Abstract
Membrane proteins comprise a third of the human genome, yet present challenging targets for reverse chemical genetics. For example, though implicated in numerous diseases including multiple myeloma, the membrane protein caveolin-1 appears to offer a poor target for the discovery of synthetic ligands due to its largely unknown structure and insolubility. To break this impasse and identify new classes of caveolae controlling lead compounds, we applied phage-based, reverse chemical genetics for the discovery of caveolin-1 ligands derived from the anti-HIV therapeutic T20. Substitution of homologous residues into the T20 sequence used a process analogous to medicinal chemistry for the affinity maturation to bind caveolin. The resultant caveolin-1 ligands bound with >1000-fold higher affinity than wild-type T20. Two types of ELISAs and isothermal titration calorimetry (ITC) measurements demonstrated high affinity binding to caveolin by the T20 variants with Kd values in the 150 nM range. Microscopy experiments with the highest affinity caveolin ligands confirmed colocalization of the ligands with endogenous caveolin in NIH 3T3 cells. The results establish the foundation for targeting caveolin and caveolae formation in living cells.
INTRODUCTION
Membrane proteins present challenging targets for structure determination, functional characterization, engineering, and drug development. Complicated by the inherent hydrophobicity of membrane proteins, such studies face numerous obstacles, starting with the poor expression yields of membrane proteins. In addition to ubiquity in all genomes, membrane proteins contribute key biological functions, and are targets for ≥50% of currently approved therapeutics.1 Despite recent advances in the structure determination of integral membrane proteins,2in vitro assays for binding and designing of ligands to membrane proteins remain difficult, idiosyncratic challenges. Thus, new approaches are required for targeting of membrane proteins, such as caveolin.
Anchored in the plasma membrane by an intramembrane domain, the monotopic subset of integral membrane proteins penetrates the leaflet of the plasma membrane, but does not span the width of the membrane. Instead, the N- and C-termini of such proteins typically remain on one side of the membrane, either the cytoplasmic or the extracellular milieu.3 Monotopic membrane proteins offer promising, yet relatively unexplored, targets for drug development. For example, caveolin, a monotopic membrane protein, remains structurally uncharacterized, but plays critical roles in cell morphology and signaling.4
Flask-shaped invaginations of the plasma membrane, termed caveolae, assist with endocytosis, lipid transport, cell differentiation, and signaling.4 The most abundant protein present in the caveolae, caveolin regulates, in part, such physiological functions.4b In complex with Polymerase Transcript Release Factor (PTRF or cavin), Serum Deprivation Protein Response (SDPR), cytoskeletal proteins, and cholesterol, caveolin forms higher order oligomeric states, often exceeding 144 subunits in size.5 The formation of such caveolin complexes shapes the caveolae. Highly conserved across vertebrate species,6 caveolin has been implicated in muscular dystrophy, Alzheimer's disease, diabetes, vascular abnormalities, urogenital disorders, and cancer.7 Though a tumor suppressor8, caveolin-1 is also over-expressed in prostate cancer, breast cancer, and Alzheimer's disease.9 The present study focuses on caveolin-1, one of three caveolin isoforms, referred to hereafter as caveolin.
The involvement of caveolin in many diseases makes the protein an attractive therapeutic target for many diseases including multiple myeloma.10 However, no natural or synthetic ligands specific to caveolin have been reported. The siRNA knockout of caveolin causes the disappearance of caveolae, which demonstrates the potential for control over caveolae formation by inhibiting caveolin.11 Current approaches to modulate caveolin transcription levels and properties target sterol regulatory elements12 and caveolar lipids (e.g., glycosphingolipids).10 For example, the sequestration of cholesterol by filipin, nystatin, or amphotericin hinders caveolae formation.13 However, cholesterol, a critical component of the plasma membrane and lipid rafts, participates in a variety of dynamic cellular processes (e.g., lipid transport).14 Thus, the cholesterol sequestration strategy hinders caveolae formation, but can also disrupt other physiological functions of the cell. New classes of caveolin targeting compounds could offer useful reagents in cell signaling studies and potential therapeutics by controlling caveolae formation.
In addition to its role in caveolae formation, caveolin binds to gp41 an envelope protein of HIV. Phage display has been used to define a caveolin binding domain (CBD1) within gp41 (residues 618 to 633).15 One class of anti-HIV therapeutics targets gp41, and blocks fusion with the host plasma membrane. For example, the FDA-approved, anti-HIV therapeutic Fuzeon (termed T20) prevents viral fusions, and consists of a 36-mer peptide derived from the gp41 CHR (C-terminal heptad repeat, residues 638 to 673).16 The potential interaction between caveolin and T20 has not been previously examined, and, in addition to CBD1, other regions of gp41 could interact with caveolin.
Here, we report low affinity binding by T20 to caveolin, and demonstrate in vitro molecular evolution of T20 variants to obtain high affinity ligands to caveolin. As previously described, the display of full-length caveolin on the phage surface requires a new type of helper phage17; this display enabled assay of the full-length caveolin interaction with T20. Having established caveolin-T20 binding, phage-displayed libraries of T20 variants were used for affinity maturation targeting a soluble, oligomeric caveolin with a deleted intramembrane domain. Following expression of the affinity-matured T20 variants as fusions to maltose binding protein (MBP), four independent assays (two different types of ELISA, ITC, and confocal microscopy) examined binding by oligomeric caveolin to the affinity-matured T20 variants.
Results and Discussion
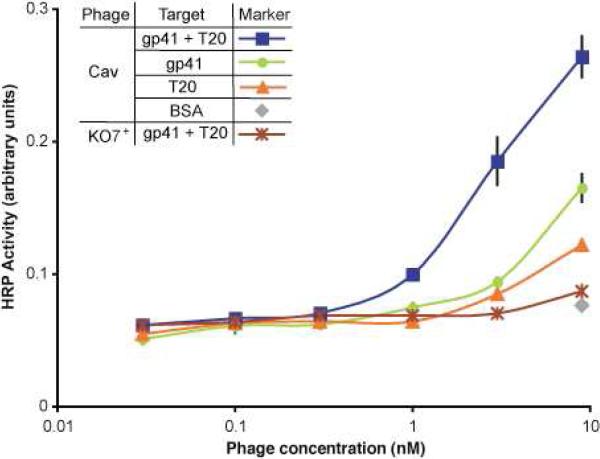
Previous analysis of the caveolin-gp41 interaction applied immunoprecipitation15a and SPR studies of 20-residues derived from caveolin that were fused to glutathione S-transferase.15 The short fragment of caveolin (20 out of 172 residues) used for the in vitro SPR study reflects the difficulty expressing and assaying full-length, membrane-bound caveolin. Recent reports of successful phage display with functional, full-length membrane proteins17 can provide a solution to this challenge. Here, for example, full-length caveolin displayed on phage could be assayed for direct binding with gp41, T20, and a 1:1 molar mixture of gp41 and T20. As shown in Figure 1, phage-displayed caveolin binds with low affinity to HIV gp41. To identify a gp41 region in addition to CBD1 critical for caveolin binding, T20 (Table 1) was also assayed. As expected for a fragment from a larger protein, T20 binds to caveolin with slightly reduced apparent binding affinity, than gp41. However, a 1:1 molar stoichiometry complex of gp41 and T20 bound to the phage-displayed caveolin with greatly enhanced relative binding affinity (Figure 1). Thus, a ternary complex of caveolin-gp41-T20 can form.
Figure 1.
Direct assay of phage-displayed full-length caveolin binding to gp41 and T20. Serial dilutions of phage-displayed, full-length caveolin (Cav-1) and KO7+ phage not displaying a protein, as a negative control, were incubated with gp41, gp41-T20 (1:1 molar ratio), or T20 coated in microtiter plate wells. The relative levels of bound proteins were quantified by anti-M13 HRP-conjugated antibody. Caveolin binds with low affinity to T20, but T20 enhances the caveolin-gp41 interaction. In ELISAs throughout this report, each data point represents the average of three experiments, and error bars indicate standard error.
Table 1.
Sequence Homology of Caveolin Ligands Selected from Phage-Displayed Librariesa
| T20 | Y | T | S | L | I | H | S | L | I | E | E | S | Q | N | Q | Q | E | K | N | E | Q | E | L | L | E | L | D | K | W | A | S | L | W | N | W | F | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gp41b | 638 | 648 | 658 | 668 | |||||||||||||||||||||||||||||||||
| Lig. | #Mut | ||||||||||||||||||||||||||||||||||||
| 1 | 15 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | H | – | K | D | I | – | D | I | E | R | L | G | T | F | – | K | C | Y |
| 2 | 9 | – | – | T | – | V | K | T | – | V | D | – | – | – | – | H | – | D | R | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 3 | 9 | – | – | – | M | V | K | – | – | – | D | D | T | – | Q | K | – | – | H | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 4 | 10 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | Q | – | K | – | – | M | – | – | E | R | – | G | – | M | L | K | C | – |
| 5 | 12 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | H | – | K | – | I | I | D | I | E | R | – | – | – | M | L | Q | L | – |
| 6 | 9 | F | S | – | – | V | K | – | – | V | – | – | – | K | H | – | – | D | N | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 7 | 5 | F | S | – | M | – | N | – | – | V | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
Hyphens indicate conservation of the wild-type residue. Gray shading indicates the CHR region of gp41.
The residue numbers correspond to the positions in the gp160 of the HIV-1 HXB2 variant. Boxed residues represent the lipid-binding pocket of wild-type T20.
Lig.= caveolin ligand, #Mut = number of mutations relative to the wild-type T20.
The observation of enhanced binding between caveolin and gp41 in the presence of T20 implicates an additional sub-domain of gp41, and also suggests a model for the interaction with caveolin. With respect to the ternary model, the gp41 used in the experiment forms a well-structured α-helix, as expected18 (Figure S1, Supporting Information). Thus, binding by the T20 peptide likely plays little role in structuring gp41, but could affect the structure of phage-displayed caveolin. The weaker binding by T20 than gp41 could indicate contributions to binding by additional regions of gp41, which do not share overlapping sequence with T20 (e.g., CBD1). Additionally, T20, unlike gp41, is unstructured in solution,18 and the entropic penalty upon binding could decrease its relative affinity for caveolin. In summary, the sequences shared by both T20 and gp41 (Table 1), corresponding to gp41 residues 638-673, must directly interact with caveolin, as both proteins bind caveolin independently.
Though modest, the caveolin-T20 interaction suggested T20 as a peptide-based lead compound for further affinity maturation using phage-displayed libraries of T20 variants. Homolog shotgun scanning libraries, either phage- or yeast-displayed, apply combinatorial libraries with a close homolog or the wild-type residue substituted into specific positions. The approach is analogous to optimization of binding by medicinal chemistry with small changes to amino acid sidechains across many positions.19 The large size (36 residues) of T20 prevented the synthesis of a single combinatorial library. Instead, two libraries of 18 residues were designed, and designated Libraries N and C for the N- and C-terminal 18-residues of T20, respectively (Table S1, Supporting Information). The diversity of the library composed entirely of 18-residues substituted with either a close homolog or the wild-type residue (218 = 2.6×105) was supplemented by additional substitutions, chosen to access potentially useful mutations for affinity maturation. The two libraries had a theoretical diversity of >108 different T20 variants. As assessed by phage titers, the practical diversity of Library N and C were 1.2×109 and 3.0×108, respectively.
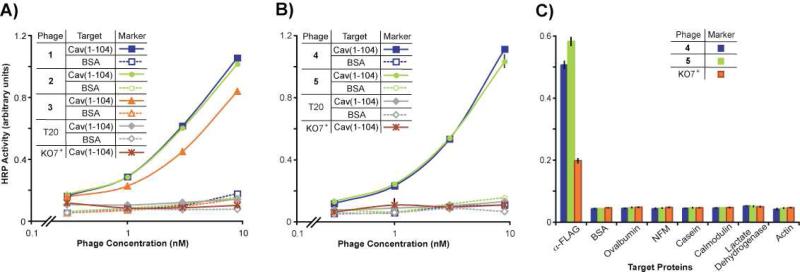
The two libraries were subjected in parallel to thermodynamic or equilibrium-based selections. Targeting a membrane protein, like caveolin, can present technical challenges to such selections. Thus, a truncated construct of caveolin (residues 1 through 104) with intramembrane and C-terminal domains deleted, termed cav(1-104), was over-expressed in E. coli, and purified to provide a robust target for the selections (Figure S2, Supporting Information). The purified protein remains functionally active, as demonstrated through binding to full-length caveolin.20 To reduce background binding between the phage scaffold and this target, KO7+ was used as a helper phage for library synthesis. With a modified phage coat, KO7+ decreases background binding to a wide range of targets.21 After bio-panning targeted to cav(1-104), functional variants of T20 from the libraries were identified by phage-based ELISA (Table 1 and Figure 2). A total of 200 selectants were screened from the different rounds of selections, and five different T20 variants were identified that bound to cav(1-104) with higher affinity than wild-type T20 (Figure 2). Ligand 1 emerged from round 5, and ligands 2-4 were isolated from round 4. The high mutation rate (9 to 15 mutations per T20 variant) reflects the large number of potential solutions for binding to caveolin, and the relatively conservative nature of mutations from a homolog-based library (Table 1).
Figure 2.
Phage-based ELISAs of representative T20 variants selected from the libraries. (A, B) In these experiments, the target cav(1-104) was coated on microtiter plates, and the ELISA developed as before. The selectants bound to cav(1-104) with much higher relative affinity than the wild-type T20 interaction with cav(1-104). (C) To demonstrate specificity for caveolin, ligands 4 and 5 (10 nM) displayed on KO7+ phage were assayed for binding to various negative control proteins coated on the plate and a positive control anti-FLAG antibody (α–FLAG), which can recognize a FLAG epitope fused to the N-terminus of the ligands. In this experiment, nonfat milk was used as a blocking agent. KO7+ without a displayed peptide provided a negative control for the assays.
Selected from Library C, the ligands 4 and 5 had 10 and 12 mutations respectively out of 18 positions altered in the wild-type T20 sequence. Interestingly, the two ligands converged on similar, but not identical, sequences for binding to caveolin with high affinity; as described previously,20 homolog shotgun scanning can improve binding affinity through small adjustments to a large number of residues responsible for a specific interaction. Overall, the two ligands accumulated more positively charged sidechains than wild-type T20. Such mutations could also assist with the solubility of the caveolin ligands. Though not a design consideration, this solubility enhancement was a welcome improvement to the poor solubility of T20.22 In addition, the lipid-binding motif of T20 (residues 670-673),23 was heavily mutated during in vitro evolution (Table 1). Here again, the directed evolution tailored ligand composition and binding specificity.
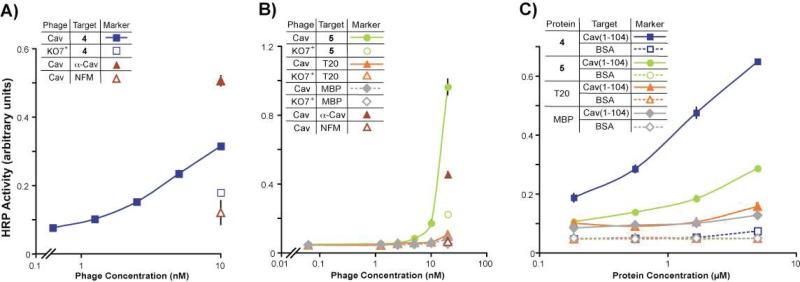
However, the selected T20 variants and wild-type, despite their largely homologous sequences, could have different display levels on the phage surface. Such differences could alter avidity effects to skew apparent binding affinity. Ligands 4 and 5, which exhibited the strongest relative affinity in the ELISA (Figure 2B), were chosen for further study. First, an additional ELISA experiment with the two phage-displayed ligands demonstrated the specificity of their binding to caveolin, as ligands 4 and 5 failed to bind to a number of control proteins (Figure 2C). For binding assays with non-phage-displayed caveolin ligands, sequences encoding the ligands 4, 5, and wild-type T20 were subcloned into a modified MBP fusion vector for protein over-expression; the modifications to the vector considerably increased the yields of over-expressed T20 variants, and are described in the Materials and Methods. The constructs were over-expressed as His6-tagged proteins in E. coli, purified to >95% homogeneity (Figure S2, Supporting Information), and first examined by ELISA for binding to various caveolin constructs. As expected, the ligands 4 and 5 fused to MBP bound to phage-displayed full-length caveolin (Figure 3A and 3B), and also to purified cav(1-104) (Figure 3C) with much higher relative affinity than wild-type T20 fused to MBP. As a negative control, MBP failed to interact with cav(1-104). Though the two ligands exhibited roughly similar binding affinities when displayed on the phage surface (Figure 2B), assays with purified ligands demonstrated the higher affinity of ligand 4, than 5, for both full-length and truncated caveolin. Such differences between the assays could result from higher levels of ligand 5 displayed on the phage surface, which would increase the apparent affinity of ligand 5 in the phage-based ELISA.
Figure 3.
ELISAs with over-expressed T20 variants removed from the phage surface. (A, B) Selected from phage-based affinity maturation, ligands 4 and 5 fused to MBP bound much more strongly to phage-displayed full-lenth caveolin than wild-type T20-MBP. (C) The ligands also bound to cav(1-104). MBP provided a negative control for the assay.
Next, isothermal titration calorimetry (ITC)24 was used to determine the thermodynamic parameters for MBP-fused ligands 4 and 5 binding to cav(1-104), which were compared to the controls MBP and T20-MBP (Table 2). The negative control, MBP, bound too weakly to cav(1-104) to measure an affinity by ITC (Figure S3, Supporting Information). The measured c-value of 0.088 for the MBP-caveolin interaction reflects the maximum solubility of around 20 μM for the cav(1-104) used as a target receptor in the ITC experiments.
Table 2.
ITC-derived Thermodynamic Binding Parameters for Interactions with cav(1-104)a.
| Protein | c-valueb | Chi2 ×105 | N1c | N2d | Kd1e | Kd2f | ΔGo̱1g | ΔGo̱2h | ΔHo̱1i | ΔHo̱2j | TΔSo̱1k | TΔSo̱2l |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (μM) | (kcal/mol) | (kcal/mol) | (kcal/(mol·K)) | |||||||||
| 4 | 45.4 | 1.23 | 0.353 | 1.85 | 0.155 | 1.96 | -9.11 | -7.64 | -20.6 | 44.8 | -11.4 | 52.4 |
| 5 | 14.3 | 3.03 | 0.249 | 1.02 | 0.348 | 5.58 | -8.67 | -6.93 | -26.1 | 83.9 | -17.3 | 90.8 |
| T20 | 0.051 | 0.371 | — | — | — | — | — | — | — | — | — | — |
| MBP | 0.088 | 0.084 | — | — | — | — | — | — | — | — | — | — |
Data for MBP-fused ligands 4, 5, and T20 binding to cav(1-104) fit a two binding sites per caveolin binding model. MBP-T20 and the negative control MBP failed to fit to a binding model, due to their very low affinity for caveolin.
C-values are for the first binding site (using Kd1).
Errors in N1 and N2 range from 1.0% to 4.0%.
Errors in N1 and N2 range from 1.0% to 4.0%.
Errors in Kd1 and Kd2 range from 5.6 to 17.2%.
Errors in Kd1 and Kd2 range from 5.6 to 17.2%.
Errors in ΔG1 and ΔG2 range from 1.0% to 13.8%.
Errors in ΔG1 and ΔG2 range from 1.0% to 13.8%.
Errors in ΔH1 and ΔH2range from 1.8 to 13.8%.
Errors in ΔH1 and ΔH2range from 1.8 to 13.8%.
Errors in TΔS1 and TΔS2 range from 1.8 to 13.8%. Hyphens indicate parameters that could not be accurately measured due to low c-values.
Errors in TΔS1 and TΔS2 range from 1.8 to 13.8%. Hyphens indicate parameters that could not be accurately measured due to low c-values.
The T20-caveolin interaction has not been reported previously, perhaps due to the weak nature of this interaction. Similar to the negative control MBP, T20 bound cav(1-104) with a very weak affinity in the ITC assays and consequently low c-values (0.051) (Figure S3, Supporting Information). Thus, the measured thermodynamic parameters for the T20-cav(1-104) interaction have very high error. With this caveat, the ITC data for the interaction fits a two site binding model, in which each T20 ligand binds weakly to two caveolin molecules (Kd in the sub-mM range). This multi-site binding is consistent with the oligomeric structure of caveolin; the cav(1-104) used in these experiments, for example, forms higher order oligomers, which can be observed and isolated by gel permeation chromatography (Figure S4, Supporting Information). T20 and variants 4 and 5 were purified as monomers, and remained monomeric under the reported conditions.
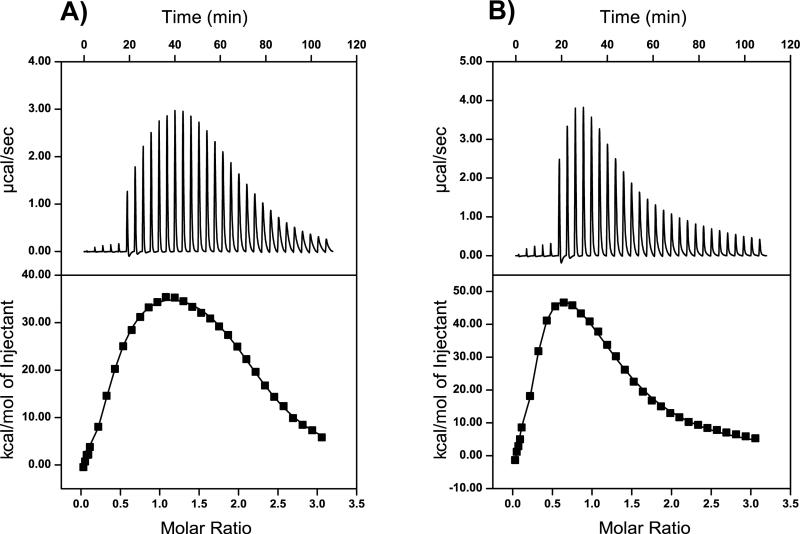
As measured by ITC, ligands 4 and 5 fused to MBP bind to cav(1-104) with a two site binding model, based upon the lowest chi-squared value for the curve fit to the isotherm (Table 2 and Figure 4). Thus, analogous to the T20-cav(1-104) interaction described above, each molecule of ligands 4 and 5 can bind to two subunits of the oligomeric cav(1-104). The stoichiometry obtained from analysis of the ITC data suggests both ligands 4 and 5 bind to the first binding site with a ratio of one ligand to approximately three caveolin receptors (N1); the second binding site has an approximately 2:1 or 1:1 ratio of ligand 4 or 5, respectively, per caveolin (N2). This estimate is subject to error inherent to the measurement and the weaker binding affinity of the second binding site; further structural analysis is necessary to assign the stoichiometry of the interaction with greater confidence.
Figure 4.
ITC measurements of caveolin ligands 4 and 5 binding to cav(1-104). (A) Ligand 4, or (B) 5 was injected into a solution of cav(1-104). The top graph depicts the calorimetric output for the interaction, which has been integrated in the lower graph. The molar ratio of the x-axis indicates the ratio of ligand to cav(1-104). The solid line represents the best least squares fit for a two binding site model. Table 2 lists the thermodynamic and kinetic parameters derived from these data.
The association with caveolin by ligands 4 and 5 is also negatively cooperative (Figure 4). Both interactions have 10-fold stronger affinity for the first caveolin binding site, than the second site (Table 2). This negative cooperativity reflects the enthalpic cost for ligands 4 and 5 interacting with the second caveolin binding site, which is counter-balanced by entropic contributions. Binding to the first caveolin site by ligands 4 and 5 is exothermic; by contrast, the interaction with the second site is strongly endothermic, and results in an overall endothermic binding for the two sites (Figure 4). The two-site binding to cav(1-104) by ligands 4 and 5 is entropically driven with a much greater ΔS2 than ΔS1 (Table 2). Such entropy driven binding could result from disruption of the caveolin oligomers upon binding to the T20 variants or increased configurational entropy due to conformational changes in the caveolin-ligand complex.
Affinity maturation by the library design described here succeeded in producing higher affinity ligands to caveolin. From the weak estimated binding affinity for the caveolin-T20 interaction (estimated Kd of in the sub mM range), both caveolin ligands 4 (Kd of 0.155 μM) and 5 protein (Kd of 0.348 μM) bound to the target with >1000-fold higher affinity. Furthermore, both ITC and ELISA data demonstrate stronger binding to caveolin by ligand 4, than ligand 5.
Next, we examined binding by the affinity-matured caveolin ligands in mammalian cells. Following cell fixation and permeabilization, ligands 4 or 5 fused to MBP were incubated for 6 hours with NIH 3T3 fibroblasts, a cell line known to express endogenous caveolin.5b The presence of MBP or MBP-fused ligand was determined using a MBP-specific primary antibody followed by addition of an Alexa Fluor 488-conjugated secondary antibody. Cells incubated with MBP not fused to the caveolin ligands served as negative controls to determine any contribution to the binding by the MBP fusion partner or the anti-MBP antibody used to label ligands 4 and 5. To visualize endogenous caveolin, Alexa Fluor 568 conjugated to a secondary anti-rabbit antibody bound to a anti-caveolin polyclonal antibody.
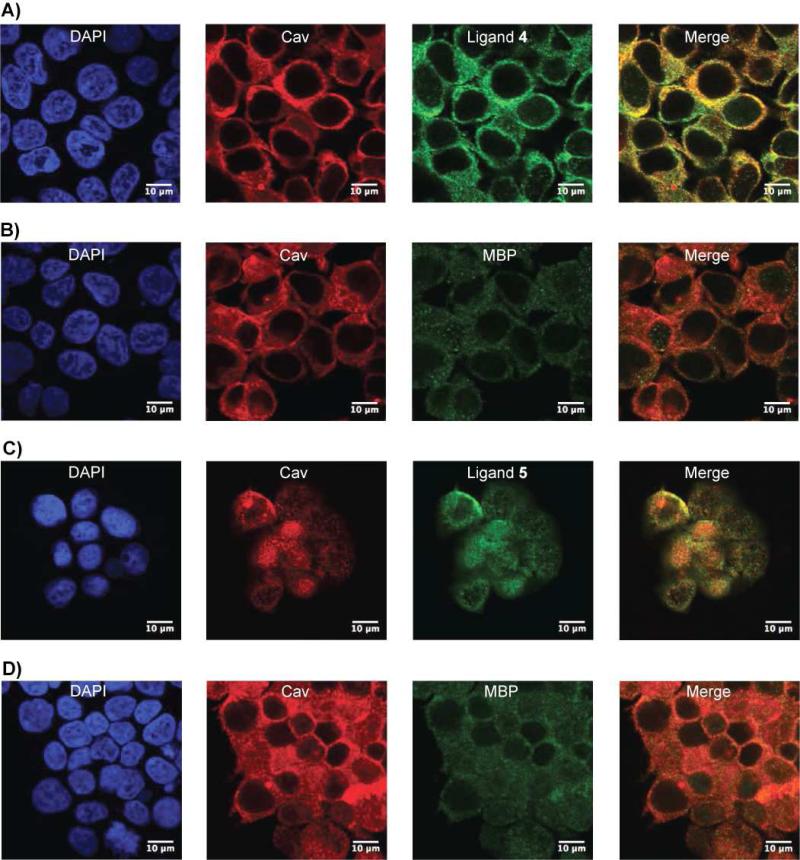
Confocal fluorescence microscopy demonstrates that both MBP-fused ligands 4 and 5 colocalize with endogenous caveolin in 3T3 cells. The yellow areas in the merged image (Figure 5A and 5C) highlight this colocalization. In the negative controls, cells were treated under identical conditions with MPB. The majority of the MBP protein and antibodies for visualization were washed out, indicating minimal binding to caveolin by MBP (Figure 5B and 5D). The colocalization of caveolin ligand 4 and caveolin was further confirmed using transient transfection of a plasmid encoding EGFP-fused to ligand 4 in NIH 3T3 cells (Figure S5, Supporting Information).
Figure 5.
Colocalization of caveolin-binding ligands 4 and 5 with endogenous caveolin in NIH 3T3 cells. Endogenously expressed caveolin is designated by the red signal. (A) MBP-fused ligand 4 or (B) the negative control, MBP, is labeled green and in the merged image, the yellow regions highlight areas of colocalization. (C) Comparable experiment showing the results with MBP-fused caveolin ligand 5 with the corresponding MBP control in panel D.
In conclusion, data from two types of ELISA, ITC, and cellular imaging demonstrate the identification of artificial, high affinity ligands to caveolin. This discovery originated with the chance observation that the anti-HIV therapeutic drug T20 binds to caveolin, albeit weakly as shown here by ITC and ELISA. Further “protein medicinal chemistry” applied a phage-based library of T20 variants with the wild-type residue, a close homolog or other substitution in every position of the initial lead. From this library, caveolin ligands with >1000-fold improvements in affinity were isolated by affinity-based selections. Further studies could examine the effects of such ligands on caveolae formation and as potential therapeutics for treating multiple myeloma. As caveolae play a critical role in endocytosis, caveolin also presents a target for the development of protein transduction reagents to assist in the cellular delivery of reagents and therapeutics.
Materials and Methods
Subcloning of Caveolin-1 and T20 into the Phage Display Vector
The human caveolin-1 cDNA was subcloned as described previously.17 The T20 cDNA (NIH AIDS Research and Reference Reagent Program, Cat #1069) was amplified by PCR to incorporate Nsi I, Afl II restriction sites, and the sequence encoding a N-terminal c-Myc epitope tag (EQKLISEEDL). Analogous to the previously described procedure, the sequence encoding the c-Myc-fused T20 was subcloned into the phage display vector.17
Bacterial Over-expression of HIV gp41, cav(1-104), and Selected Ligands
The HIV gp41 ectodomain used for this experiment was similar, though not identical (Figure S1), to the construct described by Lu and co-workers.25 The gp41 ectodomain with residues 546-578 and 624-655 were fused through a glycine-rich linker, and expressed as previously reported.17 A similar strategy was used to subclone residues 1 through 104 of caveolin-1 into the pET28c vector. Using Quikchange oligo-directed mutagenesis, the T7 epitope tag (60 base pairs) between the sequence encoding the His6 sequence and either gp41 or cav(1-104) were deleted. For expression of the T20 variants from the library, phage displaying ligands 4, 5, or wild-type T20 were subjected to a PCR using primers to introduce EcoRI and SalI restriction sites. The resulting PCR products were next subcloned into a modified pET28c vector, which included the gene encoding the maltose-binding protein (MBP) between the NheI and EcoRI restriction sites.
The vectors encoding the proteins were transformed into BL21(DE3) E. coli before growth at 37 °C for 2 h. After the cells reached an OD600 of 0.6, addition of IPTG (0.5 mM) induced protein expression during which the cells were shaken at either 37, 25, or 30 °C, respectively, for the cav(1-104), the gp41, or the MBP constructs. Following centrifugation and sonication, protein purification typically used buffers based on a common lysis buffer (300 mM NaCl, 50 mM NaH2PO4 pH 8.0) with the exception of cav(1-104), which required buffer supplemented with sarcosyl (N-lauroylsarcosine sodium salt) (0.25% w/v). The crude lysate was incubated with the nickel-NTA resin for 2 h at 4 °C in the presence of lysis buffer with 10 mM 2-mercaptoethanol, and the column was washed with wash buffer (lysis buffer containing 20 mM imidazole). The bound protein was eluted in lysis buffer containing 250 mM imidazole. Gel permeation chromatography further purified the proteins to >95% homogeneity.
Construction of the T20 Variant Libraries
Two libraries targeted consecutive regions of T20 from amino acid residues 638 to 673 (Table S1, Supporting Information). The oligonucleotides encoding Libraries N and C had theoretical diversities of 5.6 × 107 and 3.4 × 108 respectively. The actual diversities from phage titers greatly exceeded the theoretical diversity of Library N, and equaled the theoretical diversity of Library C.
Construction of the T20 variant libraries used a modified version of a previously described method.26 In brief, oligonucleotide-directed mutagenesis27 altered the sequence encoding T20 fused to the major coat protein of M13 bacteriophage (P8). To minimize high background binding to the cav(1-104) target during selections, KO7+ helper phage at a concentration of ~1.5×1010 phage/ml was used to infect the bacteria culture following recovery from electroporation. The two libraries were subjected to parallel selections.
Selection of Caveolin Binding T20 Variants
Wells (24, 12, 12, 8, 8, and 4 in six rounds of selection, respectively) of microtiter plates (Nunc) were coated with 10 μg/ml cav(1-104) in 100 μl of 50 mM Na2CO3, pH 9.6, and incubated overnight at 4 °C on a shaker. After removal of the coating solution, 400 μl of a solution of 0.2% w/v bovine serum albumin (BSA) in phosphate-buffered saline (PBS, 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 8.0) was used to block the wells for 30 min on a shaker at room temperature. In successive rounds, the blocking reagent was switched to BSA, ovalbumin, or non-fat milk (NFM). The plate was then washed three times with 200 μl per well 0.05% Tween-20 in PBS. Phage were added to the wells in a buffer containing 0.2% w/v BSA, 0.05% Tween-20 in PBS. After a 1 h incubation on a shaker at room temperature, the wells were washed with 0.05% Tween-20 in PBS. The numbers of washes increased with each round from 5, 7, and 12 for the first three rounds to 18 washes for the last three rounds. The bound phage were eluted by adding 100 μl of 0.1 M HCl and shaking vigorously at room temperature for 5 min. The phage were neutralized with 33 μl 1M Tris-HCl, pH 8.0. Before incubation for 1 h at 37 °C, 1 ml of the eluted phage was used to infect 10 ml of log phase E. coli XL-1 Blue cells. Helper phage KO7+ was added at ~1.5×1010 phage/ml, and, after 1 h of incubation, the culture was transferred to 200 ml of 2YT supplemented with 50 μg/ml carbenicillin and 20 μg/ml kanamycin, and shaken overnight at 37 °C.
Phage- and Protein-based ELISAs
In the phage-based ELISA, 96-well microtiter plates (Nunc) were coated with 10-15 μg/ml of cav(1-104), gp41, or gp41-T20 (1:1 molar ratio) in 50 mM Na2CO3 (pH 9.6), and incubated on a shaker at 4 °C overnight. The wells were blocked with 0.2% w/v solution of BSA in PBS at room temperature on a shaker for 30 min, and washed three times with wash buffer (0.05% v/v Tween-20 in PBS). Phage-displayed caveolin, T20, or T20 selectants were propagated for either one or two cycles of phage isolation, infection, and growth. The phage were then serially diluted along with a negative control (KO7+) in phage dilution buffer (0.2% w/v BSA, 0.05% v/v Tween-20 in PBS). The plates were incubated with the samples at room temperature on a shaker for 1-2 h, and then washed five times with wash buffer. Anti-M13/HRP conjugate (GE Healthcare) was diluted 1:5000 in the phage dilution buffer, added to the wells, and incubated for 30 min on a shaker at room temperature. The wells were washed five times with wash buffer and twice with PBS. 100 μl of a solution of 2 mg/ml o-phenylenediamine dihydrochloride, 0.02% w/v H2O2, in citric acid buffer (50 mM citric acid, 50 mM Na2HPO4, pH 5.0) was added to each wells. After 10 min incubation, the absorbance at 450 nm was measured using a microtiter plate reader (Bio-Tek).
For the protein-based ELISA, the target protein cav(1-104) at a concentration of 10 μg/ml was used to coat wells of a microtiter plate. After blocking with 0.2% w/v BSA in PBS, the indicated concentrations of T20, ligands 4, 5 or equivalent molar concentrations of negative control MBP were added to the wells. Anti-MBP antibody (Sigma) and horseradish peroxidase-conjugated secondary antibody (anti-Mouse-HRP from Sigma) visualized the presence of the bound protein.
Circular Dichroism
Following over-expression and purification, the 6xHis-tagged gp41 ectodomain was dialyzed overnight in 50 mM NaH2PO4, pH 8.0, 150 mM NaCl. A circular dichroism spectrum of the protein (30 μM) was acquired on a Jasco spectropolarimeter (Model J810) at 20 °C using a 2.0 nm bandwidth, 0.1 cm path length, 2.0 s response time, and a 50 nm/min scanning speed. The spectrum was corrected by subtraction of the equivalent measurements for the buffer. Thermal denaturation of gp41 (25 μM) was monitored by the change in ellipticity at 222 nm as a function of temperature every 0.2 °C with a 2 s response time. The Tm was estimated from the temperature resulting in 50% folded protein.
Isothermal Titration Calorimetry
The ITC experiments were performed using a VP-ITC instrument from Microcal (Northampton, MA). Six injections of 3 μl followed by 26 injections of 10 μl each of the ligand solution (MBP, MBP-fused T20 wild-type or variants) were added by a computer-controlled microsyringe at intervals of 210 s into the sample solution of cav(1-104) with stirring at 307 rpm. With ligands at a concentration of 300 μM and target cav(1-104) at 20 μM concentration, titrations were conducted at pH 8.0 in PBS at 20 °C. The experimental data were fit to a theoretical titration curve using software supplied by Microcal with ΔH (binding enthalpy kcal mol–1), Kd (dissociation constant), and N (number of ligands binding per caveolin), as adjustable parameters. Thermodynamic parameters were calculated from the Gibbs free energy equation, ΔG = ΔH –TΔS = –RT lnKa, where ΔG, ΔH, and ΔS are the changes in free energy, enthalpy, and entropy of binding, respectively. T is the absolute temperature, and R = 1.98 cal mol–1 K–1.
Cell Culture and Transfection
The ligand 4 fused to EGFP at the N-terminus was subcloned into a modified pEF6/Myc-His vector (Invitrogen) using the restriction sites KpnI and EcoRI. The vector included a SV40 ori and EF-1α as the promoter for expression of ligand 4 fused to EGFP inside cells. The transfection into NIH 3T3 fibroblasts (ATCC) used purified DNA (5-10 μg) and a calcium phosphate co-precipitation method (Clonetech #631312). NIH 3T3 cells were propagated in DMEM media (GIBCO #11965) supplemented with antibiotics, glutamine, non-essential amino acids, and 10% v/v fetal calf serum. Cells were grown for 36 h after transfection at 37 °C, 5% CO2 and 95% humidity.
Immunofluorescence and Microscopy
Cells were next fixed with 4% formaldehyde (Sigma) for 20 min on ice, and then permeabilized with a 1:1 mixture of cold methanol and PBS on a glass slide with incubation on ice for an additional 20 min. For the experiments with exogenously added ligands, blocking with 0.2% w/v BSA and 2% w/v NFM in PBS (1 h at room temperature) was followed by incubation with 6.5 μM MBP, MBP-fused ligand 4 or MBP-fused ligand 5 for 6 hours at 4 °C before washing three times with PBS and the addition of the pimary antibodies. To detect MBP, the primary antibody (mouse anti-MBP, Sigma) was diluted 1:800 in blocking buffer and the caveolin-specific primary antibody (rabbit anti-caveolin , Sigma) was diluted 1:500 dilution in blocking buffer) and incubated 24 h at 4 °C at a slow shaking speed. Excess antibody was removed by washing three times with wash buffer (PBS containing 0.1% Tween-20). To detect the primary antibody, the samples were incubated for 1 h with a mixture of Alexa Fluor 488-labeled goat anti-mouse (Invitrogen) and Alexa Fluor 568-labeled donkey anti-rabbi (Invitrogen) secondary antibodies, which were both diluted 1:500 in blocking buffer and incubated for 2 h at room temperature. The slides were next washed three times with wash buffer, and then sealed by a cover slip following application of DAPI-supplemented (4',6-diamidino-2-phenylindole) fluorescent mounting medium. Confocal imaging was performed with an Olympus Fluoview FV1000 laser scanning microscope using a 60x oil immersion objective (1.3 numeric aperture). The EGFP-expressed ligand 4 and the Alexa Fluor 488 were excited with an argon laser at 488 nm, and emission was monitored using a 505-530 nm bandpass filter. For Alexa Fluor 568 visualization, a HeNe laser at 543 nm and a 560-615 nm bandpass filter was used.
Supplementary Material
Acknowledgement
We acknowledge the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH for providing the gp41 cDNA (pHXB2-env) from Dr. Kathleen Page and Dr. Littman and the T20 peptide for the initial experiments; we also gratefully acknowledge Dr. Gabriel Fenteany and Dr. Alem Kahsai for supplying the EGFP vector. We thank Dr. Elizabeth Hinde from the DBC Optical Biology Core (OBC) Facility, Amanda Cinquin, Steve Chang, and Dr. Travis I. Moore for their technical help and suggestions on confocal imaging of cells; in addition, we thank Tom Yuan for assistance with the ITC analysis. This work was supported financially by the National Institute of Health, National Institute of General Medical Sciences (R01-GM078528-01 to G.A.W), NIH Virology Training Grant No. 5 T32 AI007319 - 24 to A.H.) and NIH-MARC training grant No. GM-069337 to A.S.M.
Footnotes
Supporting Information Available: Additional supporting documents and experiments, including information about the library design, CD analysis of the HIV gp41 construct, SDS-PAGE of the proteins used for ITC, ITC measurements of MBP and T20 binding to cav(1-104), gel permeation chromatography analysis of cav(1-104) and MBP-4, and confocal microscropy with EGFP-fused caveolin ligand 4, is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Russell RB, Eggleston DS. Nat. Struct. Biol. 2000;(Suppl):7, 928–930. doi: 10.1038/80691. [DOI] [PubMed] [Google Scholar]
- 2.White SH. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 3.Blobel G. Proc Natl Acad Sci U S A. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Sargiacomo M, Sudol M, Tang Z, Lisanti MP. J. Cell Biol. 1993;122:789–807. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Parton RG, Simons K. Nat. Rev. Mol. Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 5.a Murata M, Peranen J, Schreiner R, Wieland F, Kurzchalia TV, Simons K. Proc. Natl. Acad. Sci. U. S. A. 1995;92:10339–10343. doi: 10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, Walser P, Abankwa D, Oorschot VM, Martin S, Hancock JF, Parton RG. Cell. 2008;132:113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Hansen CG, Bright NA, Howard G, Nichols BJ. Nat. Cell Biol. 2009;11:807–814. doi: 10.1038/ncb1887. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Stahlhut M, van Deurs B. Mol. Biol. Cell. 2000;11:325–337. doi: 10.1091/mbc.11.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a Spisni E, Tomasi V, Cestaro A, Tosatto SC. Biochem. Biophys. Res. Commun. 2005;338:1383–1390. doi: 10.1016/j.bbrc.2005.10.099. [DOI] [PubMed] [Google Scholar]; b Tang Z, Okamoto T, Boontrakulpoontawee P, Katada T, Otsuka AJ, Lisanti MP. J. Biol. Chem. 1997;272:2437–2445. doi: 10.1074/jbc.272.4.2437. [DOI] [PubMed] [Google Scholar]
- 7.Cohen AW, Hnasko R, Schubert W, Lisanti MP. Physiol. Rev. 2004;84:1341–1379. doi: 10.1152/physrev.00046.2003. [DOI] [PubMed] [Google Scholar]
- 8.Engelman JA, Zhang XL, Lisanti MP. FEBS Lett. 1998;436:403–410. doi: 10.1016/s0014-5793(98)01134-x. [DOI] [PubMed] [Google Scholar]
- 9.a Yang G, Truong LD, Timme TL, Ren C, Wheeler TM, Park SH, Nasu Y, Bangma CH, Kattan MW, Scardino PT, Thompson TC. Clin. Cancer Res. 1998;4:1873–1880. [PubMed] [Google Scholar]; b Gaudreault SB, Dea D, Poirier J. Neurobiol. Aging. 2004;25:753–759. doi: 10.1016/j.neurobiolaging.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Podar K, Anderson KC. Cancer Lett. 2006;233:10–15. doi: 10.1016/j.canlet.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 11.Miyawaki-Shimizu K, Predescu D, Shimizu J, Broman M, Predescu S, Malik AB. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;290:L405–413. doi: 10.1152/ajplung.00292.2005. [DOI] [PubMed] [Google Scholar]
- 12.Bist A, Fielding PE, Fielding CJ. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10693–10698. doi: 10.1073/pnas.94.20.10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a Rothberg KG, Ying YS, Kamen BA, Anderson RG. J. Cell Biol. 1990;111:2931–2938. doi: 10.1083/jcb.111.6.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]; c Carozzi AJ, Ikonen E, Lindsay MR, Parton RG. Traffic. 2000;1:326–341. doi: 10.1034/j.1600-0854.2000.010406.x. [DOI] [PubMed] [Google Scholar]
- 14.Hao M, Mukherjee S, Sun Y, Maxfield FR. J. Biol. Chem. 2004;279:14171–14178. doi: 10.1074/jbc.M309793200. [DOI] [PubMed] [Google Scholar]
- 15.a Hovanessian AG, Briand JP, Said EA, Svab J, Ferris S, Dali H, Muller S, Desgranges C, Krust B. Immunity. 2004;21:617–627. doi: 10.1016/j.immuni.2004.08.015. [DOI] [PubMed] [Google Scholar]; b Huang JH, Lu L, Lu H, Chen X, Jiang S, Chen YH. J. Biol. Chem. 2007;282:6143–6152. doi: 10.1074/jbc.M607701200. [DOI] [PubMed] [Google Scholar]
- 16.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majumdar S, Hajduczki A, Mendez AS, Weiss GA. Bioorg. Med. Chem. Lett. 2008;18:5937–5940. doi: 10.1016/j.bmcl.2008.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Lu H, Niu J, Xu Y, Wu S, Jiang S. J. Biol. Chem. 2005;280:11259–11273. doi: 10.1074/jbc.M411141200. [DOI] [PubMed] [Google Scholar]
- 19.a Murase K, Morrison KL, Tam PY, Stafford RL, Jurnak F, Weiss GA. Chem. Biol. 2003;10:161–168. doi: 10.1016/s1074-5521(03)00025-5. [DOI] [PubMed] [Google Scholar]; b Cochran JR, Kim YS, Lippow SM, Rao B, Wittrup KD. Protein Eng. Des. Sel. 2006;19:245–253. doi: 10.1093/protein/gzl006. [DOI] [PubMed] [Google Scholar]; c Levin AM, Murase K, Jackson PJ, Flinspach ML, Poulos TL, Weiss GA. ACS Chem. Biol. 2007;2:493–500. doi: 10.1021/cb700055t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajduczki A, Majumdar S, Fricke M, Brown IAM, Weiss GA. 2010. Submitted for publication.
- 21.Lamboy JA, Tam PY, Lee LS, Jackson PJ, Avrantinis SK, Lee HJ, Corn RM, Weiss GA. Chembiochem. 2008;9:2846–2852. doi: 10.1002/cbic.200800366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao J, Burn A, Tolbert TJ. Bioconjug. Chem. 2008;19:1113–1118. doi: 10.1021/bc800063k. [DOI] [PubMed] [Google Scholar]
- 23.Liu S, Jing W, Cheung B, Lu H, Sun J, Yan X, Niu J, Farmar J, Wu S, Jiang S. J. Biol. Chem. 2007;282:9612–9620. doi: 10.1074/jbc.M609148200. [DOI] [PubMed] [Google Scholar]
- 24.a Wiseman T, Williston S, Brandts JF, Lin LN. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]; b Freire E, Schon A, Velazquez-Campoy A. Methods Enzymol. 2009;455:127–155. doi: 10.1016/S0076-6879(08)04205-5. [DOI] [PubMed] [Google Scholar]
- 25.Shu W, Liu J, Ji H, Radigen L, Jiang S, Lu M. Biochemistry. 2000;39:1634–1642. doi: 10.1021/bi9921687. [DOI] [PubMed] [Google Scholar]
- 26.Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Methods Enzymol. 2000;328:333–363. doi: 10.1016/s0076-6879(00)28406-1. [DOI] [PubMed] [Google Scholar]
- 27.Kunkel TA. J. Biol. Chem. 1985;260:5787–5796. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.