Abstract
Introduction
Several genes that modify risk of factor VIII inhibitors in hemophilia A patients have been identified. Aside from the underlying mutations that cause hemophilia A, inhibitor risk appears to be modified by polymorphisms in various cytokines and immunomodulators, including IL10, TNFα, and CTLA4. HLA haplotypes have not been strong determinants of inhibitor risk.
Aim
We sought to confirm previous observations on factor VIII inhibitor risk-modifying genes and to test new candidate genes encoding various otherTH1/TH2 cytokines. We also sought to determine whether normal factor VIII gene polymorphisms affect inhibitor risk in Caucasians.
Methods
We studied 915 Caucasian, severe hemophilia A patients (282 inhibitor cases and 633 non-inhibitor controls) Genes were analyzed using 368 tagging SNPs starting 20kb 5′ and ending 10kb 3′ of each gene's coding sequence; four other polymorphisms (factor V Leiden & prothrombin 20210 polymorphisms and two in HFE) were also evaluated.
Results
Haplotypes that increased inhibitor risk were found in IL10 (OR 1.33, P = 0.04), IL12 (OR 1.31, P = 0.04), and IL1α (OR 2.16, P = 0.034). Protective haplotypes were seen in IL2 (OR 0.69, P = 0.008) and IL1β (OR 0.75, P = 0.02). One rare haplotype in the factor VIII gene increased the risk of inhibitor development by nearly four-fold (OR 3.8, P = 0.004).
Conclusion
We replicate previous findings for IL10; identify new associations with IL1, IL2 and IL12; and identify a rare factor VIII haplotype in Caucasians that is associated with increased inhibitor risk.
Keywords: hemophilia A, inhibitor, factor VIII, IL10, IL2, IL12, IL1
Introduction
Treatment of hemophilia A with factor VIII has improved the life of patients with severe disease [1]. Epidemics of hepatitis and human immunodeficiency virus (HIV) from concentrates in the 1970's and 1980's were a setback, but virus-free plasma-derived concentrates and recombinant DNA products re-established the upward trend in life expectancy [2]. Patients with hemophilia A (without HIV) now experience a nearly normal life expectancy (64 years) [3].
Neutralizing antibodies to factor VIII (“inhibitors”) still complicate factor VIII therapy for approximately 20% of severe hemophilia A patients. Mutations in factor VIII are the chief risk determinants of inhibitor development predominantly in patients with severe disease [4]. However, discordance for inhibitors in related patients provides evidence that other genetic factors play a role [5]. Polymorphisms in immunoregulatory genes such as IL10, TNFα, and CTLA4 have been associated with altered risk for inhibitor development [6-8]. In contrast, HLA, IL1β and IL4 genes have shown weak or nonexistent association with inhibitor development [9, 10].
African-Americans and Latinos with hemophilia A have higher inhibitor risk than Caucasians [11]. Viel et al recently showed that inhibitor status among African-Americans was associated with factor VIII haplotypes that are rarely seen in Caucasians [12]. For the 24% of African-Americans with an uncommon factor VIII isoform (H3 or H4), the inhibitor risk was 3.6 times (95% confidence interval, 1.1 to 12.3; P= 0.04) that seen in those who had one of the common isoforms H1 or H2.
We sought to confirm association of immune response modifier gene polymorphisms on inhibitor development and to extend this work to other TH1 and TH2 immune response genes. We also looked at polymorphisms in the factor VIII gene itself to see if the findings in African-Americans with hemophilia could be found in Caucasians. We replicated previous family study findings for IL10, found association of inhibitors with other immune response modifier genes, and showed an association with an infrequent factor VIII gene haplotype in Caucasians.
Materials and Methods
Human Subjects
Subjects were non-Hispanic Caucasians with severe hemophilia A (factor VIII <1%), recruited to the Multicenter Hemophilia Cohort Studies I/II between 1982 and 2005, under IRB-approved protocols (Clinical Trial registration number NCT00341705). Subjects were from 48 hemophilia centers located primarily in North America and Europe. All subjects were at least 13 years of age and extensively treated with factor VIII.
Case-Control Study Design
Cases (n=302) were patients with factor VIII inhibitors, defined as having at least one Bethesda inhibitor assay titer of > 1.0 Bethesda Inhibitor Assay Unit. Controls (n= 633) were a simple random sample of all severe hemophilia A subjects without factor VIII inhibitors. The prevalence of HIV was 41% in Cases and 46% in Controls (P= 0.95). The prevalence of chronic HCV was 80% in Cases and 77% in Controls (P= 0.98).
DNA Purification & SNP Genotyping
Genomic DNA was purified from leukocytes using Qiagen chromatography columns (Qiagen, Valencia, CA). Tag SNP genotypes were determined by Sequenom (Boston, MA), using PCR and template-dependent single base pair extension. Extended nucleotides were identified using MALDI-TOF mass spectroscopy.
Candidate Genes
Twenty candidate genes (Table 1) were selected for analysis based on previous associations with factor VIII inhibitor development in humans [6-8], animal studies of factor VIII [13] or factor IX immunogenicity [14], effects on hemophilia A severity [15], or their role in the TH1/TH2 immune response.
Table 1. Candidate Genes Analyzed for Association with Inhibitor Risk.
| Gene | # Tag SNPs for r2 = 1.0 | # SNPs with 80% Amplification | Gene Coverage | Previous Studies |
|---|---|---|---|---|
| Previously Studied Genes | ||||
| IL10 | 28 | 24 | 86% | Astermark, 20066 |
| TNFα | 22 | 18 | 82% | Astermark, 20067 |
| IL1A+B | 32 | 26 | 81% | Astermark, 20066 |
| IL13+4 | 17 | 12 | 71% | Astermark, 20066 |
| CTLA4 | 13 | 13 | 100% | Astermark, 20068 |
| Novel Immunomodulatory Candidate Genes | ||||
| IL2 | 10 | 6 | 60% | |
| IL6 | 26 | 17 | 65% | |
| IL12A | 29 | 26 | 90% | |
| IL12B | 28 | 26 | 93% | |
| IL17A | 19 | 16 | 84% | Ettinger et al, 200829 |
| IL22 | 31 | 26 | 84% | Lozier et al, 200514 |
| IFNγ | 13 | 12 | 92% | Lozier et al, 200514 |
| TGFβ1 | 10 | 7 | 70% | |
| CD28 | 20 | 17 | 85% | |
| IL1RN | 42 | 31 | 74% | |
| AZGP1 | 13 | 7 | 54% | Lozier et al, 200513 |
| Factor VIII and Related Coagulation Factors | ||||
| Factor VIII | 15 | 10 | 67% | Viel, et al 200912 |
| FVLeiden | 1 | 1 | 100% | Kurnik et al, 200815 |
| Prothrombin20210 | 1 | 1 | 100% | Kurnik et al, 200815 |
| HFE | 2 | 2 | 100% | |
| Sum | 372 | 298 | 80% | |
Single Nucleotide Polymorphisms
Single nucleotide polymorphisms that are within blocks of DNA that do not undergo homologous recombination during meiosis are termed “tag SNPs”. Tag SNPs that define haplotypes in Caucasians were selected from the HapMap database (Public Release #26), using the Haploview program. SNPs were located between 20 kb 5′ upstream to and 10 kb 3′ downstream to the gene's coding sequence. In all, 372 SNPs were selected, consisting of tag SNPs chosen to cover all possible common (greater than 1% prevalence) haplotypes in each genomic region (r2 = 1.0) [16].
SNP-based Association Analysis
The risk of inhibitor development associated with individual SNPs was determined using unconditional logistic regression models adjusted for HIV and HCV status. SNP genotypes were evaluated using a co-dominant model with a three-level ordinal variable coded 0, 1, and 2 for the number of rare alleles. Likelihood-ratio test was used to assess the relative improvement in model fit after the inclusion of individual SNPs. False discovery rate (FDR) adjustment was used to control for the number of tests (SNPs) in each region.
Haplotype-based Association Analysis
Haplotype phasing was inferred using Haploview (version 4.1), and analyzed for significant association with inhibitor status, using the HaploStats package in R [16]. We calculated odds ratios (OR) and 95% confidence intervals (CI) estimating the relative risk of inhibitor in relation to specific haplotypes within a haplotype block region using a logistic regression model adjusted for HIV and HCV status. The global additive effect of common haplotypes (>1% prevalence) within a block was also calculated using a global score test [17].
Results
Replication of Association between IL10 Polymorphisms and Inhibitor Development
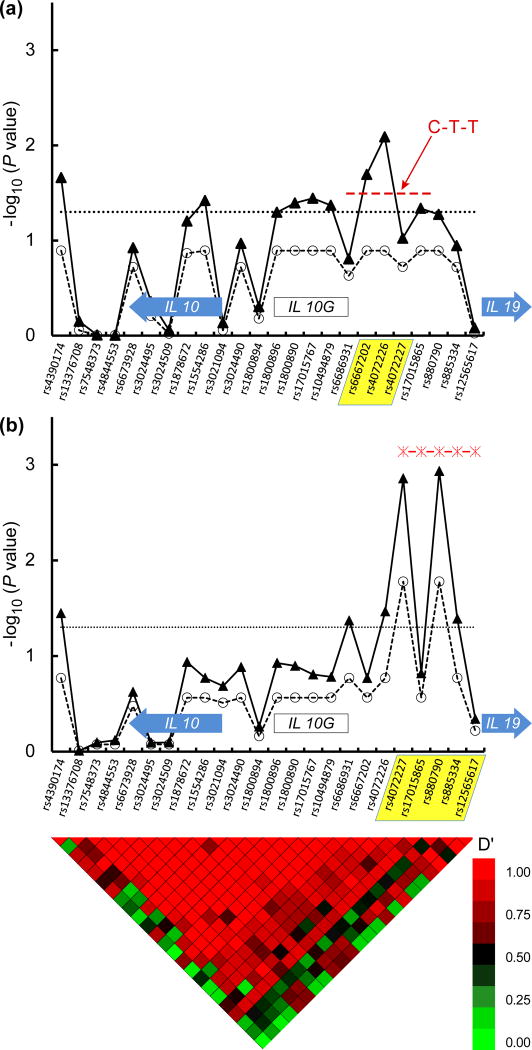
Inhibitor status was associated with six SNPs located ∼3,700 to 11,900 nucleotides 5′ to the IL10 Met translation start site (raw P values <0.05, Figure 1). This region also contains the IL10G microsatellite dinucleotide CA repeat DNA sequence [18], shown previously to be associated with inhibitor risk. [6] The global effect of haplotypes in this region was also significant (P = 0.016, Figure 1). The signal mostly reflected one common (32% prevalence) haplotype consisting of three tag SNPs (at rs6667202, rs4072226, and rs4072227) that was associated with increased inhibitor (OR = 1.23, CI 1.01-1.5, individual P value = 0.04).
Figure 1. IL10 Haplotypes are Associated with Increased or Decreased Factor VIII Inhibitor Risk.
The upper panel (A) depicts the association of IL10 tag SNPs and associated haplotypes with inhibitor risk in the overall group and the lower (B) panel depicts the association of IL10 tag SNPs and haplotypes with inhibitors in the HIV seropositive subjects. 24 tag SNPs are shown along the X axis, in their genomic order. In each panel, the position of the IL10 and IL19 genes are indicated by block arrows. The position of the IL10G microsatellite CA dinucleotide repeat is indicated by the open block as labeled. Haplotype blocks associated with inhibitor risk are indicated by yellow shading of the constituent tag SNPs. The -log10 of the raw P value for each tag SNP and their corresponding FDR values that accounts for multiple hypothesis testing are depicted with closed triangles connected with solid line and open circles connected with dashed line, respectively. The global effect of IL10 haplotypes is depicted with stars connected by a dashed red line in panel B. The dotted horizontal line indicates a significance level of P = 0.05. Nucleotides at polymorphic sites in each haplotype are indicated in red. Note that two haplotypes with odds ratios of 2.3 × 10-9 and 1.3 × 10-30 and P values of 0 (seen in 5% and 3% of the HIV positive subjects only) are not depicted on the lower panel. The heat map at the bottom of the figure denotes pairwise linkage disequilibrium (LD) values (D′) between tag SNPs. High LD (red color) indicate lack of evidence for recombination between pairs of SNPs, while low LD (green color) indicates recombination between pairs of SNPs. Tracing the squares below a tag SNP in a diagonal direction gives an indication of the LD between that SNP and its neighbors.
The global effect of IL10 haplotypes on inhibitors was stronger in HIV-positive subjects (P= 0.0007). Most of this effect came from two rare haplotypes defined by tag SNPs rs4072227, rs1701586, rs880790, rs885334, and rs12565617 (Table 2), that conferred protection against inhibitors (prevalence of 4.7-6.7% and odds ratio ≤ 2 × 10-9 for each, not shown in Figure 1).
Table 2. Haplotypes Associated with Increased/Decreased Inhibitor Risk.
| Gene (tag SNP rs #) Tag SNP Nucleotides | Cases (%) | Controls (%) | OR | 95% CI | P Value |
|---|---|---|---|---|---|
| IL10 (rs6667202,rs407226,rs4072227) | |||||
| C-T-T | 38 | 32 | 1.23 | 1.01-1.50 | 0.04 |
| IL10 (rs4072227, rs1701586, rs880790, rs885334, rs12565617) | |||||
| C-G-C-G-T* | <1 | 6.7 | 2.3 × 10-9 | NA | 0.0009 |
| T-A-C-A-T* | <1 | 4.7 | 1.3 × 10-30 | NA | 0.0048 |
| IL2 (rs10027390, rs2069772, rs2069779, rs2069778, rs2069762, rs4833248) | |||||
| T-G-C-C-T-G | 23 | 28 | 0.69 | 0.53-0.91 | 0.008 |
| IL12A (rs2243115, rs583911, rs2243131, rs568408, rs2243148, rs2243154, rs2133310) | |||||
| T-A-A-G-C-G-A | 29 | 24 | 1.31 | 1.02-1.68 | 0.041 |
| IL1α (rs3783557, rs2071373, rs6746923, rs17597976, rs11687624, rs11898680, rs6716046, rs6716046, rs7567619, rs7585707, rs11680809, rs12711742, rs12469600) | |||||
| G-T-A-G-T-T-T-T-T-C-G-T | 2.5 | 1.3 | 2.2 | 1.1-4.3 | 0.034 |
| G-T-A-G-T-C-C-T-T-C-G-T | 27 | 24 | 1.3 | 1.0-1.7 | 0.056 (NS) |
| A-T-G-G-T-T-T-T-C-A-G-C | 24 | 22 | 1.3 | 1.0-1.7 | 0.077 (NS) |
| IL1β (rs1143627, rs16944, rs1143623, rs1261220, rs13032029, rs13008855, rs6735739, rs12053091) | |||||
| T-A-G-C-C-T-C-C-G | 23 | 28 | 0.75 | 0.58-0.96 | 0.02 |
| Factor VIII (rs5945250, rs17281377, rs4898399, rs6643622, rs7053448, rs1936645, rs5945269, rs5945270, rs17281398, rs6649625) | |||||
| T-C-T-G-C-G-G-C-A-T | 3 | 0.25 | 3.8 | 1.50-9.51 | 0.004 |
HIV positive subjects
Novel Associations: IL2, IL1, and IL12A
Three SNPs in the IL2 gene had significant association with inhibitor development, two of which (rs2069762 and rs4833248) remained significant after correction for multiple hypothesis-testing (Figure 2). One haplotype with 28% prevalence among controls (Table 2) conferred protection against inhibitor development (OR 0.69, CI 0.53-0.91).
Figure 2. IL2 Tag SNPs are Associated with Decreased Factor VIII Inhibitor Risk.
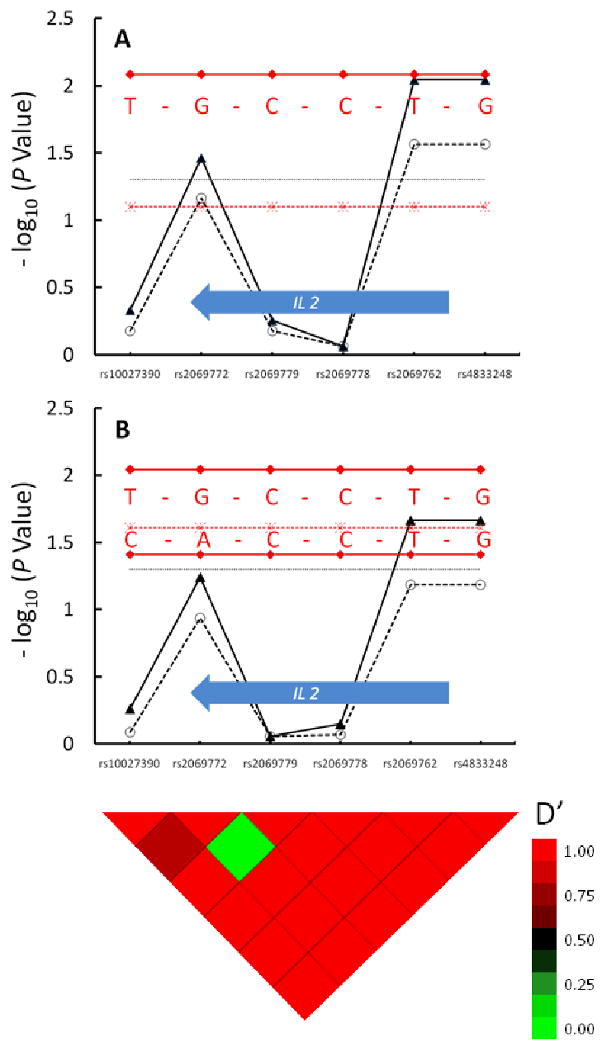
The upper panel depicts the association of IL2 tag SNPs and associated haplotypes with inhibitor risk in the overall group, and the lower panel depicts the associations in the HIV seropositive subjects. Six tag SNPs are shown along the X axis, in their genomic order. The IL2 gene is depicted by the block arrow. There is one haplotype block that encompasses the IL2 gene, based on the linkage disequilibrium “heat map” shown at the bottom. The -log10 of the raw P value for each tag SNP and their corresponding FDR values that accounts for multiple hypothesis testing are depicted with closed triangles connected with solid line and open circles connected with dashed line, respectively. The global effect of IL2 haplotypes is depicted with stars connected by a dashed red line in each panel. The dotted horizontal line indicates a significance level of P = 0.05. Nucleotides at polymorphic sites in each haplotype are indicated in red, and the global test of haplotype effect for HIV positive subjects is indicated with a dashed red line. The heat map at the bottom of the figure denotes pairwise linkage disequilibrium (LD) values (D′) between tag SNPs. High LD (red color) indicate lack of evidence for recombination between pairs of SNPs, while low LD (green color) indicates recombination between pairs of SNPs. Tracing the squares below a tag SNP in a diagonal direction gives an indication of the LD between that SNP and its neighbors.
The associations with IL2 tended to be stronger in HIV-infected patients (Figure 2). The haplotype with significant protective effect in the overall analysis increased further (OR 0.57, CI 0.38-0.86, P = 0.009), and a second haplotype (13% prevalence among controls) was also significant in the HIV-positive subgroup (OR 0.56, CI 0.32-0.98, P = 0.04). The global effect of all IL2 haplotypes was significant only in the HIV-positive subgroup (P= 0.03).
A series of SNPs in the IL12A gene were associated with inhibitor (Figure 3). One IL12A haplotype with a prevalence among controls of 24% was associated with increased risk of inhibitor development (OR = 1.31, CI 1.02-1.68, P = 0.04). The global effect of all IL12A haplotypes on inhibitor risk did not achieve significance (P = 0.10). No difference in the association with inhibitor risk was seen in HIV-infected patients.
Figure 3. IL12A Tag SNPs and Haplotypes Are Associated with Increased Factor VIII Inhibitor Risk.
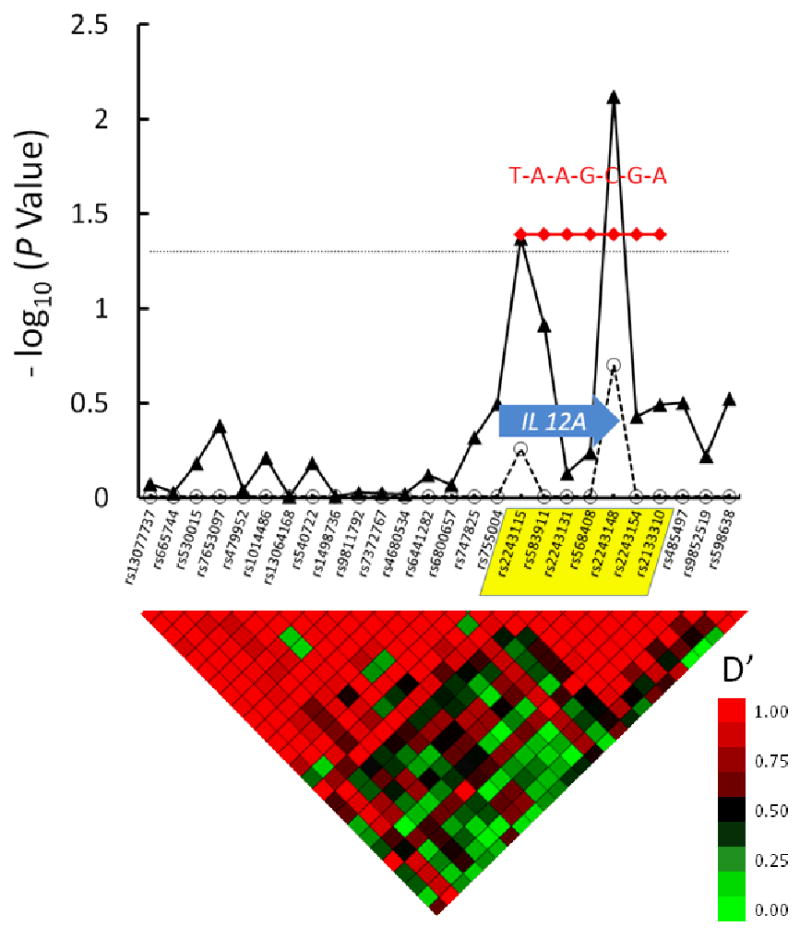
26 tag SNPs are shown along the X axis, in their genomic order. The IL12A gene is depicted as a block arrow; one haplotype block is evident in this region, based on the linkage disequilibrium “heat map” shown below the graph. The IL12A haplotype block is indicated by yellow shading of reference SNPs on the X axis. The -log10 of the raw P value for each tag SNP and their corresponding FDR values that accounts for multiple hypothesis testing are depicted with closed triangles connected with solid line and open circles connected with dashed line, respectively. The dotted horizontal line indicates a significance level of P = 0.05. Nucleotides at polymorphic sites in each haplotype are indicated in red. The heat map at the bottom of the figure denotes pairwise linkage disequilibrium (LD) values (D′) between tag SNPs. High LD (red color) indicate lack of evidence for recombination between pairs of SNPs, while low LD (green color) indicates recombination between pairs of SNPs. Tracing the squares below a tag SNP in a diagonal direction gives an indication of the LD between that SNP and its neighbors.
Multiple individual tag SNPs in the genes for the ILα and ILβ chains had marginal associations with inhibitor risk (Figure 4), but none of these was significant after correction for multiple testing. Analysis of haplotypes in these genes showed a rare haplotype (1.3% prevalence among controls) in IL1α that conferred significantly increased risk for inhibitor (OR = 2.2, CI 1.1-4.3, P=0.03). Other relatively common haplotypes in the IL1α gene were weakly associated with increased risk but were not significant, as shown in Table 2. One common IL1β haplotype with 28% prevalence among controls, was associated with decreased inhibitor risk (OR 0.75, CI 0.58-0.96, P = 0.02).
Figure 4. IL1α and ILβ Haplotypes May Increase or Decrease Factor VIII Inhibitor Risk.
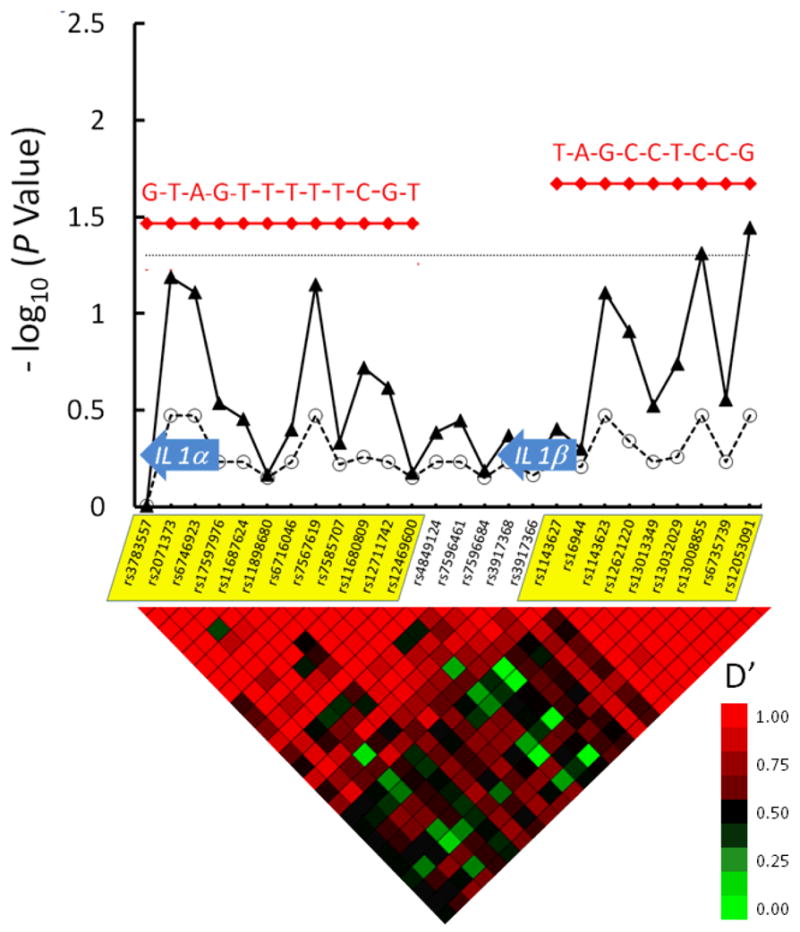
26 tag SNPs are shown along the X axis, in their genomic order. The IL1α and IL1β chain genes are depicted with block arrows. Two major haplotype blocks in this region are indicated by yellow shading of the tag SNPs on the X-axis. The -log10 of the raw P value for each tag SNP and their corresponding FDR values that accounts for multiple hypothesis testing are depicted with closed triangles connected with solid line and open circles connected with dashed line, respectively. The dotted horizontal line indicates a significance level of P = 0.05. Haplotypes are indicated in red. Nucleotides at polymorphic sites in each haplotype are indicated in red. The heat map at the bottom of the figure denotes pairwise linkage disequilibrium (LD) values (D′) between tag SNPs. High LD (red color) indicate lack of evidence for recombination between pairs of SNPs, while low LD (green color) indicates recombination between pairs of SNPs. Tracing the squares below a tag SNP in a diagonal direction gives an indication of the LD between that SNP and its neighbors.
Polymorphisms in the Factor VIII Gene
Initial analysis of 10 tag SNPs in the 216,928 bp region containing the factor VIII gene suggested no significant associations with inhibitor development (Figure 5). In light of the findings of Viel et al [12], we also analyzed factor VIII haplotypes in this Caucasian population. One rare haplotype was associated with significantly increased risk for inhibitor (OR 3.8, CI 1.5-9.5, P = 0.004), as it was seen predominantly among cases (3.1%) and was extremely rare among controls (0.25%). This haplotype was the strongest risk factor detected in our study.
Figure 5. A Rare Coagulation Factor VIII Haplotype is Associated with Factor VIII Inhibitor Risk.
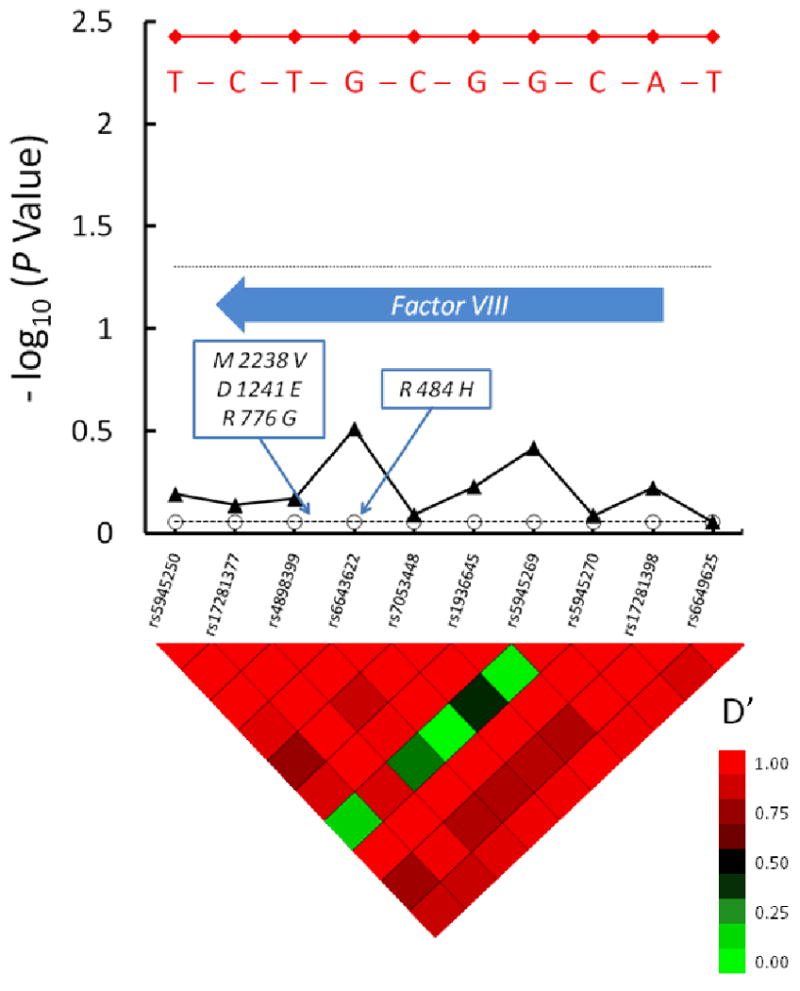
10 tag SNPs that encompass the factor VIII gene are shown along the X axis, in their genomic order. The factor VIII gene is depicted with a block arrow. One major haplotype block is evident in this region, based on the linkage disequilibrium “heat map” shown below the graph. The -log10 of the raw P value for each tag SNP and their corresponding FDR values that accounts for multiple hypothesis testing are depicted with closed triangles connected with solid line and open circles connected with dashed line, respectively. The dotted horizontal line indicates a significance level of P = 0.05. The one haplotype that was found to be associated with inhibitor risk is indicated in red, as are the nucleotides at polymorphic sites in this haplotype. The positions of amino acid coding polymorphisms in the factor VIII gene reported on by Viel et al [12] are indicated in the boxes in the Figure. M2238V = valine substitution for methionine at amino acid 2238, D1241E = glutamic acid substitution for aspartic acid, R776G = glycine substitution for arginine, R484H = histidine substitution for arginine at amino acid 484. The heat map at the bottom of the figure denotes pairwise linkage disequilibrium (LD) values (D′) between tag SNPs. High LD (red color) indicate lack of evidence for recombination between pairs of SNPs, while low LD (green color) indicates recombination between pairs of SNPs. Tracing the squares below a tag SNP in a diagonal direction gives an indication of the LD between that SNP and its neighbors.
Other Genes
Non-significant associations were seen with individual SNPs in genes previously associated with inhibitors in human (e.g., TNFα) or in vitro laboratory studies (i.e., IL17A) [7, 29]. No association was found with factor V Leiden or prothrombin 20210 gene polymorphisms that are associated with prothrombotic states thereby moderating the severity of the hemophilia A bleeding phenotype [15] and potentially decreasing intensity of factor VIII exposure and inhibitor development. The relative rarity of the factor V (4.3% prevalence) and prothrombin 20210 (3.0% prevalence) polymorphisms in our subjects limits our power to detect their effects.
Discussion
Our case-control study replicates the association of factor VIII inhibitors with polymorphisms in the IL10 promoter region seen in family studies [6]. This region is located upstream to both IL10 and IL19 promoter regions. IL19, is a member of the IL10 gene super-family [19], and may have resulted from duplication of the IL10 gene. IL 10 increases B-cell proliferation and maturation into immunoglobulin-producing plasma cells. IL19 increases expression of IL10, IL4, and IL13, cytokines critical to the TH2 immune response and antibody development [20]. The possibility that inhibitor development is partially mediated by the effects of IL19 should be investigated through in vivo and in vitro mechanistic studies.
The increased significance of the IL10 gene association in HIV seropositive subjects suggests an interplay between viral infection and the inhibitor response. Although chronic HIV infection may cause loss of factor VIII inhibitors and anti-factor VIII CD4 T-lymphocytes [21], during acute HIV infection there is increased IL10 expression from various immune cells [22]. The latter may promote the Th2/antibody response and increase factor VIII inhibitor risk. Certain IL10 promoter haplotypes are known to accelerate the progression to AIDS in HIV-infected persons of European/Caucasian ethnicity [23, 24]. Further, polymorphic microsatellite dinucleotide repeats in the same region are associated with lower concentrations of IL10 [25]. We speculate that IL10 haplotypes associated with lower expression may be critical both to AIDS progression as well as the factor VIII inhibitor response. We do not know the dates for HIV seroconversion and inhibitor development in our subjects, so we cannot say with certainty what the effect of HIV infection on inhibitor formation was in our study. As reported previously for our cohort and others, inhibitor prevalence was slightly lower among HIV+ subjects (30%, vs. 34% in HIV- subjects). The HIV+ and HIV- subjects born during the epidemic have essentially the same inhibitor rates (26-27%). We suspect that subjects born before 1977 developed inhibitors prior to HIV infection. For these subjects, the inhibitor rate is marginally higher (43%) in HIV + subjects than for HIV − subjects (39%), but we doubt the significance of that finding.
Individual SNPs and haplotypes in other immune genes modified inhibitor risk (Table 2). There was decreased risk of inhibitor associated with one haplotype encompassing the IL2 gene, and this negative association was stronger in HIV positive subjects (Figure 2), as seen for IL10. No differential effect in HIV- or chronically HCV-infected subjects was seen for any other genes that modified inhibitor risk. Inhibitors also were associated with haplotypes in the IL1β and IL1α genes. A previous study of one SNP in the IL1β gene showed no association with inhibitor risk [6], underscoring the importance of looking at haplotypes. Interestingly, there was increased inhibitor risk (OR 2.16) in one rare (1.7% prevalence) haplotype in the IL1α gene, whereas a fairly common haplotype in the IL1β gene (27% prevalence) had a protective effect (OR 0.75).
Interleukin 12, a heterodimeric cytokine expressed from the IL12A and IL12B genes, is secreted by dendritic cells. It stimulates synthesis of interferon-γ critical to the TH1 immune response [26]. IL12A polymorphisms were significantly associated with increased inhibitor risk (OR = 1.3) but IL12B polymorphisms were not (data not shown). The association of certain IL12A haplotypes with increased inhibitor risk may relate to its role in stimulating TH1 lymphocyte development [27]. Conversely, IL-12 is suppressed by the activity of IL-10, and modulation of inhibitor risk may depend on the balance between the effects of these cytokines [28].
The strongest risk factor in our study was one rare haplotype of the factor VIII gene that nearly quadrupled the risk of inhibitor development. This finding would be consistent with the hypothesis offered by Viel et al that rare haplotypes in patients of African ancestry represent “mismatches” with factor VIII product derived from predominantly Caucasian blood donors or recombinant products with “Caucasian” protein coding sequences [12]. This “factor VIII mismatch” mechanism, although more common in African Americans, presumably may occur at a lower frequency in the Caucasian population. If confirmed, this would support development of factor VIII products with amino acid sequences matching that of each patient's factor VIII sequence. Our study design differs from that of Viel et al [15], in that we chose to analyze tag SNPs, rather than polymorphisms leading to changes in the factor VIII amino acid coding sequence. However, their polymorphisms are clustered near one of our tag SNPs (rs6643622) that has a marginal signal for inhibitor risk (Figure 5).The genetic component of inhibitor risk is certainly complex, and different polymorphisms in different immune response genes may have additive or opposing effects on risk in the same patient. The factor VIII gene defect and severity of disease will likely remain the most important inherited determinant of inhibitor risk for patients with hemophilia
Our study has inherent limitations. Our subjects were treated in a unique era when HIV- and HCV- infected plasma-derived concentrates were commonly used. Recombinant products and modern, virus-free concentrates from pooled plasma make this unlikely to occur again. Another inherent limitation of our study is that genes were chosen on the basis of a priori mechanistic assumptions regarding the immune response. A genome-wide association analysis should be employed to discover genes whose association with factor VIII inhibitors is not expected on mechanistic grounds. Such an approach imposes the obligation to consider and control for false-positive findings from an enormous number of genes, SNPs, and their derivative haplotypes. Recently, Astermark and colleagues presented data from a collaborative trial involving 680 predominantly Caucasian hemophilia patients that surveyed more than 16,000 SNPs in 385 immunomodulatory and coagulation related genes [30]. They found associations of inhibitor risk with polymorphisms in the putative immunomodulatory genes DOCK2, MAPK9, PTPRR, and CD36, as well as the coagulation factor XIII gene [30]. These genes would not likely have been tested except in a systematic survey of all immunomodulatory genes. Our studies serve as stepping stones to comprehensive, genome-wide surveys of genetic and other risk factors for inhibitor development. Nonetheless, our data, together with other targeted genetic association studies to date, firmly suggest that polymorphisms that influence the TH1/TH2 balance may be key determinants of risk. The ultimate goals of these studies are to predict inhibitor risk for individuals, to identify novel targets for risk modulation, and to design lower risk recombinant factor VIII products.
Multicenter Hemophilia Cohort Study Center Locations.
United States
University of Arizona, Tucson, Arizona; University of California, San Francisco, San Francisco, California; University of California, Davis, Davis, California; Children's Hospital of Orange County, Orange, California; University of Colorado, Denver, Colorado; Christiana Hospital, Wilmington, Delaware; Children's National Medical Center, Washington, District of Columbia; Georgetown University, Washington, District of Columbia; Emory University, Atlanta, Georgia; University of Illinois, Chicago, Illinois; Children's Memorial Hospital, Chicago, Chicago, Illinois; St. Vincent's Hospital, Indianapolis, Indiana; Tulane University, New Orleans, Louisiana; Wayne State University Hutzel Hospital, Detroit, Michigan, 48201; University of Minnesota, Minneapolis, Minnesota; University of Mississippi, Jackson, Mississippi; St. Louis University, St. Louis, Missouri; University of New Mexico, Albuquerque, New Mexico; North Shore Long Island Jewish Health System, Lake Success, New York; University of Buffalo, Buffalo, New York; Cornell University, New York, New York; Mt. Sinai Medical Center, New York, New York; University of North Carolina, Chapel Hill, North Carolina; Wake Forest University, Winston-Salem, North Carolina; Wright State University, Dayton, Ohio; University of Cincinnati, Cincinnati, Ohio; Children's Hospital, Cincinnati, Cincinnati, Ohio; Ohio State University, Columbus, Ohio; University of Oklahoma, Oklahoma City, Oklahoma; Milton Hershey Medical Center, Hershey, Pennsylvania; University of Pennsylvania, Philadelphia, Pennsylvania; Thomas Jefferson University, Philadelphia, Pennsylvania; Palmetto Health Alliance, Columbia, South Carolina; University of Tennessee, Memphis, Tennessee; Vanderbilt University, Nashville, Tennessee; University of Texas, Houston, Houston, Texas; University of Texas, San Antonio, San Antonio, Texas; University of Utah, Salt Lake City, Utah; Children's Hospital of Wisconsin, Milwaukee, Wisconsin.
Brazil
University of Sao Paulo, Sao Paulo, Brazil; Hospital Brigadeiro, Sao Paulo, Brazil
Canada
South East Health Care Corporation, New Brunswick, Canada
France
Hospices Civils de Lyon, Lyons, France
Germany
University of Bonn, Bonn, Germany
Greece
Laikon General Hospital of Athens, Athens, Greece
Italy
University of Florence, Florence, Italy; University de Milano, Milan, Italy
Sweden
Karolinska Institute St. Gorans Hospital, Stockholm, Sweden
The contribution of subjects from centers in various geographic regions appear in Table 3
Table 3. Distribution of Subjects by Geographic Regions.
| Region of Center | Total Subjects | Case Subjects | Control Subjects |
|---|---|---|---|
| North America | 644 | 228 | 416 |
| Argentina | 61 | 14 | 47 |
| Brazil | 41 | 16 | 25 |
| Greece | 56 | 23 | 33 |
| Other European | 133 | 21 | 112 |
| Total | 935 | 302 | 633 |
Acknowledgments
We would like to acknowledge the National Hemophilia Foundation for funding of this project (through a Career Development Award to JNL), the NIH Clinical Center, and National Cancer Institute for salary support and additional research support. Partial support for the cohort was also provided by the National Heart, Lung and Blood Institute.
We would like to acknowledge Barbara Kroner, Susan Wilson, and Liliana Preiss of RTI International for management of research subject samples and information.
Finally, we would like to acknowledge and thank the MHCS I-II patients, investigators, and staff, without whom this research could not have been done.
Supported by the National Hemophilia Foundation Career Development Award (JNL), and the National Cancer Institute Intramural Research Program (PSR, JJG, IM)
Footnotes
Presented in part at the 51st Annual Meeting of the American Society of Hematology, New Orleans, LA, December 7, 2009
This is a US Government work; there are no restrictions on its use.
Authorship Contributions
JL secured funding for this project, designed experiments, assisted with data analysis and interpretation, and wrote the manuscript.
PSR helped to secure funding for this project, designed experiments, oversaw data analysis and interpretation, and helped write the manuscript.
JJG helped to secure funding for the Multicenter Hemophilia Cohort Studies, helped secure additional funding for this project, designed experiments, secured patient samples, helped with data analysis, and helped to edit the manuscript.
IM designed experiments, did the data analysis and most of its interpretation, and helped write the manuscript.
Disclosure of Conflicts of Interest
There are no conflicts of interest relevant to this report to disclose.
References
- 1.Ragni MV, Kessler CM, Lozier JN. Clinical Aspects and Therapy of Hemophilia, chapter 125. In: Hoffman R, Benz E, Shattil S, Furie B, Cohen H, editors. Hematology, Principles and Practice. 5th. Churchill-Livingstone; New York: 2009. [Google Scholar]
- 2.Chorba TL, Holman RC, Clarke MJ, Evatt BL. Effects of HIV infection on age and cause of death for persons with hemophilia A in the United States. Am J Hematol. 2001;66(4):229–240. doi: 10.1002/ajh.1050. [DOI] [PubMed] [Google Scholar]
- 3.Soucie JM, Nuss R, Evatt B, Abdelhak A, Cowan L, Hill H, Kolakoski M, Wilber N, the Hmophilia Surveillance System Project Investigators Mortality among males with hemophilia: relations with source of medical care. Blood. 2000;96(2):437–442. [PubMed] [Google Scholar]
- 4.Oldenburg J, El-Maarri O, Schwaab R. Inhibitor development in correlation to factor VIII genotypes. Haemophilia. 2002;8(Suppl 2):23–9. doi: 10.1046/j.1351-8216.2001.00134.x. [DOI] [PubMed] [Google Scholar]
- 5.Gill JC. The role of genetics in inhibitor formation. Thromb Haemost. 1999;82:500–504. [PubMed] [Google Scholar]
- 6.Astermark J, Oldenburg J, Pavlova A, Berntorp E, Lefvert AK, MIBS Study Group Polymorphisms in the IL10 but not in the IL1beta and IL4 genes are associated with inhibitor development in patients with hemophilia A. Blood. 2006;107(8):3167–72. doi: 10.1182/blood-2005-09-3918. [DOI] [PubMed] [Google Scholar]
- 7.Astermark J, Oldenburg J, Carlson J, Pavlova A, Kavakli K, Berntorp E, Lefvert AK, for the MIBS Study Group Polymorphisms in the TNFA gene and the risk of inhibitor development in patients with hemophilia A. Blood. 2006;108(12):3739–3745. doi: 10.1182/blood-2006-05-024711. [DOI] [PubMed] [Google Scholar]
- 8.Astermark J, Wang X, Oldenburg J, Berntorp E, Lefvert AK, on behalf of the MIBS Study Group Polymorphisms in the CTLA-4 gene and inhibitor development in patients with severe hemophilia A. J Thromb Haemost. 2007;5(2):263–5. doi: 10.1111/j.1538-7836.2007.02290.x. [DOI] [PubMed] [Google Scholar]
- 9.Hay CR, Ollier W, Pepper L, Cumming A, Keeney S, Goodeve AC, Colvin BT, Hill FG, Preston FE, Peake IR, Hay CRM. HLA class II profile: a weak determinant of factor VIII inhibitor development in hemophilia A. UKHCDO Working Party. Thromb Haemost. 1997;77(2):234–237. [PubMed] [Google Scholar]
- 10.White GC, 2nd, Kempton CL, Grimsley A, Nielson B, Roberts HR. Cellular immune responses in hemophilia: why do inhibitors develop in some, but not all hemophiliacs? J Thromb Haemost. 2005;3(8):1676–81. doi: 10.1111/j.1538-7836.2005.01375.x. [DOI] [PubMed] [Google Scholar]
- 11.Aledort LM, Dimichele DM. Inhibitors occur more frequently in African-American and Latino haemophiliacs. Haemophilia. 1998;4(1):68. doi: 10.1046/j.1365-2516.1998.0146c.x. [DOI] [PubMed] [Google Scholar]
- 12.Viel KR, Ameri A, Abshire TC, Iyer RV, Watts RG, Lutcher C, Channell C, Cole SA, Fernstrom KM, Nakaya S, Kasper CK, Thompson AR, Almasy L, Howard TE. Inhibitors of factor VIII in black patients with hemophilia A. N Engl J Med. 2009;360(16):1618–1627. doi: 10.1056/NEJMoa075760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lozier JN, Zhang Pei. Mapping of genes that control the antibody response to human FVIII in mice. Blood. 2005;106(suppl 1):536a. [Google Scholar]
- 14.Lozier JN, Tayebi N, Zhang P. Mapping of genes that control the antibody response to human factor IX in mice. Blood. 2005;105(3):1029–1035. doi: 10.1182/blood-2004-03-1126. [DOI] [PubMed] [Google Scholar]
- 15.Kurnik K, Bidlingmaier C, During C, Halimeh S, Schobess R, Nowak-Gottl U. Presence of factor V Leiden or prothrombin mutation influence inhibitor development in children with severe hemophilia A. Blood. 2008;112(11):96. [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score test for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eskdale J, Kube D, Gallagher G. A second polymorphic dinucleotide repeat in the 5′ flanking region of the human IL10 gene. Immunogenetics. 1996;45(1):82–83. doi: 10.1007/s002510050174. [DOI] [PubMed] [Google Scholar]
- 19.Gallagher G, Dickensheets H, Eskdale J, Izotova LS, Mirochnitchenko OV, Peat JD, Vazquez N, Pestka S, Donnelly RP, Kotenko SV. Cloning, expression and initial characterisation of interleukin-19 (IL-19), a novel homologue of human interleukin-10 (IL-10) Genes and Immunity. 2000;1(1):442–50. doi: 10.1038/sj.gene.6363714. [DOI] [PubMed] [Google Scholar]
- 20.Oral HB, Kotenko SV, Yilmaz M, Mani O, Zumkehr J, Blaser K, Akdis CA, Akdis M. Regulation of T cell s and cytokines by the interleukin-10 (IL-10)-family cytokines IL-19, IL-20, IL-22, IL-24 and IL26. Eur J Immunol. 2006;36(2):380–388. doi: 10.1002/eji.200425523. [DOI] [PubMed] [Google Scholar]
- 21.Bray GL, Kroner BL, Arkin S, Aledort LW, Hilgartner MW, Eyster ME, Ragni MV, Goedert JJ. Loss of high-responder inhibitors in patients with severe hemophilia A and human immunodeficiency virus type 1 infection: a report from the Multi-Center Hemophilia Cohort Study. Am J Hematol. 1993;42(4):375–379. doi: 10.1002/ajh.2830420408. [DOI] [PubMed] [Google Scholar]
- 22.Brockman MA, Kwon DS, Tighe DP, Pavlik DF, Rosato PC, Sela J, Porichis F, Le Gall S, Waring MT, Moss K, Jessen H, Pereyra F, Kavanagh DG, Walker BD, Kaufmann DE. IL-10 is up-regulated in multiple cell types during viremic HIV infection and reversibly inhibits virus-specific T cells. Blood. 2009;114(2):346–356. doi: 10.1182/blood-2008-12-191296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin HD, Winkler C, Stephens JC, Bream J, Young H, Goedert JJ, O'Brien TR, Vlahov D, Buchbinder S, Giorgi J, Rinaldo C, Donfield S, Willoughby A, O'Brien SJ, Smith MW. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc Natl Acad Sci USA. 2000;97(26):14467–72. doi: 10.1073/pnas.97.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olekysk TK, Shrestha S, Truelove AL, Goedert JJ, Donfield SM, Phair J, Mehta S, O'Brien SJ, Smith MW. Extended IL10 haplotypes and their association with HIV progression to AIDS. Genes Immun. 2009;10(4):309–322. doi: 10.1038/gene.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mörmann M, Rieth H, Hua TD, Assohou C, Roupelieva M, Hu SL, Kremsner PG, Luty AJ, Kube D. Mosaics of gene variations in the Interleukin-10 gene promoter affect interleukin-10 production depending on the stimulation used. Genes Immun. 2004;5(4):246–55. doi: 10.1038/sj.gene.6364073. [DOI] [PubMed] [Google Scholar]
- 26.Seder RA, Gazzinelli R, Sher A, Paul WE. Interleukin 12 acts directly on CD4+ T cells to enhance priming for interferon-gamma production and diminishes interleukin 4 inhibtion of such priming. Proc Natl Acad Sci USA. 1993;90(21):10188–10192. doi: 10.1073/pnas.90.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sieling PA, Wang XH, Gately MK, Oliveros JL, McHugh T, Barnes SF, Wolf L, Golkar L, Yamamura M, Yogi Y, Koichi Uyemura K, Rea TH, Modlin RL. IL-12 regulates T helper Type 1 cytokine response in human infectious disease. J Immunol. 1994;153(8):3639–3647. [PubMed] [Google Scholar]
- 28.D'Andrea A, Aste-Amezaga M, Valiante NM, Ma X, Kubin M, Trinchieri G. Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J Exp Med. 1993;178(3):1041–1048. doi: 10.1084/jem.178.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ettinger RA, James E, Kwok WW, Thompson AR, Pratt KP. IL-17 is secreted by T-cell clones isolated following an initial peak inhibitor response to factor VIIII, suggesting Th17 cells play a role in inhibitor development. Blood. 2008;112(11):96–97a. [Google Scholar]
- 30.Astermark J, Schwarz J, Donfield SM, DiMichele DM, Ewenstein BM, Gomperts ED, Nelson GW, Oldenburg J, Shapiro AD, Spotts G, Winkler CA, Berntorp E. Genetic Factors Associated with Inhibitor Development in Hemophilia A: Initial Results From the Hemophilia Inhibitor Genetics Study (HIGS) Combined Cohort. Blood. 2009;114(22):217. [Google Scholar]