Abstract
Rapid and sensitive detection of pathogens is a key requirement for both environmental and clinical settings. We report here a colorimetric enzyme-nanoparticle conjugate system for detection of microbial contamination. In this approach cationic gold nanoparticles (NP) featuring quaternary amine head-groups are electrostatically bound to an enzyme (β-galactosidase, β-Gal), inhibiting enzyme activity. Analyte bacteria bind to the NP, releasing the β -Gal, restoring activity, and providing an enzyme-amplified colorimetric read-out of the binding event. Using this strategy we have been able to quantify bacteria at 1×102 bacteria/mL in solution and at 1×104 bacteria/mL in a field-friendly test strip format.
Bacterial infection causes 300 million cases of severe illness each year,1 and is estimated to kill over 2 million children every year.2 The great majority of these deaths occur in emerging nations where bacteria are widespread in drinking water and food.3 Several techniques4,5 are available in laboratories for pathogenic bacteria detection and identification, including i) plating and culturing,6,7,8,9,10,11,12 ii) luminescence,13 iii) immunological approaches,7,8 iv) nucleic acid probe-based methods9 (PCR, LCR), v) mass spectrometry,10 vi) microarrays,11 and vii) biosensors.12 Each of these systems has its advantages; however the utility of these methods is generally limited by their high cost for use and requirement for trained operators.
Recent advances in nanotechnology have enabled the development of new diagnostic platforms14 for sensitive and rapid pathogen detection. For example, Ji et.al. 15 used positively-charged amine-terminated polyamidoamine dendrimers to capture bacteria, reporting a detection limit of 1 × 104 cells/mL.16 Functionalized gold nanoparticles (AuNPs), have likewise been used to detect bacteria,17 virus,18 cancer cells,19 and proteins.20 In 2005, Murphy et al.21 showed that CTAB (cetyltrimethylammonium bromide)-functionalized gold nanorods or nanospheres can conformally deposit to form monolayer on Bacillus cereus by strong electrostatic interaction. More recently, our group17a" demonstrated bacteria sensing through a nanoparticle-fluorescent polymer conjugate system at 2×105 cells/mL.
Two key issues can be identified in designing effective sensors for pathogen detection in the field. First, the limits of detection (LOD) required for application in either environmental testing4a,25, 22 or clinical applications25, 23 is 104–102 cells/mL, Second, readout should not require expensive instrumentation. To address these issues, we have developed a hybrid colorimetric enzymatic nanocomposite biosensor that uses enzyme amplification to provide high sensitivity for the detection of pathogens in aqueous solutions. The efficacy of this system was then demonstrated in both solution and test strip format.
Our colorimetric sensor design features three main components: a) β-Galactosidase (β-Gal),24 an anionic enzyme (pI 4.6) to provide signal amplification, b) a chromogenic substrate to provide color readout (chlorophenol-red-β-D-galactopyranoside, CPRG), and c) a cationic nanoparticle that binds reversibly to β-Gal, inhibiting the enzyme without denaturation (Figure 1a). The AuNPs used here are functionalized with quaternary ammonium ligands to provide high stability, biocompatibility, with a head group for tuning surface interactios, critical requirements for stable and sensitive biosensors (Figure 1b). Binding of anionic surface of analyte bacteria25 to the cationic particle surface displaces the β-Gal with concomitant restoration of activity. The active enzyme converts the pale yellow substrate into the red product, providing a colorimetric readout (Figure 1a)
Figure 1.
a) Enzyme-amplified sensing of bacteria, showing relative sizes of 2 nm core diameter particles and β-Gal. b) Structure of ligands used for sensing studies.
Prior to our sensing studies, we conducted activity titrations of β-Gal-catalyzed hydrolysis of the CPRG substrate using NP1–NP4 (Figure. 2). These studies were performed at 0.5 nM of β-Gal, a concentration that provided a reasonable timecourse (~10 min) for the colorimetric event. In practice, β-Gal in phosphate buffer solution (5 mM, pH = 7.4) was incubated with various concentration of NP1–NP4 for 15 minutes, and then 1.5 mM of the chromogenic substrate (CPRG, λmax = 595 nm) was added to NP-enzyme complexes. The normalized first-order rate of chromogenic substrate hydrolysis was plotted versus the molar ratio of nanoparticles to β-Gal, and decreased upon addition of nanoparticles, as shown for NP2 (Figure 2) After preliminary activity studies, NP2 was chosen as the highest affinity enzyme inhibitor (Figure S6), inhibiting the β-Gal activity at very low concentrations and providing the lowest LOD (Figure S7). The AuNP-enzyme complex solution was freshly prepared before each experiment, with no significant precipitation or color change observed during or after the experimental process. As a control, the enzyme inhibition was also studied with neutral tetraethylene glycol (NPTEG) and carboxylate (NPco2) functionalized nanoparticles, with no inhibition observed with these particles (Figure S8).
Figure 2.
Inhibition of activity assay of β-Gal (0.5 nM) with 1.5 mM substrate CPRG upon addition of NP2 (5 mM phosphate buffer), a) Enzyme inhibition upon addition of NP2. b) Inhibition of β-Gal (Vmax) before (ON) and after (OFF) addition of NP2.
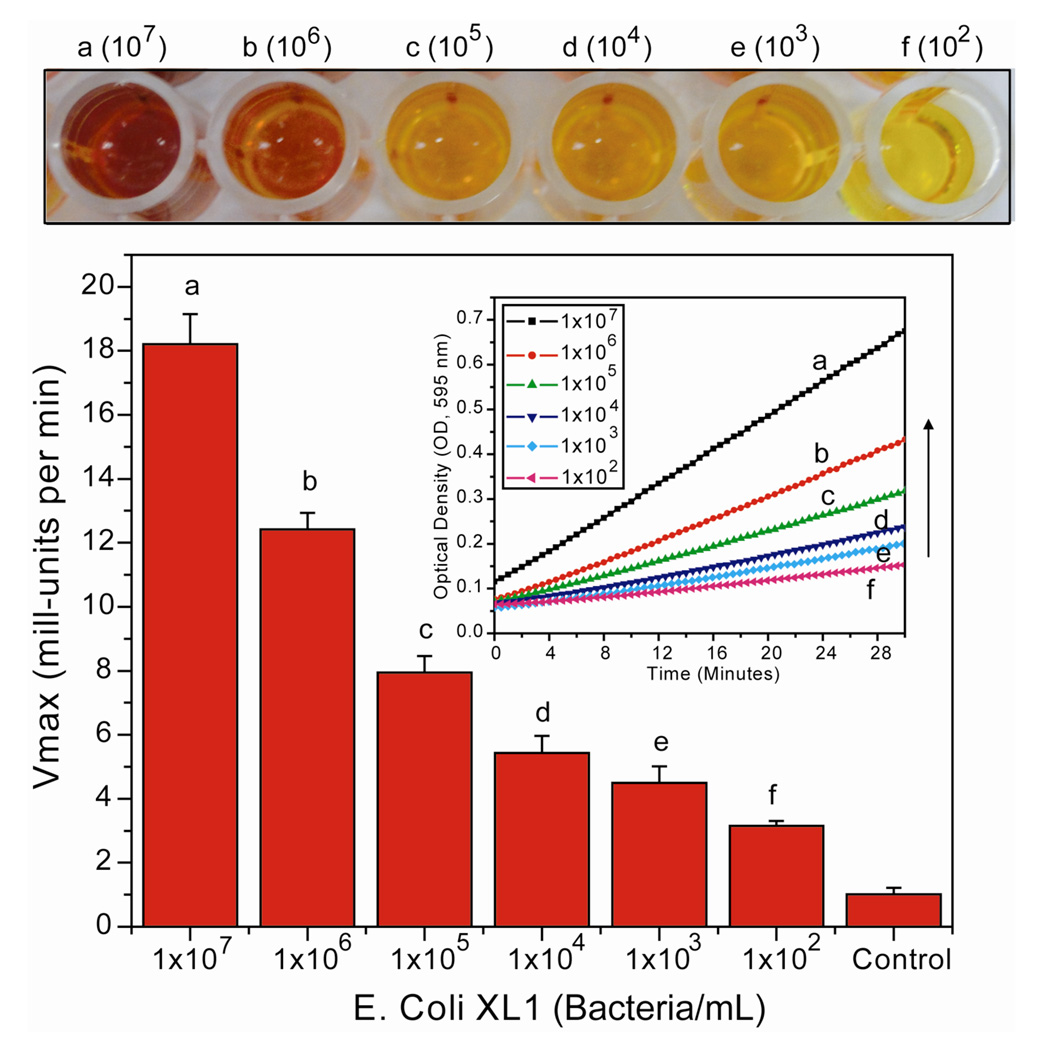
For our initial sensing studies we used E. coli (XL1) as a model analyte (Figure 3). From these studies, we can reproducibly differentiate bacterial levels as low as 100 cells/mL (three replicates were carried out for each sample, and each sample was also replicated three times). Each concentration can be discerned not only by intensity curves and the Vmax histogram but also by visible color changes: images taken immediately after reading (10 min) by an LCD camera demonstrate this colorimetric effect (Figure 3 top). Similar changes in Vmax were observed using S. griseus and B. subtilis (Gram positive, Figure Sll), indicating the generality of the system.
Figure 3.
Limit of detection of E. coli using β-Gal/NP2 nanocomposite. Kinetic absorbance response upon addition of different bacteria concentrations, as control β-Gal/NP2 nanocomposite was used without bacteria. At the top, microplate wells showing the color change upon variation of bacteria concentrations.
We next investigated the application of our design to a test strip format suitable for potential field use,26 featuring visual read-out of the originated color in comparison to a reference color scale.27 A key issue in this format is generating rapid and reproducible response times. Rapid bacterial penetration occurs on highly porous papers, while restriction of particle/enzyme conjugates to the surface occurs on less porous materials. Considering these issues, we explored a wide range of materials available to maintain the enzyme activity and the efficiency of enzyme inhibition and activity recovery process. Of the materials tested, GF/B binder-free microfiber filter was selected as the platform due to its high wet strength, high loading capacity and rapid response. The formulation of our strip sensor featured 25 mM CPRG and 15 nM β-Gal, providing conversion from yellow to dark red within 10 minutes with uninhibited enzyme. Then inhibition studies were carried out to determine the optimal concentration of cationic particle NP2 and β-Gal to form the hybrid enzymatic nanocomposite sensor [(β-Gal/NP2) complex]. NPTEG and NPco2 were also used as controls with no inhibition observed (Figure 4). The β-Gal/NP2 complex was ultimately generated by mixing β-Gal (15 nM) and NP2 (80 nM) and allowing the composite to dry for 15 minutes.
Figure 4.
Enzymatic inhibition-colorimetric assay of β-Gal (15 nM) against 25 mM substrate CPRG upon addition of cationic, anionic and neutral nanoparticles. (a) carboxylate (NPCO2), (b) hydroxyl (NPTEG), and quaternary amine (NP2) functionalized gold on a platform testing. Inset shows total inhibition for the positive nanoparticle NP2 at 80 nM, while no inhibition was observed for both the anionic and neutral AuNPs even at 160 nM.
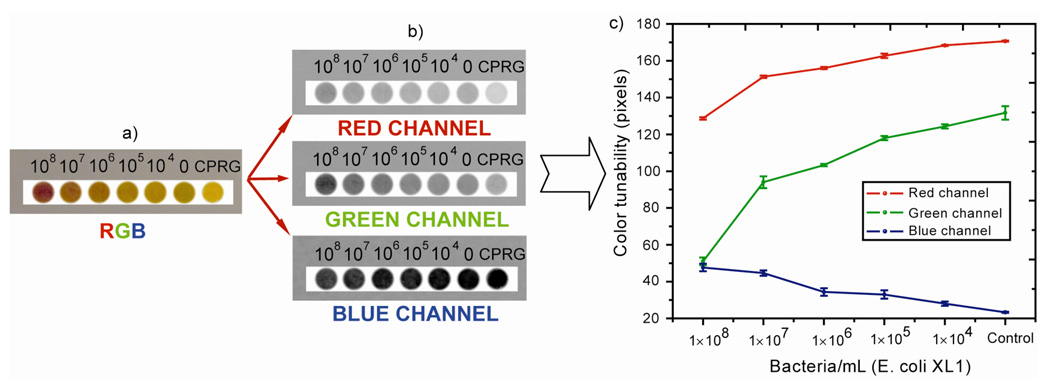
To test the performance of our system on a paper strip, 3 µL of CPRG (25 mM), complex solution and solutions from 1×108 to 1×104 bacteria/mL of E. coli (XL1) were spotted onto GF/B filter paper at pH 7.4. Images were obtained after 10 min with an LCD digital camera and appropriate lighting. As shown in Figure 5, clear visual differences were observed for concentrations ranging from 108-104 cells/mL. To provide quantitative assessment of the test strips, the RGB profile of the images were analyzed.27 The plots of RGB colorimetric channels (all values were taken at least three times) in Fig. 5c established the effectiveness of the chromogenic platform, demonstrating that 1×104 bacteria/mL can be distinguished using this method.
Figure 5.
Schematic illustration of the RGB colorimetric analysis to monitor color changes on the GF/B filter paper spot at pH 7.4. (a) imagine of the enzymatic activity response-colorimetric assay of the β-Gal-NP2 complex upon addition of E. coli (XL1) at different concentration, CPRG substrate was used as a control, (b) Red, green, and blue channels obtained from the original sample (a) to differentiate between bacteria concentration, (c) The extracted values of red, green, and blue channel from the original data (a). The measurement process is repeated at least three times for each measurement in a series of images.
In summary, we have the use of enzyme-nanoparticle assemblies to provide rapid and sensitive colorimetric sensing of bacteria. Using this system in a solution platform, bacteria concentrations as low as 100 cells/mL could be determined in a matter of minutes. Transfer of this methodology to a test strip format provided a potential tool for field applications with a visual sensitivity of 104 cells/mL. This work was conducted on the model analyte E.coli (XL1), but the detection sensitivity of different bacteria may vary from between species. Efforts are ongoing to improve the sensitivity of both formats, as well as adapting the methodology to dual detection and identification strategies for a general application.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NSF Center for Hierarchical Manufacturing at the University of Massachusetts (NSEC, DMI-0531171) and the NIH (GM077173). U.H.F.B and V.M.R. thank the Department of Energy Grant for generous financial support (DE-FG02-04ER46141).
Footnotes
Supporting Information
Experimental section, zeta potential, and dynamic light scattering, 13C NMR and 1H NMR spectra of NP1–NP4, Laser desorption/Ionization Mass Spectroscopy (LDA-MS) of NP1–NP4, Inhibition activity titration, recovery activity, control experiments, bacteria density image. These materials are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.a) Inglesby TV, et al. J. Am. Med. Assoc. 2002;287:2236–2252. [Google Scholar]; b) Brecher ME, Hay SN. Clin. Microbiol. Rev. 2005;18:195. doi: 10.1128/CMR.18.1.195-204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shorten PR, Pleasants AB, Soboleva TK. Int. J. Food Microbiol. 2006;108:369–375. doi: 10.1016/j.ijfoodmicro.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Ford EE, Colwell RR. A global decline in microbiological safety: a call for action. Washington, D.C.: A report from The American Academy of Microbiology; 1996. pp. 1–40. [Google Scholar]
- 3.Pedley S, Bartram J, Rees G, Dufour A, Cotruvo JA. Pathogenic mycobacteria in Water. London: World Health Organization (WHO); 2004. [Google Scholar]
- 4.a) Deisingh AK, Thompson M. Analyst. 2002;127:567–581. doi: 10.1039/b109895k. [DOI] [PubMed] [Google Scholar]; b) Deisingh AM, Thompson M. Can. J. Microbiol. 2004;50:69–77. doi: 10.1139/w03-095. [DOI] [PubMed] [Google Scholar]; c) Cole LA. Sci. Am. 1996;275:60–65. doi: 10.1038/scientificamerican1296-60. [DOI] [PubMed] [Google Scholar]
- 5.Straub TM, Chandler DP. J. Microbiol. Meth. 2003;53(2):185–197. doi: 10.1016/s0167-7012(03)00023-x. [DOI] [PubMed] [Google Scholar]
- 6.a) Hugo WB, Russell AD. Pharmaceutical Microbiology. 6th ed. Oxford: Blackwell; 1998. [Google Scholar]; b) Adams MR, Moss MO. Food Microbiology. 2nd ed. Guildford: Royal Society of Chemistry; 2000. [Google Scholar]
- 7.Safarik I, Safarikova M, Forsythe SJ. J. Appl. Bacteriol. 1995;78:575–585. doi: 10.1111/j.1365-2672.1995.tb03102.x. [DOI] [PubMed] [Google Scholar]
- 8.Harrigan HF. Laboratory Methods in Food Microbiology. 3rd ed. San Diego: Academic Press; 1998. [Google Scholar]
- 9.a) Newton CR, Graham A. PCR. 2nd ed. Oxford: Bios Scientific; 1997. [Google Scholar]; b) Eeles R, Sharp AC. Polymerase Chain Reaction: The technique and its applications. Austin: Landes, R.G.; 1993. [Google Scholar]
- 10.Strachan NJC, Nicholson FJ, Ogden ID. Anal. Chim. Acta. 1995;313:63–67. [Google Scholar]
- 11.a) Maughan NJ, Lewis FA, Smith V. J. Pathol. 2001;195:3–6. doi: 10.1002/path.924. [DOI] [PubMed] [Google Scholar]; b) Aitman TJ. Brit. Med. J. 2001;323:611–615. doi: 10.1136/bmj.323.7313.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Hall EAH. Biosensors. Buckingham: Open University Press; 1990. [Google Scholar]; b) Catrall RW. Chemical Sensors. Oxford: Oxford University Press; 1997. [Google Scholar]; c) Ivnitski D, Abdel-Hamid I, Atanasov P, Wilkins E. Biosens. Bioelectron. 1999;14:599–624. doi: 10.1016/s0956-5663(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 13.Hilpert K, Hancock REW. Nat. Protoc. 2007;2:1652–1660. doi: 10.1038/nprot.2007.203. [DOI] [PubMed] [Google Scholar]
- 14.a) Anslyn EV, Rotello VM. Curr. Opin. Colloid Interface Sci. 2010;14:683–684. [Google Scholar]; b) Nguyen BT, Anslyn EV. Coordin. Chem. Rev. 2006;250:3118–3127. [Google Scholar]; c) Boeneman K, Mei BC, Dennis AM, Bao G, Deschamps JR, Mattoussi H, Medintz IL. J. Am. Chem. Soc. 2009;131:3828–3829. doi: 10.1021/ja809721j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji J, Schanzle A, Tabacco MB. Anal. Chem. 2004;76:1411–1418. doi: 10.1021/ac034914q. [DOI] [PubMed] [Google Scholar]
- 16.a) Heal JS, Blom AW, Titcomb D, Taylor A, Bowker K, Hardy JRW. J. Hosp. Infect. 2003;53:136–139. doi: 10.1053/jhin.2002.1352. [DOI] [PubMed] [Google Scholar]; b) Perkins SD, Mayfield J, Fraser V, Angenent LT. Appl. Environ. Microbiol. 2009;75:5363–5372. doi: 10.1128/AEM.00658-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chen CL, Yu J-C, Holme S, Jacobs MR, Yomtovian R, McDonald CP. Transfusion. 2008;48:1550–1557. doi: 10.1111/j.1537-2995.2008.01716.x. [DOI] [PubMed] [Google Scholar]; d) Orihuela CJ, Fillon S, Tuomanen EI. Infect Immun. 2006;74:3783–3789. doi: 10.1128/IAI.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Wu Y-D, Chen L-H, Zhao Z-Y. J. Clin. Microbiol. 2008;46:2613–2619. doi: 10.1128/JCM.02237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Phillips R, Miranda O, You C, Rotello VM, Bunz U. Angew. Chem. Int. Ed. 2008;47:2590–2594. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]; b) Lee H, Yoon TJ, Weissleder R. Angew. Chem. Int. Ed. 2009;48:5657–5660. doi: 10.1002/anie.200901791. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Grossman H, Myers W, Vreeland V, Bruehl R, Alper M, Bertozzi C, Clarke J. P. Natl. Acad. Sci. USA. 2004;101:129–134. doi: 10.1073/pnas.0307128101. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhao X, Hilliard L, Mechery S, Wang Y, Bagwe R, Jin S, Tan W. P. Natl. Acad. Sci. USA. 2004;101:15027–15032. doi: 10.1073/pnas.0404806101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Bruno JG, Francis K, Ikanovic M, Rao P, Dwarakanath S, Rudzinski WE. J. Bionanosci. 2007;1:84–89. [Google Scholar]; b) Yang SY, Wang WC, Lan CB, Chen CH, Chieh JJ, Horng HE, Hong CY, Yang HC, Tsai CP, Yang CY, Cheng IC, Chung WC. J. Virol. Methods. 2010;164:14–18. doi: 10.1016/j.jviromet.2009.11.016. [DOI] [PubMed] [Google Scholar]; c) Niikura K, Nagakawa K, Ohtake N, Suzuki T, Matsuo Y, Hirofumi S, Ijiro K. Bioconjugate Chem. 2009;20:1848–1852. doi: 10.1021/bc900255x. [DOI] [PubMed] [Google Scholar]; d) Halfpenny KC, Wright DW. WIREs Nanomed. Nanobiotechnol. 2010;2:277–290. doi: 10.1002/wnan.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Bajaj A, R Miranda O, Kim IB, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. P. Natl. Acad. Sci. U.S.A. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, Rotello VM. Chem. Sci. 2010;1:134–138. [Google Scholar]
- 20.a) You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nat. Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]; b) De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Nat. Chem. 2009;1:461–465. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Miranda OR, Chen HT, You CC, Mortenson DE, Yang XC, Bunz UHF, Rotello VM. J. Am. Chem. Soc. 2010;132:5285–5289. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry V, Gole A, Kundu S, Murphy CJ, Saraf RF. J. Am. Chem. Soc. 2005;127:17600–17601. doi: 10.1021/ja056428l. [DOI] [PubMed] [Google Scholar]
- 22.a) Guidelines for Drinking Water. 3rd ed. Geneva, C: World Health Organization (WHO); 2004. [Google Scholar]; c) Gerba CP. Pathogens in the environment. New York: Academic press; 1996. pp. 279–299. [Google Scholar]; d) Chandler DP, Straub TM. J. Microbiol. Meth. 2003;53:185–197. doi: 10.1016/s0167-7012(03)00023-x. [DOI] [PubMed] [Google Scholar]; e) McFeters GA. Drinking water microbiology. New York: Springer; 1990. [Google Scholar]
- 23.a) Holme S, McAlister MB, Ortolano GA, Chong C, Cortus MA, Jacobs MR, Yomtovian R, Freundlich LF, Wenz B. Transfusion. 2005;45:984–993. doi: 10.1111/j.1537-2995.2005.04405.x. [DOI] [PubMed] [Google Scholar]; b) Hoffmann O, Keilwerth N, Bille M, Reuter U, Angstwurm K, Schumann RR, Dirnagl U, Weber JR. J. Cerebr. Blood F. Met. 2002;22:988–996. doi: 10.1097/00004647-200208000-00010. [DOI] [PubMed] [Google Scholar]; c) Wagner SJ. Vox Sang. 2004;86:157–163. doi: 10.1111/j.0042-9007.2004.00410.x. [DOI] [PubMed] [Google Scholar]; d) Wu Y-D, Chen L-H, Wu X-J, Shang S-Q, Lou J-T, Du L-Z, Zhao Z-Y. J. Clin. Microbiol. 2008;46:2613–2619. doi: 10.1128/JCM.02237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Jacobson RH, Zhang XJ, Dubose RF, Matthews BW. Nature. 1994;369:761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]; b) Verma A, Simard JM, Worrall JWE, Rotello VM. J. Am. Chem. Soc. 2004;126:13987–13991. doi: 10.1021/ja046572r. [DOI] [PubMed] [Google Scholar]; c) Fowler AV, Zabin I. J. Biol. Chem. 1978;253:5521–5525. [PubMed] [Google Scholar]
- 25.a) Kates M. Handbook of Lipid Research: Glycolipids, Phosphoglycolipids and Sulfoglycopids. Oxford: Plenum; 1990. pp. 123–234. [Google Scholar]; b) Dmitriev B, Toukach F, Ehlers S. Trends Microbiol. 2005;13:569–574. doi: 10.1016/j.tim.2005.10.001. [DOI] [PubMed] [Google Scholar]; c) Koch AL. Clin. Microbiol. Rev. 2003;16:673–687. doi: 10.1128/CMR.16.4.673-687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Schaffer C, Messner P. Microbiology. 2005;151:643–651. doi: 10.1099/mic.0.27749-0. [DOI] [PubMed] [Google Scholar]; e) Bos MP, Tefsen B, Geurtsen J, Tommassen J. P. Natl. Acad. Sci. USA. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.a) Hall M, Eldridge DB, Saunders RD, Fairclough DL, Bateman RC., Jr Food Biotech. 1995;9:39–57. [Google Scholar]; b) Patange SB, Mukundan MK, Kumar KA. Food Control. 2005;16:465–472. [Google Scholar]
- 27.a) Steiner MS, Meier RJ, Duerkop A, Wolfbeis OS. Anal. Chem. 2010;82:8402–8405. doi: 10.1021/ac102029j. [DOI] [PubMed] [Google Scholar]; b) Bonifacio LD, Puzzo DP, Breslav S, Ozin GA. Adv. Mater. 2010;22:1351. doi: 10.1002/adma.200902763. [DOI] [PubMed] [Google Scholar]; c) Janzen MC, Ponder JB, Bailey DP, Ingison CK, Suslick KS. Anal. Chem. 2006;78:3591–3600. doi: 10.1021/ac052111s. [DOI] [PubMed] [Google Scholar]; d) Filippini D, Gatto E, Alimelli A, Malik MA, Di Natale C, Paolesse R, Damico A, Lundstron I. J. Porphyr. Phthalocy. 2009;13:77–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.