Abstract
Plant chloroplasts originated from an endosymbiotic event by which an ancestor of contemporary cyanobacteria was engulfed by an early eukaryotic cell and then transformed into an organelle. Oxygenic photosynthesis is the specific feature of cyanobacteria and chloroplasts, and the photosynthetic machinery resides in an internal membrane system, the thylakoids. The origin and genesis of thylakoid membranes, which are essential for oxygenic photosynthesis, are still an enigma. Vipp1 (vesicle-inducing protein in plastids 1) is a protein located in both the inner envelope and the thylakoids of Pisum sativum and Arabidopsis thaliana. In Arabidopsis disruption of the VIPP1 gene severely affects the plant's ability to form properly structured thylakoids and as a consequence to carry out photosynthesis. In contrast, Vipp1 in Synechocystis appears to be located exclusively in the plasma membrane. Yet, as in higher plants, disruption of the VIPP1 gene locus leads to the complete loss of thylakoid formation. So far VIPP1 genes are found only in organisms carrying out oxygenic photosynthesis. They share sequence homology with a subunit encoded by the bacterial phage shock operon (PspA) but differ from PspA by a C-terminal extension of about 30 amino acids. In two cyanobacteria, Synechocystis and Anabaena, both a VIPP1 and a pspA gene are present, and phylogenetic analysis indicates that VIPP1 originated from a gene duplication of the latter and thereafter acquired its new function. It also appears that the C-terminal extension that discriminates VIPP1 proteins from PspA is important for its function in thylakoid formation.
Oxygenic photosynthesis is a feature specific to cyanobacteria and chloroplasts that developed several billion years ago in an ancestor of today's cyanobacteria. An endosymbiotic event by which a cyanobacterium was engulfed by an early eukaryote and subsequently transformed into a cell organelle transferred this capacity to plants (1). During this process many of the genes encoded by the cyanobacterial genome were transferred to the nucleus of the host cell or were lost completely (2). Many of the features of the cyanobacterium vanished. Other features (e.g., the photosynthetic machinery) remained, and homologues to many proteins involved in chloroplast biogenesis and function are also found in cyanobacteria (2, 3). The photosynthetic machinery is located in a special internal membrane system, the thylakoids, and the ability to build up and alter this membrane seems to be an important feature of oxygenic photosynthesis.
Chloroplasts develop from proplastids. It is assumed that the thylakoid membranes that are formed during the maturation process are derived from the inner envelope (4, 5). A continuation between the inner envelope and the thylakoids is discussed for the early stages of this development. No connection between these membranes can be found in later states of maturation, and thylakoids seem to be maintained by a flux of inner membrane vesicles. Thylakoids consist of a complex network of protein complexes, pigments, and other accessory components built into a membrane support. In mature chloroplasts they are continuously altered for adaptation to different environmental conditions, e.g., light or temperature. Thylakoid proteins encoded by the chloroplast are synthesized on stromal ribosomes and are co- or posttranslationally inserted into the membrane. Nuclear encoded plastidal proteins are imported into the chloroplast and inserted into thylakoid membranes by at least four different pathways (6). Whether the typical protein targeting pathways into thylakoids cooperate or act independently of the vesicular system is not known (4, 7, 8). The interdependence of the different transport systems might also be influenced by chloroplast differentiation. Despite the importance of the thylakoid membrane for oxygenic photosynthesis, a lot of questions about the processes of thylakoid formation and maintenance remain. Even less is known about how this membrane originated in the first place. No internal membrane systems are described for eukaryotic organelles other than the chloroplast. The photosynthetic machinery of purple bacteria that carry out anoxygenic photosynthesis is often located in intracytoplasmic membranes, but it remains unclear whether these are a separate entity like thylakoids or a continuum (i.e., invagination) of the plasma membrane (9, 10). It is therefore tempting to speculate that the genesis of the thylakoid membrane is directly connected to the development of oxygenic photosynthesis.
Vesicle-inducing protein in plastids 1 (VIPP1, IM30 protein of pea) was first identified in pea, where it is located in both the envelope membrane and the thylakoids (11) (GenBank accession no. M73744). A matching dual location was found in Arabidopsis thaliana (12). Disruption of the VIPP1 gene in Arabidopsis led to a high chlorophyll fluorescence phenotype indicating a reduced photosynthetic capability. Ultrathin sections showed that the mutant does not have a properly developed thylakoid system, but instead very few membrane-like structures are dispersed throughout the stroma. At the same time no vesicle budding from the inner envelope can be observed in this mutant (12).
Genes encoding VIPP1-like proteins can be found in several plants and in cyanobacteria, which is consistent with the observation that homologues to many cyanobacterial genes are found to be involved in chloroplast biogenesis and function (2, 3). Here we show that VIPP1 of Synechocystis, in contrast to higher plants, seems to be located exclusively in the plasma membrane. Disruption of the VIPP1 gene in Synechocystis leads to a phenotype similar to that of Arabidopsis, indicated by the loss of thylakoid formation and the absence of light-dependent oxygen evolution. Homologues to VIPP1 can also be found in several bacteria in the form of the phage shock protein PspA. VIPP1 proteins of plants and cyanobacteria differ from PspA by a C-terminal extension of about 30 amino acids. In two cyanobacteria, Synechocystis and Anabaena, both a VIPP1 and a pspA gene are present, and we propose from phylogenetic analysis that VIPP1 originated from a gene duplication of pspA. It thereafter acquired its new function, for which the C-terminal extension appears to be important.
Materials and Methods
Cell Growth and Transformation.
Cells of Synechocystis sp. strain PCC6803 were grown at 28°C in BG-11 medium (13) under 80 μmol⋅m−2⋅s−1 white light in glass flasks bubbled through from the bottom with 2% CO2 in air. Under photoheterotrophic growth conditions the medium was supplemented with 0.5% glucose. Under photoheterotrophic conditions the mutant cells grew well for about two or three cycles of division, but further growth was significantly slower than under heterotrophic conditions or compared with the wild type. Transformation and selection for mutant and rescue strains were done as previously described (14). In short, cells were grown photoautotrophically to an OD730 of 1.6. Cells were collected by centrifugation, washed with fresh BG-11 medium, and resuspended in BG-11 at 1.8 × 1010 cells per milliliter. After the addition of 10 μg of plasmid DNA, the cells were incubated at 28°C for 2 h before being plated onto BG-11 plates. After 20 h of growth at 28°C in light, increasing amounts of selective antibiotics were added. Cells were further grown on selective plates containing a final concentration of 20 μg/ml kanamycin or 50 μg/ml ampicillin.
Isolation of Cellular Subfractions and Protein Extraction.
For the isolation of cellular subfractions, wild-type Synechocystis cells were grown photoautotrophically at 2% CO2 and 25°C, in a 12 h/12 h light–dark regime (400 μmol⋅m−2⋅s−1). Cells were collected by centrifugation and fractionated as previously described (15). Protein extraction on total membranes was performed as described (16). In short, membranes were collected by centrifugation and resuspended in 1 M NaCl, 0.1 M Na2CO3 (pH 11), or 8 M urea. After incubation on ice for 10 min (30 min for urea), soluble and insoluble proteins were separated by centrifugation.
Sequence Analyses.
Amino acid sequences for several plant, cyanobacterial, and bacterial VIPP1 and PspA proteins were obtained from the nonredundant protein database of the National Center for Biotechnology Information. In short: Aquifex aeolicus (AE000736) (PspA-Aa), Arabidopsis thaliana (AAC27134) (VIPP1-At), Bacillus subtilis (AB007638) (PspA-Bs), Deinococcus radiodurans (AE001992) (PspA-Dr), Escherichia coli (AE000228) (PspA-Ec), Pisum sativum (M73744) (VIPP1-Ps), and Synechocystis sp. strain PCC6803 (sll0617) (VIPP1-Ss) (slr1188) (PspA-Ss). The Anabaena sp. PCC7120 sequences were obtained by blast searches against the unfinished genome at the Kazusa DNA Research Institute (http://www.kazusa.org) and were named by their contig number and order anac361a (pspA-As) and anac361b (VIPP1-As), respectively. VIPP1 sequences from Nostoc punctiforme and the marine Synechococcus sp. WH8102 were obtained from the Department of Energy Joint Genome Institut by blast searches against their unfinished genomes (http://www.jgi.doe.gov/JGI_/home.html). The sequence of the Yersinia pestis PspA protein was obtained by a blast search against the Yersinia genome at the Sanger Centre. (These sequence data were produced by the Y. pestis Sequencing Group at the Sanger Centre and can be obtained from http://www.sanger.ac.uk/Projects/Y_pestis/blast_server.shtml.) Sequence alignments were performed with the use of the multiple sequence alignment tool from the BCM Search Launcher. After sequence alignment with clustal w (17), a phylogenetic analysis was performed with the puzzle program (18) and the protml program in the molphy package (19).
Southern Blot Analysis.
Genomic DNA from wild-type and Δsynvipp1 cells was isolated from Synechocystis as described (20). DNA was digested with KpnI, separated on a 1% agarose gel, and blotted onto nylon Hybond N+ membrane (Amersham Pharmacia), following the manufacturer's instructions. A digoxigenin (DIG)-labeled probe was generated by PCR on genomic DNA with the DIG High Prime Kit (Roche Molecular Biochemicals) and the following primers: 5′-GCAAAAACATATCACGGATGCTGATAAAAG-3′ and 5′-GCCAAATTTTCTTCCCCATTGGTGAGGG-3′. Prehybridization and hybridization of the membrane were conducted at 65°C, following standard procedure (21). DIG-labeled DNA was detected with an alkaline phosphatase-conjugated anti-DIG antibody.
Western Blot Analysis.
For immunoblot analysis proteins were separated by gel electrophoresis on SDS/12.5% polyacrylamide gels (22) and blotted onto nitrocellulose membrane (Schleicher & Schüll) with the use of standard protocols. Immunoreactions were visualized with the use of secondary antibodies coupled to alkaline phosphatase. Antibodies against Synechocystis 75-kDa protein (α-syn-Toc75), pea large subunit of Rubisco (ribulose-bisphosphate carboxylase) (α-RbcL), spinach ATP synthase (α-AtpA/B), and Synechocystis nitrate reductase subunit A (α-NrtA) were used as markers for the purity of different subfractions of Synechocystis. The α-VIPP1 and α-pspA antisera were raised in a rabbit or chicken, respectively, after immunization with pea VIPP1 protein or Synechocystis PspA protein heterologously expressed and purified from E. coli.
Transmission Electron Microscopy.
Synechocystis cells were incubated for 2 h at 4°C in 0.05 M phosphate buffer (pH 7.2) followed by postfixation with 4% OsO4 containing 2% paraformaldehyde and 2.5% glutaraldehyde. After fixation the samples were dehydrated in a graded ethanol series and embedded in resin with a low-viscosity embedding kit (Ted Pella, Redding, CA), following the manufacturer's instructions. Ultrathin sections were cut with a Reichert microtome, put on single-hole grids, covered with a Formvar film, and stained with 2% aqueous uranyl acetate for 10 min, followed by lead citrate for 5 min. The samples were analyzed and photographed with a Philips CM10 electron microscope.
Oxygen Measurement.
Photosynthesis and respiration rates were measured with a YSI Clark-type electrode (23) at 28°C, with a cell density of 5 μg of chlorophyll per ml. Respiration was followed after a 5-min incubation in the dark; photosynthesis was measured at least three times for 10 min at 1400 μmol⋅m−2⋅s−1.
Results and Discussion
Phylogenetic Analysis of VIPP1.
VIPP1 (IM30 protein of pea) was first described in pea (11) (GenBank accession no. M73744) as a protein located in both the envelope and the thylakoids. Homologues to VIPP1 from pea can be found in plants, but also in several bacteria in form of the phage shock protein PspA. PspA was originally characterized in E. coli, where it is a peripherally bound inner membrane protein. It is expressed as part of the five-gene-containing psp operon (24). Expression of pspA is strongly induced upon infection of E. coli cells with the filamentous phage f1 or severe stresses such as heat, ethanol, or altered osmolarity (25). More intriguingly, pspA is also induced by conditions that block protein export via the general Sec protein export pathway (26), and under these conditions the protein seems to be involved in the protein translocation process. A homologous Sec pathway is used in chloroplasts and cyanobacteria for the translocation of proteins over and insertion into the thylakoid membrane (6).
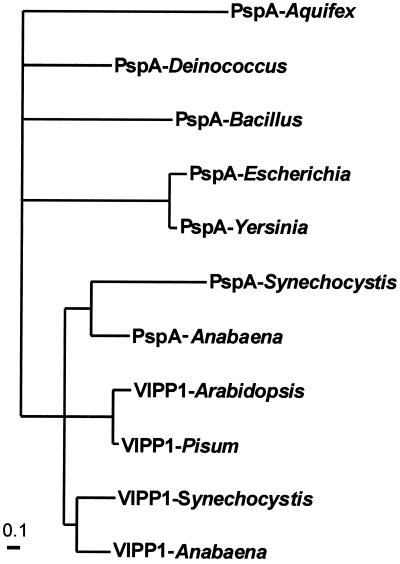
In the genome sequence of the cyanobacterium Synechocystis sp. PCC6803 (3) two open reading frames, sll0617 and slr1188, exist that have high sequence homology to VIPP1 and PspA proteins (Fig. 1). Moreover, two similar proteins, anac361a and anac361b, can be found in the deduced amino acid sequences of the Anabaena sp. PCC7120 genome (Kazusa DNA Research Institute). Two more cyanobacterial proteins with homology to VIPP1 and PsnA were identified in the partially sequenced genomes of Nostoc punctiforme and Synechococcus sp. strain WH8102 (Department of Energy Joint Genome Institute). Sequence comparison shows that the nuclear encoded plant VIPP1 proteins possess the N-terminal extension that is required as a transit sequence for import into the chloroplast, and this sequence is not present in the cyanobacterial or the bacterial proteins. N-terminal amino acid sequencing of immunoprecipitated VIPP1 protein from pea has confirmed that the mature protein starts at the methionine indicated by an asterisk in Fig. 1. This result provides an indication that the plant gene might have evolved from a bacterial gene and that the N-terminal extension was added once import into the chloroplast became necessary. More importantly, the plant VIPP1 proteins contain a C-terminal extension of about 30 amino acids that is not present in the bacterial PspA proteins. From the cyanobacterial VIPP1/PspA-like proteins only the gene products of sll0617 and anac361b and the proteins from Synechococcus and Nostoc punctiforme carry a similar C-terminal extension. Thus we conclude that these proteins are the cyanobacterial VIPP1 homologues. The gene products of slr1188 and anac361a have a degree of amino acid homology to pspA from nonphotosynthetic bacteria similar to that of the proteins encoded by sll0617 and anac361b. However, they are missing the C-terminal extension found in VIPP1 proteins and might therefore be functionally more closely related to PspA. Phylogenetic analysis reveals that the plant VIPP1 proteins cluster together with both the cyanobacterial VIPP1-like and PspA-like proteins (Fig. 2). The PspA proteins from nonphotosynthetic bacteria are positioned on a separate branch of the tree. Intriguingly, in the genome of Anabaena the gene coding for VIPP1 is positioned adjacent to the pspA gene with a spacing of only 162 bp between the stop codon of the latter and the start codon of the former. These findings give strong support to the suggestion that one evolved from the other by means of a gene duplication. We assume that VIPP1 derived from pspA, and simultaneously with or subsequent to the gene duplication acquired the C-terminal extension that is found in all VIPP1 genes. VIPP1 was then passed from the cyanobacterial endosymbiont to the plant genome. The fact that no pspA gene is found in the genome of Arabidopsis, the sequence of which is now completed, or in any other photosynthetic eukaryote, implies that it was either no longer present in the original endosymbiont or that it later was lost in plants. No homologues to VIPP1 or pspA have been found in either Rhodobacter sphaeroides or Rhodobacter capsulatus, although it must be taken into consideration that the genomes are not yet fully sequenced. Nor was a homologue found in Chlorobium tepidum, the genome of which is completed. In contrast to cyanobacteria and chloroplasts, these organisms perform photosynthesis under anaerobic conditions with the use of photosystem II only. It is conceivable that VIPP1 proteins are present only in organisms conducting oxygenic photosynthesis. The presence of the C-terminal extension in all VIPP1 proteins known so far indicates that this modification is necessary for the newly acquired function.
Figure 1.
Alignment of PspA and VIPP1 protein sequences from bacteria and plants. Amino acid residues that are conserved in at least half of the sequences are boxed in black, and conserved amino acid residue changes are marked by gray boxes. The start of the mature VIPP1 protein from pea after removal of the transit sequence is indicated by an asterisk.
Figure 2.
Phylogeny of bacterial and plant VIPP1 and PspA proteins supports the close relationship between the plant and cyanobacterial proteins. Maximum likelihood analysis of PspA and VIPP1 proteins from various organisms was performed with puzzle (17) and protml (18).
Synechocystis VIPP1 Protein Seems to Be Exclusively Located in the Plasma Membrane.
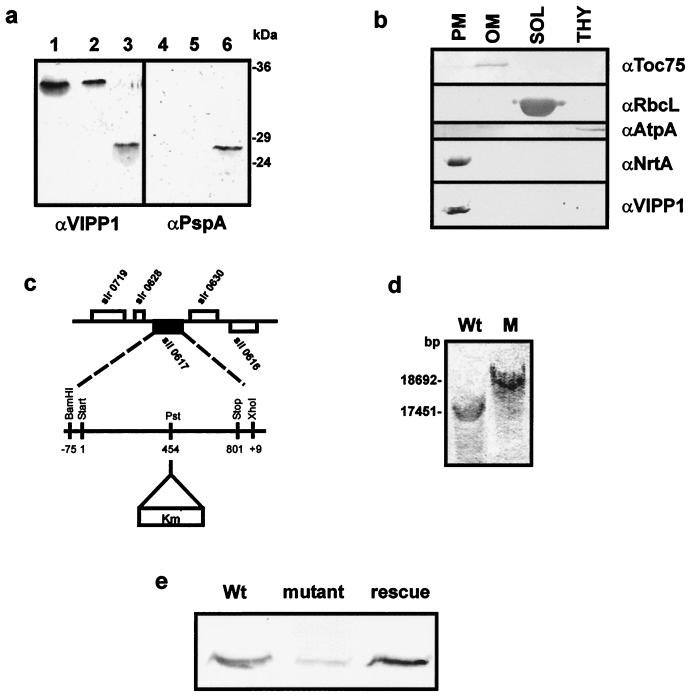
From the phylogenetic analysis it was tempting to speculate that the VIPP1 gene evolved in an ancestral cyanobacterium in concert with the evolution of thylakoids. To investigate the function of the VIPP1 protein, we first examined its intracellular location by immunoblotting. An immunoreactive band of about 34 kDa can be observed in total protein extracts from wild-type Synechocystis when analyzed with an antibody raised against the VIPP1 protein from pea (Fig. 3a, lane 2). The protein migrates in a manner similar to that of the VIPP1 protein from Arabidopsis as shown in total leaf extracts (Fig. 3a, lane 1) and significantly higher than the heterologously expressed full-length PspA (gene product of slr1188) from Synechocystis. To further ensure that the protein recognized by α-VIPP1 is indeed the gene product of sll0617 (VIPP1), we performed a Western blot with an antibody raised against the heterologously expressed PspA from Synechocystis (Fig. 3a, lanes 4–6). The two antibodies recognize PspA equally well (Fig. 3a, lanes 3 and 6). Yet, when equal amounts of protein were used and immunodecorated, no immunoreactive band could be observed in total protein from wild-type Synechocystis or in Arabidopsis leaf extracts with α-PspA (Fig. 3a; compare lanes 1 and 2 to lanes 4 and 5). Thus we conclude that most likely slr1188 is not expressed in wild-type cells under the conditions of our experiment and that the protein recognized by α-VIPP1 is the gene product of sll0617. Wild-type Synechocystis cells were then separated into different cellular fractions (15), and analysis by Western blot revealed an immunoreactive band in the plasma membrane fraction only (Fig. 3b). The localization of VIPP1 was confirmed by immunogold labeling of Synechocystis cells with α-VIPP1 (data not shown). The location in the plasma membrane is unlike the location in both pea and Arabidopsis chloroplasts, where VIPP1 has been observed in the envelope and the thylakoids (11, 12). In pea, the VIPP1 protein was shown to be tightly associated with the envelope membrane, even though the amino acid sequence gave no indication for membrane-spanning domains (11). A hydrophobicity analysis of Synechocystis VIPP1 did not indicate a membrane domain either. To elucidate whether Synechocystis VIPP1 is an integral protein of the plasma membrane we extracted total membranes of wild-type cells with NaCl, Na2CO3 (pH 11), or 8 M urea (16). No extraction of VIPP1 from the membrane is observed by treatment with 1.0 M NaCl, and only a minor fraction was extracted with high pH. Incubation with 8 M urea extracted about 50% of the protein (data not shown). These results confirm that also in Synechocystis VIPP1 is not a membrane-spanning protein, but like the plant protein it is tightly associated with membranes.
Figure 3.
sll0617 encodes a cyanobacterial VIPP1 homologue, and it seems to be located exclusively in the plasma membrane. (a) Immunoblotting with α-VIPP1 and α-PspA antisera identifies VIPP1 in Synechocystis. Lanes 1–3, immunoblot performed with α-VIPP1; lanes 4–6, immunoblot performed with α-PspA; lanes 1 and 4, total leaf extract from A. thaliana; lanes 2 and 5, total protein extract from Synechocystis; lanes 3 and 6, PspA, heterologously expressed in and purified from E. coli. (b) Synechocystis wild-type cells were separated into outer membrane (OM), plasma membrane (PM), cytoplasm (SOL), and thylakoids (THY). Each fraction was analyzed by immunoblotting for the presence of VIPP1. Antisera against the 75-kDa outer membrane protein (α-syn-Toc75), large subunit of Rubisco (α-RbcL), ATP synthase (α-AtpA/B), and nitrate reductase subunit A (α-NrtA) were used as markers for the purity of the different subfractions. (c) sll0617 was disrupted by the insertion of a kanamycin-resistance cassette at position 454 of the coding region. (d) Southern blot analysis was carried out with wild-type and Δsynvipp1 genomic DNA to confirm the disruption of sll0617. (e) Immunoblotting with α-VIPP1 protein establishes an extreme reduction of the level of VIPP1 protein in the Δsynvipp1 mutant compared with wild-type and rescue cells.
VIPP1 Disruption in Synechocystis Leads to Loss of Thylakoid Formation.
To further investigate the function of VIPP1 in cyanobacteria we decided to generate a VIPP1 disruption mutant of Synechocystis (Δsynvipp1) by insertion of a kanamycin-resistance cassette into the alleged VIPP1 gene sll0617 (Fig. 3c). The gene was cloned from Synechocystis genomic DNA including 75 bp of the 5′ noncoding region, a kanamycin cassette was inserted, and the disrupted gene was cloned into pUC18, creating pUC18-Δsynvipp1. Wild-type Synechocystis cells were transformed with this vector, and a Δsynvipp1 mutant strain was obtained by homologous recombination and kanamycin resistance selection. Mutant cells grew well on plates supplemented with glucose, but only very slow growth was detected on minimal plates. To confirm the disruption of VIPP1 in the mutant cells, Southern blot hybridization was performed with the use of part of the 5′ coding region of sll0617 as a probe. Signals of the expected size were observed with mutant and wild-type DNA (Fig. 3d). The strongest signal in Δsynvipp1 DNA reflected the insertion of the kanamycin cassette into sll0617 at the PstI site (Fig. 3 c and d). However, a very weak reaction could also be observed in the position of the undisrupted VIPP1 gene, indicating that segregation was not complete. Despite several efforts, no entire segregation could be achieved, suggesting that loss of VIPP1 protein is lethal to the cells. To investigate the expression of the sll0617 gene product in the mutant we performed Western blot analysis with VIPP1 antiserum on total protein from wild-type and mutant cells. Only a very weak immunoreactive band was seen in the mutant cells compared with the wild type when identical protein amounts were analyzed (data not shown). To ensure that the lack of VIPP1 reaction was not due to an overall change in protein content, we also analyzed protein extracted from an identical amount of cell mass as determined by OD750 and counted cell numbers. Again only minor amounts of VIPP1 protein were detected in the mutant compared with the wild-type level (Fig. 3e). The significant reduction of immunoreactive protein further confirms that sll0617 in fact encodes the Synechocystis VIPP1 protein and that this gene locus is disrupted in the Δsynvipp1 mutant.
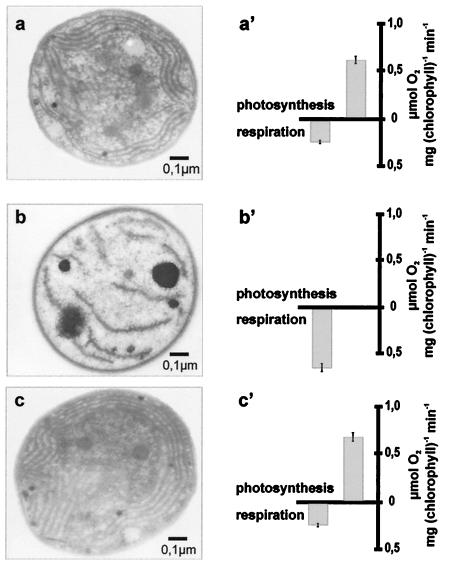
Δsynvipp1 cells were still greenish in appearance, and therefore we asked whether thylakoids were present and whether cells were still able to carry out light-dependent oxygen evolution. Ultrathin sections of wild-type and mutant cells show significant changes in the ultrastructure of the mutant (Fig. 4 a and b). In wild-type Synechocystis the thylakoids are tidily organized in structures paralleling the contour of the surface. In the mutant cells no organized thylakoids can be observed. Instead membrane-like structures seem to be dispersed in low numbers throughout the cytoplasm of the cell. At the same time the number of high-density carboxysomes is increased (27). A very similar phenotype, i.e., a nearly complete loss of thylakoids, had been observed for hcf155, the vipp1 mutant of Arabidopsis thaliana (12). This mutant is characterized by a high chlorophyll fluorescence phenotype, indicating a reduced photosynthetic capability. Thus we measured the photosynthesis rate of Synechocystis wild-type and Δsynvipp1 cells as light-dependent oxygen evolution. Δsynvipp1 cells showed no light-dependent oxygen evolution in comparison with wild-type cells, which produced around 0.6 μmol of O2 per mg of chlorophyll per min. Consequently oxygen consumption by respiratory processes was increased in mutant cells (Fig. 4 a′ and b′). To ensure that the observed phenotype is caused by the reduction of VIPP1 protein, we proceeded to rescue the Δsynvipp1 mutant by plasmidal expression of the wild-type VIPP1 gene. sll0617, including about 450 bp of the 5′ noncoding region, was amplified by PCR from Synechocystis wild-type DNA and cloned into the pCR2.1 vector (Invitrogen). Mutant rescue was achieved by transformation of the Δsynvipp1 mutant strain with this plasmid. Cells transformed with pTOPO-VIPP1 grew well on minimal medium and again displayed well-ordered thylakoid structures when analyzed by electron microscopy (Fig. 4c). Light-dependent oxygen evolution rates were similar to those of wild-type cells, indicating that the thylakoid membranes were able to support photosynthesis (Fig. 4c′). Furthermore, Western blot analysis of total protein from rescued cells revealed wild-type levels of VIPP1 protein (Fig. 3e). These results indicate that the VIPP1 protein in cyanobacteria as well as in plants is essential for thylakoid formation.
Figure 4.
Ultrathin sections reveal that Δsynvipp1 mutant cells contain no orderly structured thylakoids but instead have very few membrane-like structures dispersed throughout the cytoplasm. They are unable to carry out photosynthesis. The phenotype can be rescued by plasmidal expression of VIPP1. (a–c) Ultrathin sections of wild-type, Δsynvipp1 mutant, and rescue cells. (a′–c′) Analysis of oxygen evolution and consumption in wild-type, Δsynvipp1, and rescue cells.
Conclusions
Oxygenic photosynthesis developed 2–3 billion years ago in prokaryotic organisms, where it is still prominent today. These organisms also achieved the capacity to build up specialized membranes, i.e., the thylakoids, where the oxygenic photosynthesis machinery is localized. However, very little is known about how this membrane system developed and which ancestral components were used to facilitate the initial process in the rise of a new membrane system. VIPP1 is a protein required in plants and cyanobacteria for the formation of thylakoids. Phylogenetic evidence points toward the evolution of VIPP1 from an ancient bacterial protein involved in phage shock response, PspA. The proteins encoded by the bacterial phage shock operon and especially PspA seem to be involved in stabilizing and maintaining membrane integrity upon phage invasion and osmotic stress (25, 26, 28). Although the exact mechanism of PspA function is not well understood, it is tempting to propose that the role of PspA in sustaining membrane function was exploited to generate a protein that became essential for the formation of a new internal membrane system, the thylakoids. This event, however, seemed to require changes in the functional capacity of the PspA protein. These changes were most likely achieved by a gene duplication of the pspA gene and simultaneous or subsequent addition of a novel C-terminal domain resulting in an ancestral VIPP1 gene. Today, VIPP1 is essential for thylakoid formation in both cyanobacteria and eukaryotic chloroplasts. The exact function of VIPP1 in thylakoid genesis has yet to be elucidated.
Acknowledgments
We thank Prof. Dr. W. Martin for his help with the phylogenetic analysis. This work was supported by grants for J.S. from the Deutsche Forschungsgemeinschaft (SFB TR1), the Human Frontier Science Program, and the Fonds der Chemischen Industrie.
Abbreviations
- VIPP1
vesicle-inducing protein in plastids 1
- DIG
digoxigenin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 3633.
References
- 1.Margulis L. Origin of Eukaryotic Cells. New Haven, CT: Yale Univ. Press; 1970. [Google Scholar]
- 2.Martin W, Herrmann R G. Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;30:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 4.Carde J P, Joyard J, Douce R. Biol Cell. 1982;44:315–324. [Google Scholar]
- 5.Douce R, Joyard R. In: Chloroplast Biogenesis. Baker N R, Barber J, editors. Amsterdam: Elsevier Science; 1984. pp. 71–132. [Google Scholar]
- 6.Robinson C, Woolhead C, Edwards W. J Exp Bot. 2000;51:369–374. doi: 10.1093/jexbot/51.suppl_1.369. [DOI] [PubMed] [Google Scholar]
- 7.Morré D J, Selldén G, Sundqvist C, Sandelius A S. Plant Physiol. 1991;97:1558–1564. doi: 10.1104/pp.97.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoober J K, Boyd C O, Paavola L G. Plant Physiol. 1991;96:1321–1328. doi: 10.1104/pp.96.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dierstein R, Schumacher A, Drews G. Arch Microbiol. 1981;128:376–383. [Google Scholar]
- 10.Drews G, Golecki J R. In: Advances in Photosynthesis. Blankenship R E, Madigan M T, Bauer C E, editors. Vol. 2. Dordrecht, The Netherlands: Kluwer; 1995. pp. 231–257. [Google Scholar]
- 11.Li H-M, Kaneko Y, Keegstra K. Plant Mol Biol. 1994;25:619–632. doi: 10.1007/BF00029601. [DOI] [PubMed] [Google Scholar]
- 12.Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht U C, Soll J, Westhoff P. Proc Natl Acad Sci USA. 2001;98:4238–4242. doi: 10.1073/pnas.061500998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rippka R, Keruelles J, Waterbury J B, Herdman M, Stanier R Y. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 14.Chauvat F, De Vries L, Van der Ende A, Van Arkel G A. Mol Gen Genet. 1986;204:185–191. [Google Scholar]
- 15.Bölter B, Soll J, Schulz A, Hinnah S, Wagner R. Proc Natl Acad Sci USA. 1998;95:15831–15836. doi: 10.1073/pnas.95.26.15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiki Y, Hubbard A L, Fowler S, Lazarow P B. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson J D, Higgins T J, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 19.Adachi J, Hasegawa M. Computer Science Monographs. 1996. No. 28 (Institute of Statistical Mathematics, Tokyo). [Google Scholar]
- 20.Ohad N, Hirschberg J. Plant Cell. 1992;4:273–282. doi: 10.1105/tpc.4.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Fork D C. Methods Enzymol. 1980;69:113–122. doi: 10.1016/0076-6879(72)24061-7. [DOI] [PubMed] [Google Scholar]
- 24.Brisette J L, Weiner L, Ripmaster T L, Model P. J Mol Biol. 1991;220:35–48. doi: 10.1016/0022-2836(91)90379-k. [DOI] [PubMed] [Google Scholar]
- 25.Brissette J L, Russel M, Weiner L, Model P. Proc Natl Acad Sci USA. 1990;87:862–866. doi: 10.1073/pnas.87.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleerebezem M, Tommassen J. Mol Microbiol. 1993;7:947–956. doi: 10.1111/j.1365-2958.1993.tb01186.x. [DOI] [PubMed] [Google Scholar]
- 27.Codd G A. In: Advances in Microbial Physiology. Ross A H, Tempest D W, editors. Vol. 29. London: Academic; 1988. pp. 115–164. [DOI] [PubMed] [Google Scholar]
- 28.Kleerebezem M, Crielaard W, Tommassen J. EMBO J. 1996;15:162–171. [PMC free article] [PubMed] [Google Scholar]