Abstract
Vibrio mimicus, the species most similar to V. cholerae, is a microbe present in the natural environmental and sometimes causes diarrhea and internal infections in humans. It shows similar phenotypes to V. cholerae but differs in some biochemical characteristics. The molecular mechanisms underlying the differences in biochemical metabolism between V. mimicus and V. cholerae are currently unclear. Several V. mimicus isolates have been found that carry cholera toxin genes (ctxAB) and cause cholera-like diarrhea in humans. Here, the genome of the V. mimicus isolate SX-4, which carries an intact CTX element, was sequenced and annotated. Analysis of its genome, together with those of other Vibrio species, revealed extensive differences within the Vibrionaceae. Common mutations in gene clusters involved in three biochemical metabolism pathways that are used for discrimination between V. mimicus and V. cholerae were found in V. mimicus strains. We also constructed detailed genomic structures and evolution maps for the general types of genomic drift associated with pathogenic characters in polysaccharides, CTX elements and toxin co-regulated pilus (TCP) gene clusters. Overall, the whole-genome sequencing of the V. mimicus strain carrying the cholera toxin gene provides detailed information for understanding genomic differences among Vibrio spp. V. mimicus has a large number of diverse gene and nucleotide differences from its nearest neighbor, V. cholerae. The observed mutations in the characteristic metabolism pathways may indicate different adaptations to different niches for these species and may be caused by ancient events in evolution before the divergence of V. cholerae and V. mimicus. Horizontal transfers of virulence-related genes from an uncommon clone of V. cholerae, rather than the seventh pandemic strains, have generated the pathogenic V. mimicus strain carrying cholera toxin genes.
Introduction
Vibrio mimicus occasionally causes sporadic diarrhea and extraintestinal infections [1]. Although it was previously recognized as a biotype of V. cholerae, it has now been reclassified as an independent species because of differences in a number of biochemical characteristics; e.g., V. mimicus is negative for sucrose fermentation, Voges-Proskauer, lipase (corn oil) activity, and Jordan tartrate reactions [2]. However, in terms of pathogenesis, V. mimicus and V. cholerae are similar due to sharing virulence factors, such as enterotoxins or hemolysins [3], [4], [5], [6], [7]. Although V. mimicus has not yet been reported to produce a severe epidemic of diarrhea, it is often isolated from sporadic diarrheal patients, sea water and food, indicating that it may be a potential source of the emergence of a new pathogen, as increasing numbers of genetic elements and virulence factors are exchanged by acquisition of foreign DNA from V. cholerae or other bacteria [8].
The naturally occurring strain of V. mimicus lives in aquatic ecosystems, and its hosts are phytoplankton and crustaceans [9]. A number of virulence factors have been reported related to human infections [10]. V. mimicus is known to produce three types of hemolysins [11]. V. mimicus hemolysin (VMH) is heat labile and immunologically similar to V. cholerae El Tor hemolysin [5]. Vm-TDH is a heat stable hemolysin that is closely related to the thermostable direct hemolysin (TDH) produced by V. parahaemolyticus [6]. Finally, V. mimicus produces a novel hemolysin, designated HLX, but little is known regarding its function [11]. Additionally, some clinical isolates of V. mimicus have been found to carry a heat-stable enterotoxin (ST) gene identical to that of V. cholerae non-O1/non-O139 (nag-st) [4]. However, the determinants of the virulence factors of V. mimicus and the molecular mechanisms of the differences in biochemical metabolism between V. mimicus and V. cholerae, have not been well characterized.
The CTX element, which encodes cholera toxin (CT) in V. cholerae [12], is a virulence gene that is well known to be associated with horizontal transfer of the filamentous bacteriophage CTXФ. This element is widely found in epidemic V. cholerae but is rarely seen in environmental non-O1/non-O139 isolates [13]. It has been reported that V. mimicus isolates from the natural environment can be infected by CTXФ experimentally [14], and four clinical strains, from Bangladesh, India, Japan and the United States, were also reported to carry CTX [8].The receptor for CTXФ, toxin-coregulated pilus (TCP), which is encoded by the vibrio pathogenicity island (VPI) [15], was also present in these V. mimicus isolates [8]. This suggests that V. cholerae is not the only host of CTXФ and that contemporary horizontal transfer between species has occurred [16]. The potential ability of such strains to cause an epidemic is unclear, but this threat may rise.
Four draft genomic sequences of V. mimicus isolates have recently become available, which were produced using the Roche-454 pyrosequencing technology, including strains VM603 (GenBank: NZ_ACYU00000000), VM573 (NZ_ACYV00000000), MB-451 (NZ_ADAF00000000) and VM223 (NZ_ADAJ00000000) [17]. Additionally, 36 published genomes of V. cholerae have also been widely studied. Extensive genomic sequencing of V. mimicus isolates may increase our understanding of the taxonomy and ecology, as well as the pathogenicity of V. mimicus. In this study, we analyzed the variation dynamics of a V. mimicus genome carrying the CTX element based on comparative genome analysis with V. cholerae. This provided a global view of the genome of V. mimicus and a clear map of the phylogenetic relationships with other Vibrio spp. Additionally, Instead of making pairwise comparisons among different species, the variations identified were further examined using a combinatorial approach and confirmed for available V. mimicus strains. The obtained data provided detailed information of a close relationship and potential horizontal gene transfer between V. mimicus and V. cholerae, as well as showing the genomic mutations of metabolic pathways used for the identification of these two species.
Results
Genome features of V. mimicus strain SX-4
We sequenced the genome of V. mimicus strain SX-4 with whole genome shotgun method and the total genome sequences is estimated to 4,391,932 bp (Text S1). 51 IS elements, 102 tRNA, and 25 rRNA clusters were found in our genome. The general features of the genome were summarized in Table 1 and Figure 1.
Table 1. General feature of the V. mimicus SX-4 genome.
| N16691 | Simulated N16961 | SX-4 | Complete SX-4 (Simulated) | |
| Genome | ||||
| Size | 4033464 | 3924560 | 4273349 | 4391931.67 |
| Contig Number (>1K) | - | 57 | 35 | - |
| N50 | - | 335321 | 253803 | - |
| G+C percentage | 47.49 | 47.5 | 46.25 | - |
| Scaffold Number (1K) | 2 | 54 | 31 | - |
| Paired read coverage (#) | - | 73.65 (3905736) | 69.86 (4034050) | - |
| Single read coverage (#) | - | 66.11 (3546891) | 61.14 (3572283) | - |
| Unmap read coverage(#) | - | 2.91 (153706) | 80.67 (4617101) | - |
| Aver. Gap size | - | 39.33 | 88 | - |
| Predicted CDS | ||||
| CDS Numbers | 3836 | 3558 | 3903 | 4207.96 |
| Average Size | 914.94 | 961.6 | 960.49 | 913.88 |
| Percentage of Coding | 87.01% | 87.18% | 87.72% | 0.88 |
| Truncated CDS | - | 351 | 384 | - |
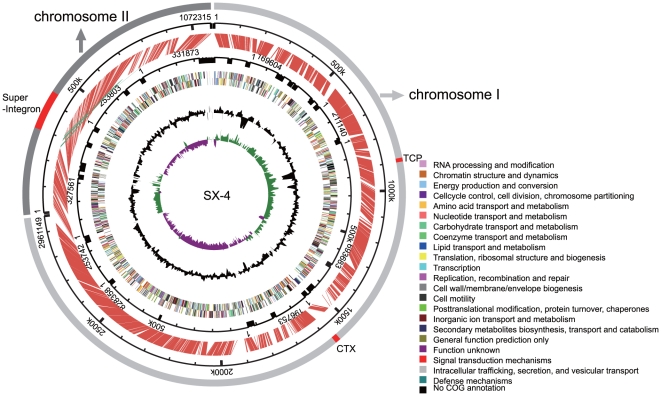
Figure 1. Schematic circular diagram of the V. mimicus strain SX-4 genome and synteny relationships between SX-4 and V. cholerae strain N16961.
Key for the circular diagram (outer to inner): (1) reference genome of V. cholerae N16961 and the backbone of our genome as well as their alignment with red lines. (2) SX-4 COG categories on the forward strain (+) and the reverse strand (-). (3) G+C content and GC skew (G-C/G+C) of SX-4 respectively with a window size of 10 kb. Superintegron, CTX and TCP were marked in the figure.
Based on our genome, we got 3903 predicted CDS in V. mimicus, among which152 did not return a significant match to the protein database in BLAST searches, indicating potentially unique V. mimicus proteins. V. mimicus shared 2490 common genes with V. cholerae, and these genes are associated with 2332 functions. Genes from the super-integron of V. cholerae have also been identified in strain SX-4, as well as some important gene clusters, including those of the O-antigen, hemolysin, CTX, TCP pathogenic islands, polysaccharide complex protein and general metabolism pathway-associated genes, which allow this strain to present a similar pathogenicity to V. cholerae. The Vibrio seventh pandemic island-II (VSPII) gene cluster, which was only found in the 7th pandemic strains of cholera, was not found in SX-4. Hemolysin represents a potential virulence factor family; in our study, we identified three typical genes, vmh, tdh, and hlx, in a hemolysin gene cluster, as well as an additional hemolysin-associated gene, tlh (thermolabile hemolysin). The tlh gene has also been identified in other Vibrio spp., such as V. parahaemolyticus, which suggests that V. mimicus is a potential epidemic pathogen, similar to other Vibrio species.
Genome-based evolutionary analysis showed that V. mimicus has a large number of mutations compared to its nearest neighbor, V. cholerae
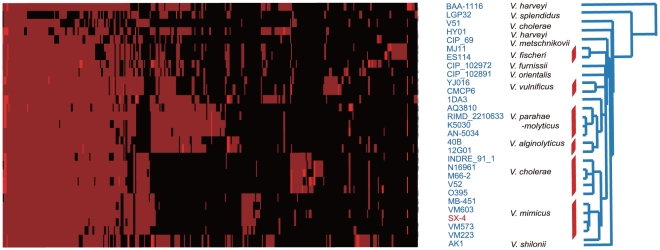
In this study, we intended to shed light on the relationships among Vibrio species that are important for the establishment and maintenance of the taxonomy of these species, as well as to construct an identification and diagnosis system. Previously, genomic sequences for V. mimicus, V. alginolyticus, V. fluvailis, and V. vurnissii, as well as V. cholerae were obtained using a whole-genome shotgun approach (summarized in Table S1). Phylogenetic analysis based on ∼8,872 kb of orthologous protein-coding regions of 29 Vibrio strains revealed that V. mimicus had recently diverged from the other species, and that V. cholerae was the closest species to V. mimicus (Figure 2). With the exception of strains 1DA3, BAA1116, and HY01, which belong to V. harveyi, the other isolates belong to different species, as determined based on the recently determined genetic diversity of Vibrionaceae. The four newly sequenced V. mimicus genomes (MB-451, VM223, VM573 and VM603 [18]) were used for subsequent analysis. In addition to SX-4, all V. mimicus strains revealed a similar genome size, GC content, and number of predicted genes (See Table S2).
Figure 2. Phylogenetic tree of Vibrio.
A cluster of 8916 orthologues grouped into 28 strains was screened for the presence (red) or absence (black) of genes. A phylogenetic tree was constructed for all strains, and the red line region represents the same category. On the right nucleotide sequences of homologs were used to construct a phylogenetic tree.
The entire set of homologous genes of the V. mimicus strains was compared in all possible pairwise combinations using BLAST analysis. The percent identity between pairs ranged from 23 to 100%, with no particular bias among Vibrio species, except V. cholerae. The results of all permutations of the genes and the gene sequences together indicated the relationships among the Vibrio species. Comparative analysis of the strains showed that V. mimicus shares a higher similarity of genes to V. cholerae than do the strains from the other two species (Figure 1). This is similar to the results of previous studies [17], which places V. mimicus and V. cholerae as the nearest neighbors based on comparative genome analysis results. Consistent with this, the global genome comparisons at the nucleotide level show that 2025 gene families were conserved in these two species, with 2111 and 2802 genes in V. cholerae and V. mimicus, respectively.
When we mapped the sequenced reads to all of the Vibrio genomes, the plot of the similarity was fitted well by the phylogenetic tree, but the percentage of mapped reads showed a large divergence between V. mimicus and V. cholerae (4.5%∼5.6%). Even among the V. mimicus strains, the 52.5%, 63.9%, 64.5% and 97.8% high quality V. mimicus reads mapped to VM223, VM603, MB-451 and VM573, indicating that V. mimicus has a large number of single nucleotide mutations among its strains with more than a 2% difference, considering that genome matching tolerated 2 bp of mutations per read.
To investigate the divergence between V. mimicus and V. cholerae with respect their basic life-cyle and metabolism, genes belonging to core gene families, classified according to their predicted functions, were used to construct the core genome carrying housekeeping functions and to perform comparative analysis. The sequenced V. cholerae and V. mimicus genomes revealed a high level of similarity with respect to homologous genes, which are clustered into 3201 homologous gene families (HGF). Among these HGF, 1014 belonged only to V. mimicus, while 323 belonged only to V. cholerae. These HGF proteins were mostly involved in essential functions related to the vegetative functions of the bacteria, such as DNA replication and repair elements, protein synthesis and carbohydrate metabolism, as well as fundamental functional categories, such as cell wall synthesis and protein folding (Figure S1).
It has been reported that the two sequenced V. mimicus genomes (strains VM573 and VM603) are closest to that of V. cholerae El Tor strain N16961[18]. Here, we also compared the genomic content of V. mimicus SX-4 with N16961 to reveal the genetic basis for the functional distinction of the bacteria. The isolates shared a high degree of HGF synteny, but we found a great deal of divergence in their genetic variants, indicating these are early divergence strains, although their phenotype and evolutionary distance are close. We found 1479 more genes in V. mimicus than in N16961. The chromosomal genes were categorized into common genes and specific genes, as compared to N16961. The analysis clearly identified monophyletic clades that appeared to be lineage specific with ancestral support (Figure S1). These different genes were present or absent in each category and played similar roles in metabolism with similar functions; therefore, this represents a possible reason that the two species show similar biological characteristics and adaptations to their natural environments.
The characteristic biochemical test differences used in the species identification of V. mimicus and V. cholerae are mainly caused by gene deletion
V. mimicus is very similar to V. cholerae in its phenotype, though biochemical tests can be used to discriminate between the two species for the purpose of identification, including a sucrose fermentation test, Voges-Proskauer test and lipase activity test [2]. In contrast to V. cholerae, V. mimicus shows negative results for these three tests. A comprehensive analysis of genomes and pathways revealed a deletion of one or more genes that was consistently different between SX-4 and V. cholerae N16961 in each of these pathways (Table S3). To investigate the timing of these deletions, we also compared all of the 50 (Table S3) genes to the associated regions in the genomes described in the literature; these sites were found to be accurate and consistent with each other.
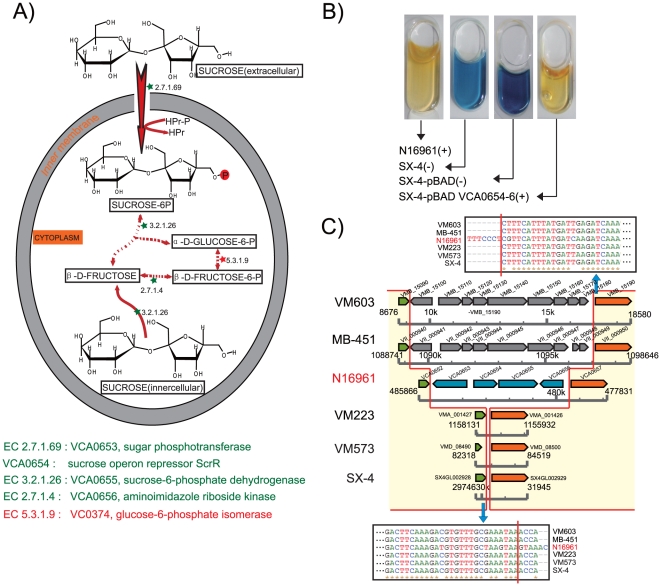
The sucrose fermentation test is associated with ability to metabolize starch and sucrose (Figure 3A). In V. cholerae N16961, 19 genes are included in this pathway according to the KEGG database (Table S3). In strain SX-4, we found 16 of these genes on the chromosome, with three genes being absent (VCA0654, VCA0655 and VCA0656 corresponding to N16961 genome). A complementation experiment of these genes in strain SX-4 showed that the complementary strain presented similar positive results for sucrose fermentation to what is seen in V. cholerae N16961 (Figure 3B), indicating that VCA0654, VCA0655 and VCA0656 (sucrose operon repressor ScrR, sucrose-6-phosphate dehydrogenase and fructokinase) are responsible for the differences in sucrose metabolism observed in V. cholerae, and their deletion in V. mimicus strain SX-4 causes negative sucrose fermentation. We further investigated the available genome sequences of V. mimicus strains VM223, VM603, MB-451 and VM573 for these genes. For strains VM223 and VM573, a quite similar deletion of these genes was found (Figure 3C), suggesting this deletion probably occurred before the separation of V. cholerae and V. mimicus. However, an additional genome fragment insertion was identified in VM603 and MB-451, indicating that the split site is also an active indel region in the genome.
Figure 3. Sucrose fermentation of SX-4 and SX-4-pBADVCA0654-6.
(A) Sucrose fermentation in V. mimicus; the red portion shows the function of the enzyme translated from VCA0654 to VCA0656 in the pathway. (B) Complement test. The gene cluster of VCA0654 to VCA0656 from V. cholerae strain N16961 was complemented into the V. mimicus strain SX-4. The strain was grown under LB in the presence of 0.1% arabinose, and then tested for sucrose fermentation (API-20E). Only the strain bearing plasmid pBADVCA0654-6 presented a positive result, like that of V. cholerae N16961. (C) Genome structure of the VCA0654-VCA0655-VCA0656 operon. Positions and orientations of ORFs are indicated with different color arrows. Homologue genes are presented with the same color. Homologue regions have been showed with red background, also with their flanking sequences. Insertions in VM603 and MB-451 are homologues presenting with the same color.
In addition to the sucrose fermentation pathway, the genes (and all of the operons) associated with the other key biochemical tests used for species identification, the Voges-Proskauer test and lipase activity test, were also identified in V. cholerae and V. mimicus. VC1590, encoding acetolactate synthase, which catalyses the transformation of acetymethyl carbinol into 2,3-bytaylene cylycol in the V. cholerae N16961 genome, was lost in V. mimicus SX-4, as well as strains MB-451, VM223, VM603 and VM573. The lack of this gene may be reflected in a defect in the catabolism of acetyl-lactic acid and cause the negative result in the Voges-Proskauer test. The lipase activity test is used to detect the pathway for catalyzing and hydrolyzing lipids, forming fat acids, glycerin and monoglycerides or diacylglycerol. In V. cholerae, six genes are associated with lipase metabolism, which encode the putative esterase/lipase YbfF (VC2097), lactonizing lipase (VCA0221), a putative lipase activator protein (VCA0222), a lipase of the GDXG family (VCA0490), a lipase-related protein (VCA0754) and a putative lipase (VCA0863). VCA0221 and VCA0222 were found to be absent in V. mimicus, which is probably the reason for the difference between V. mimicus and V. cholerae with respect to the lipase reaction.
The pathogenic characteristics of V. mimicus are highly associated with lateral gene transfer
1. Polysaccharides and lipopolysaccharides
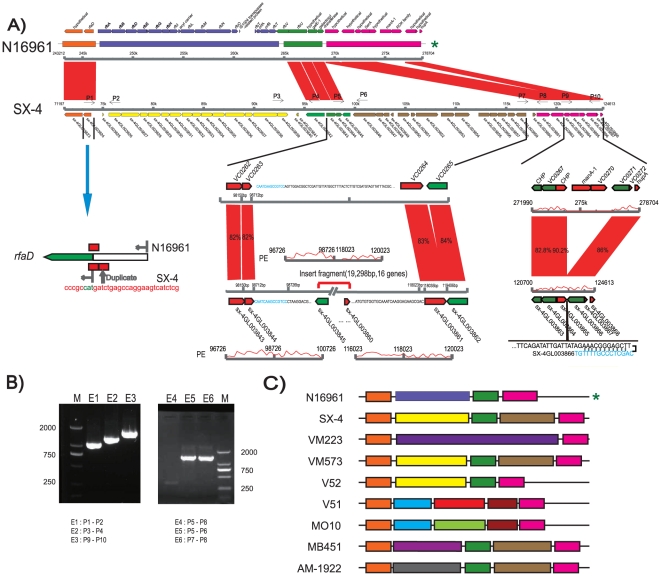
In V. cholerae, the polysaccharide gene cluster is flanked by rfaD (encoding D-glycero-D-manno-heptose 1-phosphate guanosyltransferase, which is involved in lipopolysaccharide core biosynthesis), and rjg (encoding a conserved hypothetical protein with similarities to the mRNA 3′-end processing factor) [19]. However, the structure of this cluster in V. mimicus is unclear. Fortunately, in SX-4, a similar clustered organization of its genes was observed, which contained a backbone of two genes, SX-4GL003824 (homolog to rfaD) and SX-4GL003861 (homolog to rjg) (Figure 4A). Between these two genes, SX-4 contains a ∼22 kb specific antigen encoding region, which differ to that of V. cholerae O1 and O139 (Figure S2). This region contained 17 genes and one IS element (Figure 4A), following 13 bp of repeat sequences in the rfaD gene, through the end of the rfb cluster (rfbU), which were predicted to belong to the O37 antigen cluster-associated genes in V. cholerae V52 and MZO-3, encoding enzymes or proteins of unknown function, similar to Rickettsia bellii OSU 85-389. In another conserved end region, we found a ∼19 kb insertion between VC0263 and VC0264, corresponding to the N16961 genome, encoding 16 genes in strain SX-4, the majority of which were specific and hypothetical genes. However, two loci were identified in the cluster that were similar to the bacterial sugar transferase and wca protein in Aggregatibacter aphrophilus NJ8700, indicating that the region was part of the O-antigen or a complementary region. The following two conserved genes in N16961 have a hairpin structure (AAACGGGAGCT-TC-AGCTCCCGTTT) in SX-4, causing three genes deleted compared to V. cholerae N16961 (Figure 4A). It appears that the polysaccharide produced by SX-4 has a different structure than that of N16961. Therefore, this complex mosaic structure of polysaccharides is likely to have been caused by genomic exchanges between different species.
Figure 4. Comparison of the O-antigen biosynthesis cluster from V. mimicus strain SX-4 and V. cholerae strain N16961.
(A) The gene clusters are presented according to the chromosome with different color arrows. The colors of arrows are different units in genome rearrangement between V. mimicus and V. cholerae. P1 to P10, with black arrows, indicate primers designed to validate the structure. The left bottom figure shows the duplication in SX-4 in this region. The middle bottom figure presents the region and structure associated with genome exchange, as well as the flank. The red curve represents the pair-ends sequence relationship. The right bottom figure shows the downstream region of chromosome with a hairpin structure. (B) PCR results for structure validation. Primers were designed at the location marked in Figure 4A. (C) Scheme of clustering in other available genomes. For better describe the pattern of genome rearrangement in O-antigen region, the cluster were divided into different parts according to their similarity. The similar regions were showed with the same colors, and genes presented with the color bar are consistent with those in Figure 4A. * N16961 has been used as reference genome. PE curve is drawn by supporting pair-ends SOLEXA reads coverage.
Genetic exchange in the ‘O’ antigen synthesis region (Figure 4A) has been widely reported in V. cholerae and plays important roles in the evolution of many pathogenic bacteria, as shown for the O139 cholera outbreak [20], [21]. Analysis of the schema structure in this region from four additional available draft genomes of V. mimicus suggested that the six genomes might encode different polysaccharides and present different serotypes, except for VM573, which was similar to the SX-4 strain sequenced here (Figure 4B,4C, Figure S2). Notably, V52, which is a serogroup O37 V. cholerae strain, presented a similar structure to SX-4 at the beginning of the conserved region, apart from the above mentioned component region at the end of this region, it showed a similar structure to N16961. These opposite divergences in strain V52 showed that the insertion at the end of the conserved region might not be necessary for the bacteria but may complement a better-constructed surface protein, indicating that the mosaic structure of this region may be generated by DNA shuffling individually.
2. CTX element
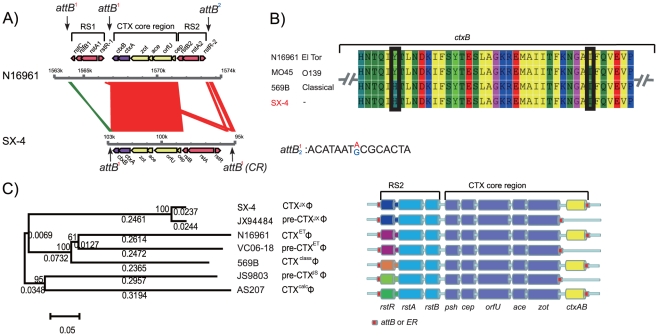
The highly invasive characteristics of V. cholerae that cause fulminate diarrhea in humans are associated with the CT that resides in epidemic V. cholerae isolates. Strain SX-4 was isolated from a watery diarrheal patient and carries ctxAB. By the real-time PCR method [22] and genome analysis, we found SX-4 contains a single copy and a typical genetic organization of the CTX element, like that of V. cholerae N16961, but no RS1 was found (Figure 5A). This result is consistent with the validation with the SOLEXA reads coverage in the genome. Also, the flanking sequences of CTX element in SX-4 are highly homologue to that of V. cholerae N16961 chromosome I, suggesting that the CTX element in SX-4 are potentially located in large chromosome. The element contains the typical RS2 and core regions of CTX. In V. cholerae, the core region plays a vital role in the morphogenesis of CTXФ, and the RS2 region is necessary for integration, replication and regulation of the CTX prophage genome. This suggests that SX-4 is able to be infected by CTXФ through its lysogenic conversion mechanism and can cross species. Recently, Kumar et al [23] found a new type of mutation in ctxB. We investigated this gene sequence and observed the same sequence as in the classical biotype sequence of ctxB (Figure 5B).
Figure 5. CTXΦ alleles and the phylogenetic tree for different strains of serogroups and biotypes.
(A) The CTX gene cluster compared to its associated region between SX-4 and N16961, as well as their flanking sequences. (B) Partial ctxB sequences show different mutations among the O1 and O139 serogroups. (C) The phylogenetic tree based on alignment results of rstR-ig2 sequences from the seven typical strains. The right portion is the modeled map for different types of CTXФ genomes.
rstR, identified as a biomarker for CTXФ in the RS region, divides the CTX element into different alleles in the classic and El Tor biotypes, which are CTXclassФ, CTXETФ, CTXcalcФ, and their pre-phages, as well as other special types, such as pre-CTXJSФ (AF302794) and pre-CTXJXФ (AF511002). Other alleles, including rstR4*, rstR4**, rstR5, rstR6 and rstR232 from O1/O139 and non-O1/non-O139 strains, have been reported [24], [25], [26]. For strain SX-4, sequence comparison showed its rstR has high similarity to that of the O139 strain JX94484 (Figure 5C), which is also known as rstR4** [26]. However, these environmental non-O1/non-O139 strains and the O139 strain JX94484 possess pre-CTXФ. Based on the rstR and ctxB sequences, it may show that CTX prophage on SX-4 genome was not obtained from the common 7th pandemic El Tor clones which carrying El Tor type ctxB and El Tor type rstR genes in CTX element.
In V. cholerae, two important toxin-associated components, TLC and the RTX cluster, locate on the flanks of the CTX element [27], [28]. Commonly, TLC has been found to be located in the 5′ region of the CTX gene cluster and is associated with CTX cluster integration, while the RTX cluster is located in the 3′ region of CTX. Both of these elements were absent in V. mimicus SX-4, as well as in MB-451 and VM223, which are also CTX prophage-positive strains. These data suggest that it might not be strictly necessary for CTX and TLC to co-occur in V. mimicus, or in V. cholerae.
In SX-4, the CTX element integrated into the chromosome via the characteristic attachment site (attB) on the flank of the CTX element (Figure 5A). The 5′-end attB sequence harbored additional pseudogenes, which probably encodes a para-aminobenzoate synthase glutamine amidotransferase component II protein, similar to V. cholerae V52 (gb|EAX60338.1|), whereas the 3′-end attB is in the intergenic regions associated with recognition sites for insertion. This attB is the same as attB1 in N16961, but differ with 1 bp to attB2 in N16961 (Figure 5A).
3. TCP pathogenicity island
TCP, the specific receptor of CTXФ in V. cholerae, is an integral part of the VPI. Analysis of the complete sequence of SX-4 identified all of the TCP genes typically found in Vibrio species, with a similarity of 87∼100% for each gene. When compared to the El Tor strain N16961, a putative att-like 20-bp attachment site on the flank of the TCP gene cluster, including complete gene clusters, such as tcpI, toxR, toxT and tagA-tagD, suggested interference with microbial attachment to host tissues. Additionally, among V. cholerae isolates, the sequence of the tcpA locus is more divergent compared to other loci in the VPI. Most likely, this diversity is a reflection of diversifying selection during adaptation to the host immune response or to CTX susceptibility [29]. TCP is a receptor used by the CTX phage to infect V. cholerae, except for in a few strains with the CTX element but no tcp genes [29]. Therefore, in our analysis, the tcpA gene was used to construct a phylogenetic tree of the V. cholerae cluster for a CTX acquisition event that occurred in a TCP-dependent manner; the tcpA gene of SX-4 is identical to that of the V. cholerae serogroup O115 strain [25].
Discussion
In this study, we sequenced the genome of a V. mimicus strain carrying an intact CTX element and compared it with the available genomes of other Vibrio spp. Analysis of the phylogenetic relationships among these species provided important insight into the genomic history related to the Vibrio genus and the rules for evaluating Vibrio evolutionary distances. Our experimental results and computational analysis, along with evolutionary clues and bacterial phenotype coupling among homologous genes, suggest that the genomes of V. mimicus are quite different from the pandemic V. cholerae strains. Considering the complexity of the serogroup structure of V. cholerae, genome sequencing in other serogroups of V. cholerae may help to reconstruct more accurate evolutionary relationships between V. mimicus and V. cholerae.
Species identification is commonly based on biochemical tests in V. cholerae and V. mimicus. Our analysis of the key biochemical test-related differences in the genomes of these species revealed the molecular mechanisms underlying the biochemical test differences between them and provided clues to link the genome features and classical biochemical tests. Based on operon structures, we uncovered the deletion of gene sequences related to these three biochemical tests. All of the operons shared clear insertion sites, which indicates that the differences related to these biochemical tests are caused by ancient events in evolution that took place before the divergence of V. cholerae and V. mimicus. It is interesting that there are different modes of the mutation of the gene cluster involved in the sucrose fermentation in different V. mimicus strains, including insertion and deletion. However, why sucrose utilization is prevented in V. mimicus is unknown. It could represent an important event in the evolution of V. mimicus that benefited these strains in adapting to a special micro-ecosystem in a particular period.
In this study we sequenced the genome of a V. mimicus isolate carrying an intact CTX element from a cholera-like patient. Strain SX-4 also carries a number of hemolysin genes, including the vmh, tdh, hlx and tlh genes, which indicates a possible pathogenic character. Additionally, this strain carries CTX, which integrated into its chromosome through an attB site, similar to what is seen in the toxigenic V. cholerae. Notably, this strain carries the rstR4** type of CTXФ, which has also been found in an O139 serogroup strain (our unpublished data) and non-O1/non-O139 strains [26]. In these V. cholerae strains pre-CTXФ prophage carrying rstR4** were found, whereas in V. mimicus SX-4, it is the intact genome of CTXФ carrying rstR4**. It can be assumed that the intact CTXФ prophage genome in SX-4 was obtained from a V. cholerae strain carrying such an element. Additionally, the ctxB gene in SX-4 is of the classical type, which was found in strains of the classical biotype worldwide and in U.S. gulf coast strains [30], Based on these two characters of its CTX prophage genome, strain SX-4 may not be transferred from the typical 7th pandemic El Tor strains, but rather, from a different toxigenic clone. These data also show the abundance of this lysogenic phage and its active transference within V. cholerae and its sister species.
Strain SX-4 also includes the TCP island, suggesting a closer relationship to the toxigenic species V. cholerae and that it in terms of colonizing and pathogenic characteristics compared to other V. mimicus strains. The tcpA sequence of SX-4 is identical to that of a V. cholerae serogroup O115 strain [25], but not the classical or El Tor type. Additionally, genome comparison showed that the LPS gene cluster is expected to have been recombined in Vibrio genome evolution. These islands have been proven to be associated with virulence [31], [32], [33], [34]. Here, we provide more detailed genomic information related to their exchange among different Vibrio spp. The sequence identity of both the CTX and VPI genes derived from V. mimicus and V. cholerae strongly suggests that successive horizontal transfer of bacteriophages from different clones or serogroups to V. mimicus has occurred.
In summary, we exploited genome information to investigate the properties of a V. mimicus virulent strain, SX-4, and the evolutionary relationships among Vibrio spp. Horizontal gene transfer may be proposed to be common between V. mimicus and V. cholerae, especially for virulence-related genes. In V. mimicus, the common gene mutations observed in a number of metabolic pathways related to the biochemical tests used for species identification may suggest different adaptations and evolution tendency in these species. The SX-4 isolate described in this study is not the only strain of V. mimicus carrying CTX that can cause diarrheal disease in humans, but the evidence presented here indicates that this may be forming a potential environmental reservoir for CTXФ and other virulence factors and may play an important role in the emergence of new toxigenic clones. It may also suggest that caution is necessary regarding a possible epidemic caused by V. mimicus strains carrying cholera toxin genes.
Materials and Methods
Strains and culture conditions
V. mimicus strain SX-4, isolated from a 55-year-old male patient who suffered from cholera-like diarrhea in 2009 in Shanxi, China, was used to construct shotgun libraries for genomic sequencing. According to a number of biochemical characteristics, including being negative for sucrose fermentation, Voges-Proskauer, and lipase tests, combined with the results of the API20E/NE test, the isolate was confirmed to be V. mimicus. PCR was used to test the V. mimicus SX-4 isolate for the presence of ctxA and then confirmed again with the genome sequencing.
SX-4 was routinely cultured using Luria-Bertani (LB) medium at 37°C. Genomic DNA was extracted from a 10-ml overnight culture using the TIANamp Bacteria DNA Kit (TIANGEN Biotech, Beijing, China). Genomic DNA was quantified on a 0.7% agarose gel, stained with ethidium bromide and spectrophotometrically assessed. The stock DNA solution was separated into two aliquots, one of which was used for sequencing, while the other was stored at −20°C for further PCR gap closing and structure validation.
The other genomic sequences used in this study were obtained from GenBank. The strain names and their accession numbers are summarized in Table S1.
DNA sequencing and annotation
An initial V. mimicus shotgun genome sequence (accession number. ADOO01000000) was obtained from a mixture of Solexa sequences from pair-ends Solexa sequencing of a 500-bp insert size library to 130X coverage. The draft genome sequence was assembled into contigs using the novel short-read assembler SOAPdenovo [35]. The Solexa library produced a total of 4,034,252 reads, providing ∼63-fold sequences coverage and was de novo assembled into 35 contigs, of which 4 contigs were linked by paired ends, with contigs N50 253,803 bp. The 39 contigs were aligned to the whole genome sequence of V. cholerae to identify the gaps to be sequenced in the genome of V. mimicus. All genome structure variations have been carefully checked to avoid assemble errors, the prediction was verified by PCR methods. The genomic structure of CTX element and major virulence factor mutations in SX-4 was one of the main aim in this study, although we did not close all of the gaps in the genome and determine the order of the contigs, we closed all of the gaps in three major lateral gene transfer regions, as well as the potential virulence genes by PCR walking. Amplicons were obtained by PCR using the genomic DNA of strain SX-4 as the template and sequenced. These new sequences, together with the contigs will be used to determine the whole-genome sequence in future analyses.
Putative CDS were identified by the Glimmer gene prediction program [36], and their functions were automatically annotated using BLASTP (e-value <1e-10 identity>80 and >100 aa) against GenBank and UniProt (version 47). For each CDS, the ORF sequences were further categorized based on Interpro, GO, and COG (Clusters of Orthologous Genes). Functional pathways were annotated based on KEGG. tRNA genes and repeats were predicted with tRNAscan-SE and Repeatmasker [37], respectively. The genome sequences of V. mimicus have been deposited in GenBank.
Comparative genome analysis and phylogenetic tree construction
Twenty-seven genomes of Vibrionaceae species available from GenBank (Table S1), representing 14 different species, were used in our analysis. Homologous genes were identified by comparisons using OrthoMCL [38] with BLASTP (e-value <1e-5) at the genome level. The genomes were automatically compared for missed gene calls by Perl scripts based on homolog relationships. Housekeeping genes were used in the construction of the phylogenetic trees for Vibrio by the neighbor-joint method in Paup [39]. Consistent with this, the homologous genes in each complete genome were clustered using a hierarchical cluster method. Their nucleotide sequences were also used to uncover evolutionary clues by Paup. The nucleotide alignment of the tcpA gene, which occurs widely in different serogroups, and the component of the CT element receptor, which presents the possible evolutionary relationships of the CT element among the different serogroups, then used to construct a phylogenetic tree by the neighbor-joining method in Mega4. The rstR gene was also used to construct the phylogenetic tree to specify the source of the CT element in different V. cholerae local strains by the neighbor-joining method in Mega4. The sequence alignment of ctxB was edited by Mega4.
To identify the potential genes that differ between the CT-carrier strain V. mimicus SX-4 and V. cholerae N16961, pairwise and reciprocal comparisons were performed by aligning the predicted genes of one strain to the whole-genome sequence of the other and vice-versa. The genome sequences were compared for missing genes, and homologous hits were identified in the second genome. The similar regions were annotated on a synteny line map or circle map with red blocks. Detailed comparative analysis of duplicated sequences and differences between the two strains in their genome structure were performed by Perl scripts. Solexa read copies along the genome in the typical region were calculated using SOAP mapping results. SNPs were searched using the software package Mummer and then annotated to the genome of V. cholerae.
Verification of genome transfer and biochemical pathway complement test
To verify the insertions and deletions in the genome of SX-4, four sets of unique primers were designed to perform a PCR-based examination of the DNA of SX-4. The primers are indicated as arrows in Figure 4A. Routing PCR was carried out with a 50 µl reaction. Amplification was carried out for 30 cycles, with initial denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 59°C for 45 s, extension at 72°C for 1 min per kb, and a final step at 72°C for 5 min.
Genes associated with three key biochemical pathways: sucrose fermentation, Voges-Proskauer and lipase (corn oil) activity, were screened and listed according to the KEGG database (http://www.genome.jp/kegg/) (Table S3). All of these genes were aligned to V. mimicus with BLAST, with overlap of less than 80%. A set of unique primers was designed to amplify fragments with missing genes (VCA0654, VCA0655 and VCA0656) in SX-4 using V. cholerae N16961 as DNA template. The amplified fragments were ligated to the expression vector pBAD24 [40], and the constructed plasmid was introduced into Escherichia coli DH5α, resulting in plasmid pBAD VCA00654-6. The plasmid was then transformed into the SX-4 strain. To induce production of VCA00654-6, the SX-4 strain was grown under LB in the presence of 0.1% arabinose with appropriate antibiotics at 37°C for 4 hours and then tested for sucrose fermentation following manufacturer's instructions (API-20E, bioMerieux, Inc.).
Copy number determination of CTX element on strain SX-4 genome
The copy number of CTX in SX-4 genome was determined according to a real-time quantitative PCR method we used previously [22] and also SOLEXA pair-ends reads coverage esitmation (Figure 1). And also, we check the assembled sequences linked with CTX element and compared the sequences to the published V. mimicus and V. cholerae genome using BLAST. In V. cholerae, their chomosome contains a single copy of house keeping gene thyA, which was used as the reference in our study. The CTX element gene zot was used to determine the copy number of CTX. By real-time quantitative PCR, the copy number(CN) of zot in different strains was determined with CN = 2△Ct , where △Ct means the difference Ct value between thyA and zot. The specific copy number of CTX was obtained according to CN with the integral function. V. cholerae O1 strains N16961 (one copy of CTX), GD93284 (two copies of CTX) and 40–42 (zero copy of CTX) have been used as positive and negative control.
Supporting Information
Genome sequencing, assembling and gene content prediction.
(DOC)
Gene function for different categories in V. mimicus.
(EPS)
Genome variation in polysaccharides among all available genomes.
(EPS)
Simulation genome and its gap distribution according to V. cholerae genome.
(EPS)
Genomes of V. cholerae and V. mimicus used in our studies.
(XLS)
Comparative genome size, GC content, and number of predicted genes of V. mimicus and V. cholerae.
(XLS)
Comparative of metabolic gene sets in three key biochemical tests.
(XLS)
Acknowledgments
We thank the Beijing Institutes of Genomics for sequencing SX-4 strains and making the result available to the public.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the National Basic Research Priorities Program (2009CB522604), the National High Technology Research and Development Program of China (863 Program) (2006AA02Z425), the National Natural Science Foundation of China (No. 30872260) and a special grant for Prevention and Treatment of Infectious Diseases (2008ZX10004-012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Shandera WX, Johnston JM, Davis BR, Blake PA. Disease from infection with Vibrio mimicus, a newly recognized Vibrio species. Clinical characteristics and edipemiology. Ann Intern Med. 1983;99:169–171. [PubMed] [Google Scholar]
- 2.Davis BR, Fanning GR, Madden JM, Steigerwalt AG, Bradford HB, et al. Characterization of biochemically atypical Vibrio cholerae strains and designation of a new pathogenic species, Vibrio mimicus. J Clin Microbiol. 1981;14:631–639. doi: 10.1128/jcm.14.6.631-639.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi L, Miyoshi S, Hiura M, Tomochika K, Shimada T, et al. Detection of genes encoding cholera toxin (CT), zonula occludens toxin (ZOT), accessory cholera enterotoxin (ACE) and heat-stable enterotoxin (ST) in Vibrio mimicus clinical strains. Microbiol Immunol. 1998;42:823–828. doi: 10.1111/j.1348-0421.1998.tb02357.x. [DOI] [PubMed] [Google Scholar]
- 4.Vicente AC, Coelho AM, Salles CA. Detection of Vibrio cholerae and V. mimicus heat-stable toxin gene sequence by PCR. J Med Microbiol. 1997;46:398–402. doi: 10.1099/00222615-46-5-398. [DOI] [PubMed] [Google Scholar]
- 5.Kim GT, Lee JY, Huh SH, Yu JH, Kong IS. Nucleotide sequence of the vmhA gene encoding hemolysin from Vibrio mimicus. Biochim Biophys Acta. 1997;1360:102–104. doi: 10.1016/s0925-4439(97)00008-2. [DOI] [PubMed] [Google Scholar]
- 6.Uchimura M, Koiwai K, Tsuruoka Y, Tanaka H. High prevalence of thermostable direct hemolysin (TDH)-like toxin in Vibrio mimicus strains isolated from diarrhoeal patients. Epidemiol Infect. 1993;111:49–53. doi: 10.1017/s0950268800056661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spira WM, Fedorka-Cray PJ. Enterotoxin production by Vibrio cholerae and Vibrio mimicus grown in continuous culture with microbial cell recycle. Appl Environ Microbiol. 1983;46:704–709. doi: 10.1128/aem.46.3.704-709.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd EF, Moyer KE, Shi L, Waldor MK. Infectious CTXPhi and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect Immun. 2000;68:1507–1513. doi: 10.1128/iai.68.3.1507-1513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acuna MT, Diaz G, Bolanos H, Barquero C, Sanchez O, et al. Sources of Vibrio mimicus contamination of turtle eggs. Appl Environ Microbiol. 1999;65:336–338. doi: 10.1128/aem.65.1.336-338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinoda S, Nakagawa T, Shi L, Bi K, Kanoh Y, et al. Distribution of virulence-associated genes in Vibrio mimicus isolates from clinical and environmental origins. Microbiol Immunol. 2004;48:547–551. doi: 10.1111/j.1348-0421.2004.tb03551.x. [DOI] [PubMed] [Google Scholar]
- 11.Shi L, Miyoshi S, Bi K, Nakamura M. Presence of hemolysin genes(vmh, tdh and hlx) in isolates of Vibrio mimicus determinated by Polymerase Chain Reaction. JHealth Science. 2000;46:63–65. [Google Scholar]
- 12.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karaolis DK, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, et al. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci U S A. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque SM, Rahman MM, Asadulghani, Nasirul Islam KM, Mekalanos JJ. Lysogenic conversion of environmental Vibrio mimicus strains by CTXPhi. Infect Immun. 1999;67:5723–5729. doi: 10.1128/iai.67.11.5723-5729.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 16.Srividhya KV, Alaguraj V, Poornima G, Kumar D, Singh GP, et al. Identification of prophages in bacterial genomes by dinucleotide relative abundance difference. PLoS One. 2007;2:e1193. doi: 10.1371/journal.pone.0001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasan NA, Grim CJ, Haley BJ, Chun J, Alam M, et al. Comparative genomics of clinical and environmental Vibrio mimicus. Proc Natl Acad Sci U S A. 2010;107:21134–21139. doi: 10.1073/pnas.1013825107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson CC, Vicente AC, Souza RC, Vasconcelos AT, Vesth T, et al. Genomic taxonomy of Vibrios. BMC Evol Biol. 2009;9:258. doi: 10.1186/1471-2148-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidelberg, F J, A. Eisen J, C. Nelson W, A. Clayton R, L. Gwinn M, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albert MJ, Siddique AK, Islam MS, Faruque AS, Ansaruzzaman M, et al. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. doi: 10.1016/0140-6736(93)90481-u. [DOI] [PubMed] [Google Scholar]
- 21.Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Nair GB, et al. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 22.Liu G, Yan M, Liang W, Gao S, Kan B. The copy number of the gene in chromosome of strain detected by the real time quantitative PCR. Chin J Microbiol Immunol. 2006;26:379–382. [Google Scholar]
- 23.Kumar P, Jain M, Goel AK, Bhadauria S, Sharma SK, et al. A large cholera outbreak due to a new cholera toxin variant of the Vibrio cholerae O1 El Tor biotype in Orissa, Eastern India. J Med Microbiol. 2009;58:234–238. doi: 10.1099/jmm.0.002089-0. [DOI] [PubMed] [Google Scholar]
- 24.Maiti D, Das B, Saha A, Nandy RK, Nair GB, et al. Genetic organization of pre-CTX and CTX prophages in the genome of an environmental Vibrio cholerae non-O1, non-O139 strain. Microbiology. 2006;152:3633–3641. doi: 10.1099/mic.0.2006/000117-0. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Kotetishvili M, Chen Y, Sozhamannan S. Comparative genomic analyses of the vibrio pathogenicity island and cholera toxin prophage regions in nonepidemic serogroup strains of Vibrio cholerae. Appl Environ Microbiol. 2003;69:1728–1738. doi: 10.1128/AEM.69.3.1728-1738.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukhopadhyay AK, Chakraborty S, Takeda Y, Nair GB, Berg DE. Characterization of VPI pathogenicity island and CTXphi prophage in environmental strains of Vibrio cholerae. J Bacteriol. 2001;183:4737–4746. doi: 10.1128/JB.183.16.4737-4746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubin EJ, Lin W, Mekalanos JJ, Waldor MK. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol Microbiol. 1998;28:1247–1254. doi: 10.1046/j.1365-2958.1998.00889.x. [DOI] [PubMed] [Google Scholar]
- 28.Lin W, Fullner KJ, Clayton R, Sexton JA, Rogers MB, et al. Identification of a vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc Natl Acad Sci U S A. 1999;96:1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faruque SM, Kamruzzaman M, Meraj IM, Chowdhury N, Nair GB, et al. Pathogenic potential of environmental Vibrio cholerae strains carrying genetic variants of the toxin-coregulated pilus pathogenicity island. Infect Immun. 2003;71:1020–1025. doi: 10.1128/IAI.71.2.1020-1025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balakrish Nair G, Firdausi Qadri, Jan Holmgren, Ann-Mari Svennerholm, Ashrafus Safa, et al. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol. 2006;44:4211–4213. doi: 10.1128/JCM.01304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sack DA, Sack RB, Nair GB, Siddique AK. Cholera. Lancet. 2004;363:223–233. doi: 10.1016/s0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 32.Waldor MK, Colwell R, Mekalanos JJ. The Vibrio cholerae O139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinants. Proc Natl Acad Sci U S A. 1994;91:11388–11392. doi: 10.1073/pnas.91.24.11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saunders J. Vesicles in virulence. Nat Rev Microbiol. 2004;2:86–87. doi: 10.1038/nrmicro830. [DOI] [PubMed] [Google Scholar]
- 34.Lee SH, Hava DL, Waldor MK, Camilli A. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 35.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 36.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swofford DL. Version 4. Sinauer Associates, Sunderland, Massachusetts; 2003. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). [Google Scholar]
- 40.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome sequencing, assembling and gene content prediction.
(DOC)
Gene function for different categories in V. mimicus.
(EPS)
Genome variation in polysaccharides among all available genomes.
(EPS)
Simulation genome and its gap distribution according to V. cholerae genome.
(EPS)
Genomes of V. cholerae and V. mimicus used in our studies.
(XLS)
Comparative genome size, GC content, and number of predicted genes of V. mimicus and V. cholerae.
(XLS)
Comparative of metabolic gene sets in three key biochemical tests.
(XLS)