25 ± 1. Their mean weight was 69.4 ± 4.8 kg. Baseline circulating levels of hemoglobin A1c (HbA1c), triglycerides (TG), thyroid-stimulating hormone (TSH), free thyroxine (free T4), C-reactive protein (CRP) and tumor necrosis factor-α (TNF-α) were within normal limits (Figure 1A).
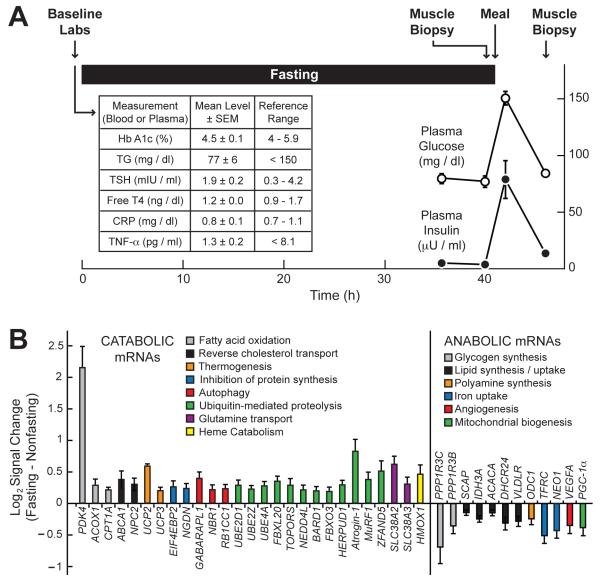
Figure 1. Effect of Fasting on Skeletal Muscle mRNA Expression in Healthy Human Adults.
(A) Study design. The table (insert) shows baseline circulating metabolic and inflammatory markers. The graph shows plasma glucose and insulin levels. Data are means ± SEM from the seven study subjects. In some cases, the error bars are too small to see. (B) Representative fasting-responsive human skeletal muscle mRNAs, and the effect of fasting on their log2 hybridization signals, as assessed by Affymetrix Human Exon 1.0 ST arrays. In each subject, the fasting signal was normalized to the nonfasting signal from the same subject. Data are means ± SEM from 7 subjects. P ≤ 0.02 by paired t-test for all mRNAs shown. The complete set of 558 fasting-responsive mRNAs is shown in Table S1. See also Figure S1.
While staying in our Clinical Research Unit, the subjects fasted for 40 h by forgoing food but not water. The mean weight loss during the fast was 1.7 ± 0.1 kg (3 ± 0 % of the initial body weight). After the 40 h fast, we obtained a biopsy from the subjects’ vastus lateralis (VL) muscle. Immediately after the muscle biopsy, the subjects ate a mixed meal. Five hours later (six hours after the first biopsy), we obtained a second muscle biopsy from their contralateral VL muscle. Thus, each subject had a muscle biopsy under fasting and nonfasting conditions. As expected, plasma glucose and insulin levels were low at the end of the 40 h fast, rose after the meal, and returned to baseline by the time of the second biopsy (Figure 1A). These data indicate comparable levels of plasma glucose and insulin at the times of the first (fasting) and second (nonfasting) muscle biopsies.
To determine the effect of fasting on skeletal muscle mRNA expression, we isolated RNA from the paired muscle biopsies and then analyzed it with exon expression arrays. Using P ≤ 0.02 (by paired t-test) as criteria for statistical significance, we found that 281 mRNAs were higher in the fasting state and 277 were lower (out of > 17,000 mRNAs measured). A complete list of these fasting-responsive mRNAs is shown in Table S1. Most of the mRNAs that were altered by fasting did not have known roles in muscle atrophy. However, fasting increased several mRNAs that encode proteins with known roles in catabolic processes such as fat oxidation, reverse cholesterol transport, thermogenesis, inhibition of protein synthesis, autophagy, ubiquitin-mediated proteolysis, glutamine transport and heme catabolism (Figure 1B). Of these, atrogin-1, MuRF1 and ZFAND5 mRNAs encode proteins known to be required for skeletal muscle atrophy in mice (Bodine et al., 2001a; Hishiya et al., 2006). Conversely, fasting significantly decreased several mRNAs encoding proteins with known roles in anabolic processes such as glycogen synthesis, lipid synthesis and uptake, polyamine synthesis, iron uptake, angiogenesis, and mitochondrial biogenesis (Figure 1B). Of these, PGC-1α mRNA encodes a protein that inhibits atrophy-associated gene expression and skeletal muscle atrophy in mice (Sandri et al., 2006). We used qPCR to validate several fasting-responsive mRNAs from human skeletal muscle (Figure S1). Taken together, these data established an mRNA expression signature of fasting in human skeletal muscle.
Identification of Ursolic Acid as an Inhibitor of Fasting-Induced Muscle Atrophy
The Connectivity Map describes the effects of > 1300 bioactive small molecules on global mRNA expression in several cultured cell lines, and contains search algorithms that permit comparisons between compound-specific mRNA expression signatures and mRNA expression signatures of interest (Lamb et al., 2006). We hypothesized that querying the Connectivity Map with the mRNA expression signature of fasting would identify inhibitors of atrophy-associated gene expression and thus, potential inhibitors of muscle atrophy. We also reasoned that increasing the specificity of our query would enhance the output. To this end, we determined an evolutionarily conserved mRNA expression signature of fasting by comparing the effect of fasting on human skeletal muscle to the effect of a 24 h fast on mouse skeletal muscle. Our mouse studies were described previously (Ebert et al., 2010). Altogether, we identified 35 mRNAs that were increased by fasting and 40 mRNAs that were decreased by fasting in both human and mouse skeletal muscle (Table S2). Of these, 63 were represented on the HG-U133A arrays used in the Connectivity Map (Figure 2A). We used these mRNAs (31 increased by fasting and 32 decreased by fasting) to query the Connectivity Map for candidate small molecule inhibitors of muscle atrophy.
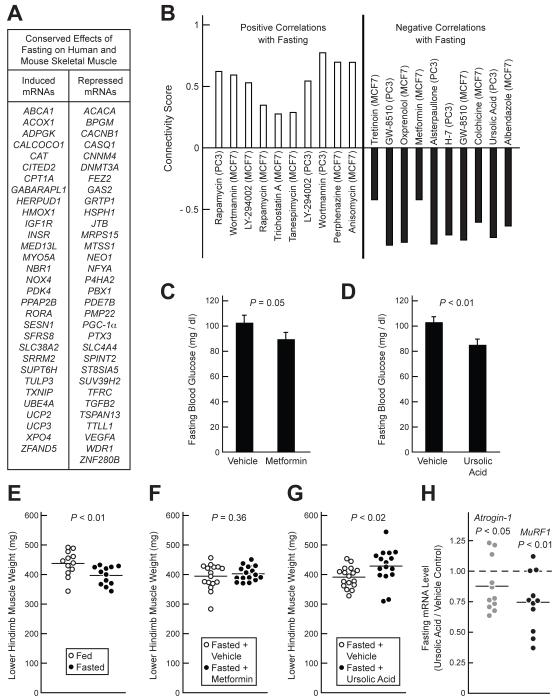
Figure 2. Identification of Ursolic Acid as an Inhibitor of Fasting-Induced Skeletal Muscle Atrophy.
(A) Fasting-regulated mRNAs common to human and mouse muscle that were used to query the Connectivity Map. Inclusion criteria were: P ≤ 0.02 in fasted human muscle (by t-test), P ≤ 0.05 in fasted mouse muscle (by t-test), and the existence of complimentary probes on HG- U133A arrays. (B) Connectivity Map instances (or datasets), sorted by compound and cell line, with the most significant positive and negative correlations to the effect of fasting in human and mouse muscle. The connectivity score, represented on the y-axis, is a measure of the strength of the correlation (Lamb et al., 2006). P < 0.004 for all compounds shown. (C-D and F-H) Mice were administered metformin (250 mg / kg), ursolic acid (200 mg / kg) or an equivalent volume of vehicle alone via i.p. injection, and then fasted. After 12 hours of fasting, a second injection of metformin, ursolic acid or vehicle was administered. After 24 hours of fasting, the blood glucose was measured and muscles were harvested. (C-D) Effect of metformin (C) and ursolic acid (D) on fasting blood glucose. Data are means ± SEM from 16 mice. (E) Effect of 24 h fast (relative to ad lib feeding) on wet weight of lower hindlimb skeletal muscle (bilateral tibialis anterior (TA), gastrocnemius, and soleus). (F-G) Effect of metformin (F) and ursolic acid (G) on fasted lower hindlimb muscle weight. (H) Effect of ursolic acid on atrogin-1 and MuRF1 mRNA levels in the TA muscles of fasted mice. The data are normalized to the levels in vehicle-treated mice, which were set at 1. (E-H) Each data point represents one mouse and the horizontal bars denote the means. (C-H) P-values were determined using unpaired t-tests. See also Table S2.
The left side of Figure 2B shows the 10 Connectivity Map instances (or data sets) with the most significant positive correlations to the effect of fasting in skeletal muscle. Of these, 6 involved wortmannin or LY-294002 (inhibitors of phosphoinositide 3-kinase (PI3K)) or rapamycin (an inhibitor of the mammalian target of rapamycin complex 1 (mTORC1)). Since PI3K and mTORC1 mediate effects of insulin and IGF-I, and since insulin/IGF-I signaling inhibits muscle atrophy and atrophy-associated changes in skeletal muscle mRNA expression (Bodine et al., 2001b; Sandri et al., 2004), these results lent confidence that the Connectivity Map might be used to identify potential inhibitors of muscle atrophy.
The right side of Figure 2B shows the 10 Connectivity Map instances with the most significant negative correlations to the effect of fasting in skeletal muscle. These compounds, whose effects on cultured cell lines were opposite to the effect of fasting on muscle, included metformin (an insulin-sensitizing agent widely used to treat type 2 diabetes), as well as ursolic acid. Interestingly, ursolic acid was the only compound identified by both this query and a second, independent query for potential inhibitors of muscle atrophy (described below). Thus, we chose to focus further experiments on metformin and ursolic acid.
To test the hypothesis that metformin and ursolic acid might reduce fasting-induced muscle atrophy, we administered each compound, or vehicle alone, via i.p. injection to C57BL/6 mice. We then fasted the mice, and after 12 hours of fasting, the mice received a second dose of the compound or vehicle. After 24 hours of fasting, the mice were examined. Both metformin (250 mg / kg) and ursolic acid (200 mg / kg) significantly reduced fasting blood glucose (Figures 2C and 2D). We next examined the effects of metformin and ursolic acid on fasting-induced muscle atrophy. In the absence of metformin and ursolic acid, fasting reduced muscle weight by 9 % (Figure 2E). Although metformin did not alter muscle weight in fasted mice (Figure 2F), ursolic acid increased it by 7 ± 2 % (Figure 2G). Moreover, consistent with its predicted inhibitory effect on fasting-induced gene expression, ursolic acid reduced fasting levels of atrogin-1 and MuRF1 mRNAs (Figure 2H). Thus, ursolic acid, but not metformin, decreased fasting-induced muscle atrophy.
Ursolic Acid Reduces Denervation-Induced Muscle Atrophy
We asked whether a different mRNA expression signature of muscle atrophy might also identify ursolic acid. To test this, we queried the Connectivity Map with human skeletal muscle mRNAs that were induced or repressed by fasting and also by spinal cord injury (SCI). Our studies of the effects of SCI on human skeletal muscle gene expression were described previously (Adams et al., 2011). Altogether, we identified 18 mRNAs that were increased by fasting and SCI, and 17 mRNAs that were decreased by fasting and SCI (Table S3). Of these, 29 were represented on the HG-U133A arrays used in the Connectivity Map (Figure 3A), but only 10 were common to the 63 mRNAs used in our first Connectivity Map query (IGF-IR, NOX4, SUPT6H, MRPS15, PDE7B, PGC-1a, TSPAN13, TTLL1, VEGFA and ZNF280B). When we queried the Connectivity Map with this second signature of muscle atrophy, the results partially overlapped with the results of our first search: both search strategies identified LY-294002, wortmannin and rapamycin as predicted mimics of atrophy-inducing stress, and ursolic acid (but not metformin) as a predicted inhibitor (Figure 3B). Strikingly, ursolic acid was the only predicted inhibitor identified by both search strategies (compare Figures 2B and 3B).
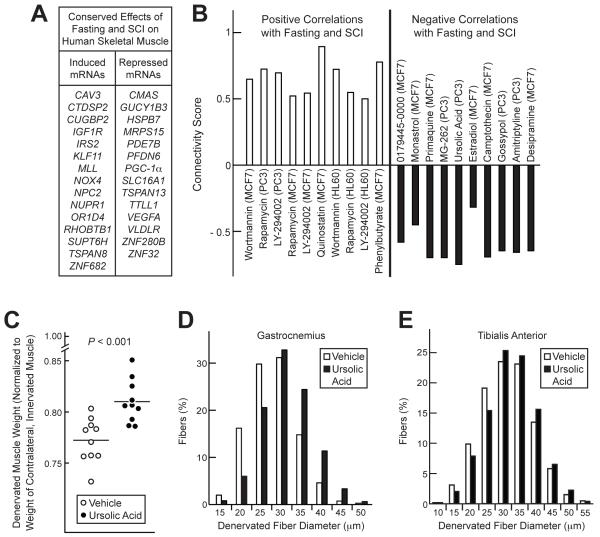
Figure 3. Identification of Ursolic Acid as an Inhibitor of Denervation-Induced Skeletal Muscle Atrophy.
(A) mRNAs altered by both fasting and SCI in human muscle that were used to query the Connectivity Map. Inclusion criteria were: P ≤ 0.02 in fasted human muscle (by t-test), P ≤ 0.05 in untrained, paralyzed muscle (by t-test), and the existence of complimentary probes on HG-U133A arrays. (B) Connectivity Map instances with the most significant positive and negative correlations to the effect of fasting and SCI in human muscle. P < 0.005 for all compounds shown. (C-E) On day 0, the left hindlimbs of C57BL/6 mice were denervated by transsecting the left sciatic nerve. Mice were then administered ursolic acid (200 mg / kg) or an equivalent volume of vehicle alone (corn oil) via i.p. injection twice daily for seven days. On day 7, muscles were harvested for analysis. (C) Weights of the left (denervated) lower hindlimb muscles were normalized to weights of the right (innervated) lower hindlimb muscles from the same mouse. Each data point represents one mouse, and horizontal bars denote the means. P-value was determined using an unpaired t-test. (D-E) Effect of ursolic acid on skeletal muscle fiber diameter in denervated gastrocnemius (D) and TA (E) muscles. Data are from > 2500 muscle fibers per condition; P < 0.0001 by unpaired t-test. See also Table S3.
Because our second strategy utilized data from SCI subjects, we hypothesized that ursolic acid might reduce denervation-induced muscle atrophy. To test this, we selectively denervated the left hindlimb muscles of mice by transsecting the left sciatic nerve. This allowed us to use the right hindlimb as an intra-subject control. We then administered ursolic acid or vehicle twice daily for the next seven days, while the mice continued to have ad libitum access to food. Seven days after denervation, we compared the left (denervated) and right (innervated) hindlimb muscles in both groups. Ursolic acid significantly decreased denervation-induced muscle loss (Figure 3C). Histologically, this effect of ursolic acid was reflected as an increase in the size of denervated skeletal muscle fibers (Figures 3D and 3E). Thus, ursolic acid reduced denervation-induced muscle atrophy.
Ursolic Acid Induces Skeletal Muscle Hypertrophy
The finding that ursolic acid reduced muscle atrophy led us to hypothesize that it might promote muscle hypertrophy in the absence of an atrophy-inducing stress. To test this, mice were allowed ad libitum access to either control chow or chow containing 0.27 % ursolic acid for 5 weeks. Compared to mice on the control diet, mice receiving ursolic acid possessed larger skeletal muscles (Figures 4A – 4C), larger skeletal muscle fibers (Figure 4D), and increased grip strength (Figure 4E). Moreover, dietary ursolic acid increased the specific force generated by muscles ex vivo (Figure S2). These data provided morphological and functional evidence that ursolic acid induced skeletal muscle hypertrophy.
Figure 4. Ursolic Acid Induces Skeletal Muscle Hypertrophy.
Mice were provided ad lib access to either standard chow (control diet) or standard chow supplemented with 0.27% ursolic acid (ursolic acid diet) for 5 weeks before grip strength was measured and tissues were harvested. (A-C) Effect of ursolic acid on lower hindlimb muscle weight (A), quadriceps weight (B), and upper forelimb muscle (triceps and biceps) weight (C). Each data point represents one mouse, and horizontal bars denote the means. (D) Effect of ursolic acid on skeletal muscle fiber size distribution. Each distribution represents measurements of > 800 triceps muscle fibers from 7 animals (> 100 measurements / animal); P < 0.0001. (E) Effect of ursolic acid on peak grip strength, normalized to body weight. Each data point represents one mouse, and horizontal bars denote the means. Non-normalized grip strength data were 157 ± 9 g (control diet) and 181 ± 6 g (ursolic acid diet) (P = 0.04). See also Figure S2.
Ursolic Acid Induces Trophic Changes in Skeletal Muscle Gene Expression
Our discovery process suggested that ursolic acid might alter skeletal muscle gene expression. To test this hypothesis, we took an unbiased approach, using exon expression arrays to analyze gastrocnemius muscle mRNA expression in mice that had been fed diets lacking or containing ursolic acid for 5 weeks. Using stringent criteria for ursolic acid-induced effects on mRNA levels (P < 0.005), and disregarding mRNAs with low levels of expression (log2 hybridization signal < 8), we found that ursolic acid decreased 18 mRNAs and increased 51 mRNAs (out of > 16,000 mRNAs analyzed; Table S4).
As discussed above, atrogin-1 and MuRF1 are transcriptionally up-regulated by atrophy-inducing stresses ((Sacheck et al., 2007) and Figure 1B), and they are required for muscle atrophy (Bodine et al., 2001a). Moreover, in our studies of fasted mice, we found that ursolic acid reduced atrogin-1 and MuRF1 mRNAs (Figure 2H). Consistent with that finding, the arrays indicated that dietary ursolic acid reduced atrogin-1 mRNA, which was the most highly repressed mRNA measured on the array (Figure 5A). Although MuRF1 mRNA was not measured by the arrays used in these experiments, qPCR analysis confirmed that dietary ursolic acid repressed both atrogin-1 and MuRF1 mRNAs (Figure 5B).
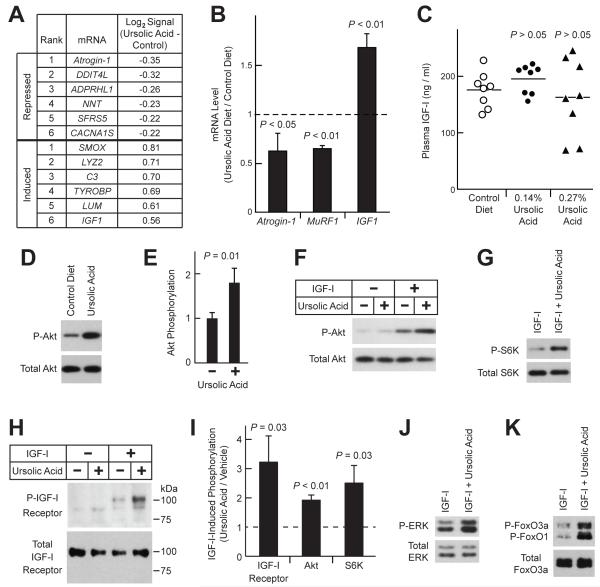
Figure 5. Ursolic Acid Promotes Muscle Growth by Repressing Atrophic Gene Expression, Inducing Trophic Gene Expression, and Enhancing Skeletal Muscle IGF-I Signaling.
(A-B) Mice were provided ad lib access to either standard chow (control diet) or standard chow supplemented with 0.27% ursolic acid (ursolic acid diet) for 5 weeks before tissues were harvested. (A) Ursolic acid-induced changes in the log2 hybridization signals of skeletal muscle mRNAs, as assessed by Affymetrix Mouse Exon 1.0 ST arrays. n = 4 arrays per diet; each array assessed gastrocnemius RNA pooled from two mice. Data were filtered for P ≤ 0.005 by unpaired t-test and log2 hybridization signal ≥ 8. Table shows the top 6 mRNAs most induced or repressed by dietary ursolic acid. (B) Effect of ursolic acid on IGF1, atrogin-1 and MuRF1 mRNA levels, as assessed by qPCR. Data are means ± SEM. (C) Mice were provided ad lib access to either standard chow (control diet) or standard chow supplemented with the indicated concentration of ursolic acid for 7 weeks before plasma IGF-I levels were measured. Each data point represents one mouse, and horizontal bars denote the means. P-values were determined by one-way ANOVA with Dunnett’s post-test. (D-E) Mice were provided ad lib access to either standard chow (control diet) or standard chow supplemented with 0.27% ursolic acid for 16 weeks. Total protein extracts from quadriceps muscles were subjected to SDS-PAGE, followed by immunoblot analysis for phosphorylated and total Akt, as indicated. (D) Representative immunoblot. (E) In each mouse, the level of phospho-Akt was normalized to the level of total Akt. These ratios were then normalized to the average phospho-Akt/total Akt ratio from control mice. Data are means ± SEM from 9 mice per diet. P-value was determined by unpaired t-test. (F-K) Serum-starved C2C12 myotubes were treated in the absence or presence of ursolic acid (10 μM) and/or IGF-I (10 nM), as indicated. For studies of the IGF-I receptor, cells were harvested 2 min later, and protein extracts were subjected to immunoprecipitation with anti-IGF-I receptor β antibody, followed by immunoblot analysis with anti-phospho-tyrosine or anti-IGF-I receptor β antibodies to assess phospho- and total IGF-I receptor, respectively. For other studies, cells were harvested 20 min after addition of ursolic acid and/or IGF-I, and immunoblot analyses were performed using total cellular protein extracts and antibodies specific for the phosphorylated or total proteins indicated. (F-H) Representative immunoblots showing effect of ursolic acid on IGF-I-mediated phosphorylation of Akt (F), S6K (G) and IGF-I receptor (H). (I) Quantification of the effect of ursolic acid on IGF-I-dependent phosphorylation of the IGF-I receptor, Akt and S6K. Levels in the presence of ursolic acid and IGF-I are normalized to levels in the presence of IGF-I alone, which were set at 1 and are indicated by the dashed line. Data are means ± SEM from ≥ 3 experiments. (J-K) Ursolic acid enhances IGF-I-mediated phosphorylation of ERK (J) and FoxO3a/FoxO1 (K). See also Table S4 and Figure S3.
Interestingly, one of the most highly up-regulated muscle mRNAs was IGF1 (Figures 5A and 5B), which encodes insulin-like growth factor-I (IGF-I), a locally generated autocrine/paracrine hormone. IGF1 mRNA is known to be transcriptionally induced in hypertrophic muscle (Adams and Haddad, 1996; Gentile et al.; Hameed et al., 2004). In addition, increased skeletal muscle IGF1 expression reduces denervation-induced muscle atrophy (Shavlakadze et al., 2005), and stimulates muscle hypertrophy (Barton-Davis et al., 1998; Musaro et al., 2001). Moreover, by stimulating skeletal muscle insulin/IGF-I signaling, IGF-I represses atrogin-1 and MuRF1 mRNAs (Frost et al., 2009; Sacheck et al., 2004), as well as DDIT4L mRNA (Frost et al., 2009; Sacheck et al., 2004), which, after atrogin-1 mRNA, was the second most highly repressed mRNA in muscle from ursolic acid-treated mice (Figure 5A). Thus, 5 weeks of dietary ursolic acid altered skeletal muscle gene expression in a manner known to reduce atrophy and promote hypertrophy, and muscle-specific IGF1 induction emerged as a likely contributing mechanism in ursolic acid-induced muscle hypertrophy.
Of note, our exon expression arrays indicated that ursolic acid increased levels of all measured IGF1 exons (exons 2-6; Figure S3). However, ursolic acid did not alter levels of mRNAs encoding myostatin (which reduces muscle mass (Lee, 2004)), or twist or myogenin (which are induced by IGF-I during development (Dupont et al., 2001; Tureckova et al., 2001)). We also measured the effect of ursolic acid on plasma IGF-I levels, which primarily reflect growth hormone-mediated hepatic IGF-I production (Yakar et al., 1999). Although diets containing 0.14% or 0.27% ursolic acid increased muscle mass (described in greater detail below; Figure 6A), neither increased plasma IGF-I (Figure 5C). Moreover, ursolic acid did not alter the amount of IGF1 mRNA in adipose tissue (Figure S3). These data suggest that ursolic acid-mediated IGF1 induction may be localized to skeletal muscle.
Figure 6. Ursolic Acid Reduces Adiposity.
(A-B) Mice were provided ad lib access to standard chow supplemented with the indicated concentration of ursolic acid for 7 weeks before tissues were harvested for analysis. Data are means ± SEM from 10 mice per diet. (A) Effects of ursolic acid on weights of skeletal muscle (quadriceps + triceps), epididymal fat, retroperitoneal fat and heart. P-values (determined by one-way ANOVA with post-test for linear trend) were < 0.001 for muscle; 0.01 and 0.04 for epididymal and retroperitoneal fat, respectively; and 0.46 for heart. (B) Total body weights before and after 7 weeks of the indicated concentration of dietary ursolic acid. P-values were 0.71 and 0.80 for initial and final weights, respectively. (C-F) Mice were provided ad lib access to either standard chow (control diet) or standard chow supplemented with 0.27% ursolic acid (ursolic acid diet) for 5 weeks, as in Figure 4. (C) Relationship between skeletal muscle weight (quadriceps, triceps, biceps, TA, gastrocnemius and soleus) and retroperitoneal adipose weight. Each data point represents one mouse. P < 0.001 for both muscle and adipose by unpaired t-test. (D) Representative H&E stain of retroperitoneal fat. (E) Effect of ursolic acid on average retroperitoneal adipocyte diameter. Each data point represents the average diameter of ≥ 125 retroperitoneal adipocytes from one mouse. (F) Effect of ursolic acid on retroperitoneal adipocyte size distribution. Each distribution represents combined adipocyte measurements (> 1000 per diet) from (E). (G-H) Mice were fed as in (A-B). (G) Plasma leptin. Each data point represents one mouse, and horizontal bars denote the means. P-values were determined by t-test. (H) Relationship between adipose weight and plasma leptin. Each data point represents one mouse. (I-J) Mice were fed as in (C-F) before plasma triglycerides (I) and cholesterol (J) were measured. Each data point represents one mouse, and horizontal bars denote the means. P-values were determined by unpaired t-test. See also Figure S4.
Ursolic Acid Enhances Skeletal Muscle Insulin/IGF-I Signaling
Although muscle-specific IGF1 induction is characteristic of, and contributes to, muscle hypertrophy, it may be a relatively late event that promotes hypertrophy after it has been initiated by other stimuli (Adams et al., 1999). We hypothesized that ursolic acid might have a more proximal effect on insulin/IGF-I signaling. In a previous study of non-muscle cell lines (CHO/IR and 3T3-L1 cells), ursolic acid enhanced insulin-mediated Akt activation (Jung et al., 2007). To determine whether ursolic acid might have a similar effect in skeletal muscle, we first assessed the level of phosphorylated Akt in quadriceps muscles of mice fed diets lacking or containing ursolic acid. In quadriceps, ursolic acid increased Akt phosphorylation by 1.8-fold (Figures 5D and 5E).
We next examined whether ursolic acid might increase Akt activation in C2C12 skeletal myotubes, a well-established in vitro model of skeletal muscle (Sandri et al., 2004; Stitt et al., 2004). Use of an in vitro system circumvented potentially confounding effects from non-muscle tissues, and allowed us to test if IGF-I or insulin was required for ursolic acid’s effect. The latter consideration was important because circulating IGF-I and insulin are always present in healthy animals. Use of an in vitro system also allowed us to test a clearly defined concentration of ursolic acid (10 μM, similar what was used in the Connectivity Map (8.8 μM)) for a clearly defined time of incubation (20 min). These considerations were important because the in vivo pharmacokinetic properties of ursolic acid are not yet known. When serum-starved myotubes were treated with ursolic acid alone, Akt phosphorylation did not increase (Figure 5F). However, in the presence of IGF-I, ursolic acid increased Akt phosphorylation by 1.9-fold (Figures 5F and 5I). Ursolic acid also increased Akt phosphorylation in the presence of insulin (Figure S3). Thus, ursolic acid enhanced IGF-I-mediated and insulin-mediated Akt phosphorylation.
The finding that ursolic acid enhanced muscle Akt activity in vivo and in vitro was consistent with the finding that ursolic acid’s mRNA expression signature negatively correlated with the mRNA expression signatures of LY-294002 and wortmannin (Figures 2B and 3B), which inhibit insulin/IGF-I signaling upstream of Akt. However, ursolic acid’s signature also negatively correlated with the signature of rapamycin, which inhibits insulin/IGF-I signaling downstream of Akt. We therefore asked whether ursolic acid might enhance S6 kinase (S6K) phosphorylation, which occurs distal to the effect of rapamycin. Although ursolic acid alone did not increase S6K phosphorylation (Supplemental Figure 5), it enhanced IGF-I-mediated and insulin-mediated S6K phosphorylation (Figures 5G, 5I and S3).
To further investigate the mechanism, we examined the IGF-I receptor. Ursolic acid increased IGF-I receptor phsophorylation in the presence but not the absence of IGF-I (Figure 5H and 5I). Similarly, ursolic acid increased insulin receptor phosphorylation in the presence but not the absence of insulin (Figure S3). Both of these effects were rapid, occurring within 2 minutes after the addition of ursolic acid and either IGF-I or insulin. Consistent with enhanced signaling at the level of the IGF-I and insulin receptors, ursolic acid also enhanced IGF-I-mediated and insulin-mediated ERK phosphorylation (Figures 5J and S3). Moreover, ursolic acid enhanced IGF-I-mediated phosphorylation (inhibition) of FoxO transcription factors, which activate transcription of atrogin-1 and MuRF1 mRNAs (Figure 5K, (Sandri et al., 2004; Stitt et al., 2004)). Taken together, these data indicate that ursolic acid represses atrophy-associated gene expression and promotes muscle hypertrophy by increasing activity of the IGF-I and insulin receptors.
In Contrast to Its Effect on Skeletal Muscle, Ursolic Acid Reduces Adiposity
Figure 6A shows that 7 weeks of dietary ursolic acid increased skeletal muscle weight in a dose-dependent manner, with a peak effect at 0.14%. Interestingly, although ursolic acid increased muscle weight, it did not increase total body weight (Figure 6B). Because ursolic acid increased Akt activity (Figure 5), and because muscle-specific increases in Akt activity reduce adiposity as a secondary consequence of muscle hypertrophy (Izumiya et al., 2008; Lai et al., 2004), we hypothesized that ursolic acid-treated mice might contain less fat. Indeed, 7 weeks of dietary ursolic acid reduced the weight of epididymal and retroperitoneal fat depots, with a peak effect at 0.14% (Figure 6A). Similar results were obtained when mice were fed a diet containing 0.27% ursolic acid for 5 weeks (Figure 6C). Thus, muscle and fat weights were inversely related.
Ursolic acid reduced adipose weight by reducing adipocyte size (Figures 6D - 6F). This was accompanied by a significant reduction in plasma leptin levels, which correlated closely with adipose weight (Figures 6G and H). Although ursolic acid reduced leptin, it did not alter food intake (Figure S4). Importantly, ursolic acid also significantly reduced plasma triglyceride (Figure 6I) and cholesterol (Figure 6J). However, ursolic acid did not alter weights of heart (Figure 6A), liver or kidney (Figure S4), nor did it elevate plasma markers of hepatotoxicity or nephrotoxicity (alanine aminotransferase, bilirubin and creatinine) (Figure S4). Thus, dietary ursolic acid had two major effects: skeletal muscle hypertrophy and reduced adiposity.
DISCUSSION
Landmark studies using rodent models showed that diverse atrophy-inducing stresses (including fasting, systemic illness and muscle disuse) generate similar patterns of changes in skeletal mRNA expression (Sacheck et al., 2007). We translated those findings to humans by determining the effect of fasting on global skeletal muscle mRNA expression in human skeletal muscle. Our fasting protocol was well tolerated by subjects, relatively simple to execute, and required only a few subjects to discern important changes in skeletal muscle mRNA expression. For example, it allowed us to demonstrate, for the first time, fasting-mediated induction of atrogin-1 and MuRF1 mRNAs in human skeletal muscle.
In human skeletal muscle, fasting altered levels of > 500 skeletal muscle mRNAs (approximately 3% of the total mRNAs examined). However, only a few of these mRNAs are known to play central roles in muscle atrophy in mice (including atrogin-1, MuRF1, ZFAND5 and PGC-1 ). Likewise, most mRNAs that were altered by fasting in both human and mouse muscle (which formed the basis for our first Connectivity Map query), and most mRNAs that were altered by both fasting and SCI in human muscle (which formed the basis for our second Connectivity Map query) have undefined roles in muscle atrophy. Although we do not yet know the functional roles of most mRNAs whose levels are altered by fasting or SCI, or the precise mechanisms that regulate them in the setting of an atrophy-inducing stress, we used these data to query the Connectivity Map. This unbiased approach singled out one compound as a predicted inhibitor of atrophy-inducing stress: ursolic acid.
A water-insoluble pentacyclic triterpenoid, ursolic acid is the major waxy component in apple peels (Frighetto et al., 2008). It is also found in many other edible plants. Interestingly, because it exerts beneficial effects in animal models of diabetes and hyperlipidemia (Liu, 1995; Wang et al., 2009), ursolic acid is thought to be the active component in a variety of folkloric anti-diabetic herbal medicines (Liu, 1995, 2005). As predicted by the Connectivity Map, we found that ursolic acid reduced skeletal muscle atrophy in the setting of two distinct atrophy-inducing stresses (fasting and muscle denervation). A major strength of the Connectivity Map is that it takes into account positive and negative changes in mRNA expression that together constitute an authentic mRNA expression signature. Thus, by querying the Connectivity Map with signatures of muscle atrophy, we were, in effect, querying with the reciprocal signature of muscle hypertrophy. Indeed, ursolic acid not only reduced muscle atrophy, but also induced muscle hypertrophy.
The strategy that led us to ursolic acid implied that ursolic acid might increase muscle mass by inhibiting atrophy-associated skeletal muscle gene expression. Indeed, we found that acute ursolic acid treatment of fasted mice reduced atrogin-1 and MuRF1 mRNAs in association with reduced muscle atrophy. Similarly, chronic ursolic acid treatment of unstressed mice reduced atrogin-1 and MuRF1 mRNAs, and induced muscle hypertrophy. Interestingly, ursolic acid-induced muscle hypertrophy was also associated with induction or repression of > 60 other skeletal muscle mRNAs, including IGF1 mRNA (which was induced). Although previous studies showed that increased skeletal muscle IGF1 expression is sufficient to inhibit atrophy and promote hypertrophy, we noted that, following a hypertrophic stimulus such as mechanical loading, increased IGF1 gene expression is a late event (Adams et al., 1999). We therefore asked whether ursolic acid might stimulate earlier events in insulin/IGF-I signaling by examining skeletal muscle Akt activation, a critical node in the insulin and IGF-I signaling cascades. Indeed, in muscles that had hypertrophied secondary to chronic ursolic acid treatment, Akt phosphorylation was increased. Interestingly, this increase in skeletal muscle Akt activity can potentially account for many of ursolic acid’s effects, including reduced atrophy-associated gene expression, reduced muscle atrophy, increased muscle hypertrophy and reduced adiposity (Izumiya et al., 2008; Lai et al., 2004). However, additional studies will be needed to determine whether Akt is required for the effects of ursolic acid, and whether other pathways (such as the calcineurin/NFAT and MAP kinase pathways) might also be involved.
Although ursolic acid increased skeletal muscle Akt phosphorylation in vivo, those experiments could not determine if ursolic acid acted directly on skeletal muscle, how quickly ursolic acid acted, and if the effect of ursolic acid required IGF-I or insulin, which are always present in healthy animals, even during fasting. To address these questions, we studied serum-starved skeletal myotubes, and found that ursolic acid rapidly stimulated IGF-I receptor and insulin receptor activity, but only if IGF-I or insulin was also present. Taken together, our data suggest that ursolic acid first enhances the capacity of pre-existing IGF-I and insulin to activate skeletal muscle IGF-I receptors and insulin receptors, respectively. This activates Akt, S6K and ERK, and alters skeletal muscle gene expression in a manner that reduces atrophy and promotes hypertrophy. Specific changes in downstream gene expression include induction of IGF1 (a feed-forward mechanism that likely contributes to ursolic acid-mediated hypertrophy), repression of atrogin-1 and MuRF1, and induction or repression of many other genes whose contributions to muscle atrophy or hypertrophy remain to be determined. Some of these changes in skeletal muscle gene expression (such as repression of atrogin-1 and MuRF1) can be explained by our finding that ursolic acid enhances IGF-I-mediated inhibition of FoxO transcription factors. However, ursolic acid might also inhibit other transcription factors that promote atrophy, such as NF-kB, and this is an important area for future investigation.
Importantly, ursolic acid alone was not sufficient to increase phosphorylation of the IGF-I receptor or the insulin receptor. Rather, its effects also required IGF-I or insulin, respectively. This suggests that ursolic acid either facilitates hormone-mediated receptor autophosphorylation, or it inhibits receptor dephosphorylation. The latter possibility is supported by previous in vitro data that ursolic acid directly inhibits PTP1B (Zhang et al., 2006), a tyrosine phosphatase that dephosphorylates (inactivates) the IGF-I and insulin receptors (Kenner et al., 1996). However, neither global nor muscle-specific PTP1B knockout mice were found to possess increased muscle mass (Delibegovic et al., 2007; Klaman et al., 2000). This may suggest the existence of another receptor for ursolic acid, which might be closely related to PTP1B. Identifying the receptor(s) for ursolic acid is an important area for future investigation that may elucidate important mechanisms of metabolic control. Pharmacokinetic studies of ursolic acid will also be critical for fully understanding its in vivo effects.
Given the current lack of therapies for skeletal muscle atrophy, we speculate that ursolic acid might be investigated as a potential therapy for illness- and age-related muscle atrophy. It may be useful as a monotherapy or in combination with other strategies that have been considered, such as myostatin inhibition (Zhou et al., 2010). Given its capacity to reduce adiposity, fasting blood glucose and plasma lipid levels, ursolic acid might also be investigated as a potential therapy for obesity, metabolic syndrome and type 2 diabetes. A systematic search for ursolic acid derivatives that are more potent and/or efficacious could also be undertaken. Further work in this area may lead to new medical therapies for increasingly common metabolic diseases that reduce the absolute or relative amount of skeletal muscle.
EXPERIMENTAL PROCEDURES
Human Subjects
Our study was approved by the Institutional Review Board at the University of Iowa, and involved seven healthy adults who gave their informed consent before participating. One week prior to the fasting study, subjects made one visit to the Clinical Research Unit (CRU) for anthropometric measurements, a dietary interview that established each subject’s routine food intake and food preferences, and baseline determinations of blood hemoglobin A1c (by turbidimetric immunoinhibition using the Hitachi 911 analyzer; Boehringer Mannheim, Indianapolis, IN), plasma triglycerides, plasma free T4 and TSH (by electrochemiluminescence immunoassay using the Elecsys system; Roche Diagnostics, Indianapolis, IN), plasma CRP (by immuno-turbidimetric assay; Roche Integra high-sensitivity assay), and plasma TNF-α levels (by the Quantikine kit from R&D Systems Inc., Minneapolis, MN). To ensure that subjects were eating their routine diet prior to the fasting study, subjects ate only meals prepared by the CRU dietician (based on the dietary interview) for 48 hours before the fasting study. The fasting study began at t = 0 hours, when subjects were admitted to the CRU and began fasting. While fasting, subjects remained in the CRU and were encouraged to maintain their routine physical activities. Water was allowed ad libitum, but caloric intake was not permitted. At t = 40 hours, a percutaneous biopsy was taken from the vastus lateralis muscle using a Temno biopsy needle (T1420, CardinalHealth) under ultrasound guidance. Subjects then ate a CRU-prepared mixed meal, and at t = 46 hours, a muscle biopsy was taken from the contralateral vastus lateralis muscle. Plasma glucose and insulin levels were measured at t = 36, 40, 42 and 46 hours; the Elecsys system was used to quantitate plasma insulin. Our study protocol of humans with spinal cord injury was described previously (Adams et al., 2011).
Mouse Protocols
Male C57BL/6 mice, ages 6-8 weeks, were obtained from NCI, housed in colony cages with 12h light/12h dark cycles, and used for experiments within 3 weeks of their arrival. Unless otherwise indicated, mice were maintained on standard chow (Harlan Teklad formula 7013). Metformin (Sigma) was dissolved in 0.9% NaCl at a concentration of 25 mg / ml. Ursolic acid (Enzo Life Sciences) was dissolved in corn oil at a concentration of 20 mg / ml (for i.p. injections), or custom added to chow formula 7013 by Harlan Teklad. Mice were fasted by removing food, but not water, for 24 hours. Fasting blood glucose levels were obtained from the tail vein with an Accucheck Aviva glucose meter. Unilateral hindlimb muscle denervation was performed by transsecting the sciatic nerve under anesthesia, and was followed by administration of ursolic acid (200 mg / kg) or vehicle alone (corn oil) via i.p injection twice daily for 7 days. Forelimb grip strength was determined using a grip strength meter equipped with a triangular pull bar (Columbus Instruments). Each mouse was subjected to 5 consecutive tests to obtain the peak value. Plasma IGF-I and leptin levels were measured by RIA at the Vanderbilt University Hormone Assay Core Facility. Plasma cholesterol, triglyceride, creatinine, bilirubin and ALT were measured using the VITROS 350 Chemistry System. All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Iowa.
Microarray Analysis of Skeletal Muscle mRNA Levels
Following harvest, skeletal muscle samples were immediately placed in RNAlater (Ambion) and stored at −80 °C until further use. Total RNA was extracted using TRIzol solution (Invitrogen), and microarray hybridizations were performed at the University of Iowa DNA Facility, as described previously (Ebert et al, 2010). Our reported log2 hybridization signals reflect the mean signal intensity of all exon probes specific for an individual mRNA. To determine which human skeletal muscle mRNAs were significantly altered by fasting (P ≤ 0.02), we used paired t-tests to compare fasted and fed log2 signals. To determine which mouse skeletal muscle mRNAs were significantly altered by ursolic acid (P ≤ 0.005), we used unpaired t-tests to compare log2 signals in mice fed control diet or diet supplemented with ursolic acid; then, to focus on highly expressed mRNAs, we selected for significantly altered mRNAs that were repressed from or induced to a log2 signal > 8. These raw microarray data from humans and mice have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE28016 and GSE28017, respectively. Exon array studies of the effects of fasting on mouse skeletal muscle, and the effects of spinal cord injury on human skeletal muscle were described previously (Adams et al., 2011; Ebert et al., 2010).
Other Methods
Additional information on qPCR, histological and immunoblot methods may be found in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Michael Welsh and Peter Snyder for invaluable advice and critical review of the manuscript, and Drs. Daryl Granner, John Stokes and Allyn Mark for helpful discussions. This work was supported by a Clinical Scientist Development Award from the Doris Duke Charitable Foundation, a Career Development Award from the Department of Veterans Affairs, a Junior Faculty Award from the American Diabetes Association, Grant Number 1R01AR059115-01 from NIAMS/NIH, NIH T32 GM073610, and grants from the University of Iowa Institute for Clinical and Translational Science, the University of Iowa Research Foundation, and the Fraternal Order of Eagles Diabetes Research Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes four figures, four tables and experimental procedures.
REFERENCES
- Adams CM, Suneja M, D.-J. S, Shields RK. Alterations in skeletal muscle mRNA expression following long-term soleus training in humans with spinal cord injury. Muscle Nerve. 2011;43:65–75. doi: 10.1002/mus.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams GR, Haddad F. The relationships among IGF-1, DNA content, and protein accumulation during skeletal muscle hypertrophy. J. Appl. Physiol. 1996;81:2509–2516. doi: 10.1152/jappl.1996.81.6.2509. [DOI] [PubMed] [Google Scholar]
- Adams GR, Haddad F, Baldwin KM. Time course of changes in markers of myogenesis in overloaded rat skeletal muscles. J. Appl. Physiol. 1999;87:1705–1712. doi: 10.1152/jappl.1999.87.5.1705. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001a;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001b;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Delibegovic M, Bence KK, Mody N, Hong EG, Ko HJ, Kim JK, Kahn BB, Neel BG. Improved glucose homeostasis in mice with muscle-specific deletion of protein-tyrosine phosphatase 1B. Mol. Cell. Biol. 2007;27:7727–7734. doi: 10.1128/MCB.00959-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont J, Fernandez AM, Glackin CA, Helman L, LeRoith D. Insulin-like growth factor 1 (IGF-1)-induced twist expression is involved in the anti-apoptotic effects of the IGF-1 receptor. J. Biol. Chem. 2001;276:26699–26707. doi: 10.1074/jbc.M102664200. [DOI] [PubMed] [Google Scholar]
- Ebert SM, Monteys AM, Fox DK, Bongers KS, Shields BE, Malmberg SE, Davidson BL, Suneja M, Adams CM. The transcription factor ATF4 promotes skeletal myofiber atrophy during fasting. Mol. Endocrinol. 2010;24:790–799. doi: 10.1210/me.2009-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frighetto RTS, Welendorf RM, Nigro EN, Frighetto N, Siani AC. Isolation of ursolic acid from apple peels by high speed counter-current chromatography. Food Chemistry. 2008;106:767–771. [Google Scholar]
- Frost RA, Huber D, Pruznak A, Lang CH. Regulation of REDD1 by insulin-like growth factor-I in skeletal muscle and myotubes. J. Cell. Biochem. 2009;108:1192–1202. doi: 10.1002/jcb.22349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile MA, Nantermet PV, Vogel RL, Phillips R, Holder D, Hodor P, Cheng C, Dai H, Freedman LP, Ray WJ. Androgen-mediated improvement of body composition and muscle function involves a novel early transcriptional program including IGF1, mechano growth factor, and induction of {beta}-catenin. J. Mol. Endocrinol. 44:55–73. doi: 10.1677/JME-09-0048. [DOI] [PubMed] [Google Scholar]
- Hameed M, Lange KH, Andersen JL, Schjerling P, Kjaer M, Harridge SD, Goldspink G. The effect of recombinant human growth hormone and resistance training on IGF-I mRNA expression in the muscles of elderly men. J. Physiol. 2004;555:231–240. doi: 10.1113/jphysiol.2003.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiya A, Iemura S, Natsume T, Takayama S, Ikeda K, Watanabe K. A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J. 2006;25:554–564. doi: 10.1038/sj.emboj.7600945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya Y, Hopkins T, Morris C, Sato K, Zeng L, Viereck J, Hamilton JA, Ouchi N, LeBrasseur NK, Walsh K. Fast/Glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 2008;7:159–172. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SH, Ha YJ, Shim EK, Choi SY, Jin JL, Yun-Choi HS, Lee JR. Insulin-mimetic and insulin-sensitizing activities of a pentacyclic triterpenoid insulin receptor activator. Biochem. J. 2007;403:243–250. doi: 10.1042/BJ20061123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenner KA, Anyanwu E, Olefsky JM, Kusari J. Protein-tyrosine phosphatase 1B is a negative regulator of insulin- and insulin-like growth factor-I-stimulated signaling. The J. Biol. Chem. 1996;271:19810–19816. doi: 10.1074/jbc.271.33.19810. [DOI] [PubMed] [Google Scholar]
- Klaman LD, Boss O, Peroni OD, Kim JK, Martino JL, Zabolotny JM, Moghal N, Lubkin M, Kim YB, Sharpe AH, Stricker-Krongrad A, Shulman GI, Neel BG, Kahn BB. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol. Cell. Biol. 2000;20:5479–5489. doi: 10.1128/mcb.20.15.5479-5489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KM, Gonzalez M, Poueymirou WT, Kline WO, Na E, Zlotchenko E, Stitt TN, Economides AN, Yancopoulos GD, Glass DJ. Conditional activation of akt in adult skeletal muscle induces rapid hypertrophy. Mol. Cell. Biol. 2004;24:9295–9304. doi: 10.1128/MCB.24.21.9295-9304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lee SJ. Regulation of muscle mass by myostatin. Annu. Rev. Cell. Dev. Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- Liu J. Pharmacology of oleanolic acid and ursolic acid. J. Ethnopharmacol. 1995;49:57–68. doi: 10.1016/0378-8741(95)90032-2. [DOI] [PubMed] [Google Scholar]
- Liu J. Oleanolic acid and ursolic acid: research perspectives. J. Ethnopharmacol. 2005;100:92–94. doi: 10.1016/j.jep.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. AJP Endocrinol. Metab. 2004;287:E591–601. doi: 10.1152/ajpendo.00073.2004. [DOI] [PubMed] [Google Scholar]
- Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc. Natl. Acad. Sci. USA. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavlakadze T, White JD, Davies M, Hoh JF, Grounds MD. Insulin-like growth factor I slows the rate of denervation induced skeletal muscle atrophy. Neuromuscul. Disord. 2005;15:139–146. doi: 10.1016/j.nmd.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- Tureckova J, Wilson EM, Cappalonga JL, Rotwein P. Insulin-like growth factor-mediated muscle differentiation: collaboration between phosphatidylinositol 3-kinase-Akt-signaling pathways and myogenin. J. Biol. Chem. 2001;276:39264–39270. doi: 10.1074/jbc.M104991200. [DOI] [PubMed] [Google Scholar]
- Wang ZH, Hsu CC, Huang CN, Yin MC. Anti-glycative effects of oleanolic acid and ursolic acid in kidney of diabetic mice. Eur. J. Pharmacol. 2009;628:255–260. doi: 10.1016/j.ejphar.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, LeRoith D. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc. Natl. Acad. Sci. USA. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Hong D, Zhou Y, Zhang Y, Shen Q, Li JY, Hu LH, Li J. Ursolic acid and its derivative inhibit protein tyrosine phosphatase 1B, enhancing insulin receptor phosphorylation and stimulating glucose uptake. Biochim. Biophys. Acta. 2006;1760:1505–1512. doi: 10.1016/j.bbagen.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang JL, Lu J, Song Y, Kwak KS, Jiao Q, Rosenfeld R, Chen Q, Boone T, Simonet WS, Lacey DL, Goldberg AL, Han HQ. Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell. 142:531–543. doi: 10.1016/j.cell.2010.07.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.