Abstract
Purpose of review
To review the recently discovered genetic risk loci in rheumatoid arthritis (RA), the pathways they implicate, and the genetic architecture of RA.
Recent findings
Since 2008 investigators have identified many common genetic variants that confer disease risk through single nucleotide polymorphism genotyping studies; the list of variants will no doubt continue to expand at a rapid rate as genotyping technologies evolve and case–control sample collections continue to grow. In aggregate, these variants implicate pathways leading to NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) activation, the interluekin-2 signaling pathway, and T-cell activation.
Summary
Although the effect of any individual variant is modest and even in aggregate considerably less than that of the major histocompatability complex, discovery of recent risk variants suggests immunological processes that are involved in disease pathogenesis.
Keywords: association study, genetics, genomics, rheumatoid arthritis
Introduction
Rheumatoid arthritis (RA) is a complex disease involving many different pathways within the innate and the adaptive immune system [1]; in addition to an inflammatory polyarthritis that predominantly affects the hands, RA can affect multiple other tissues, including the lungs [2]. The possibility that RA might have a genetic component was considered as long ago as 1806 by William Heberden in his book Commentaries on the History and Cure of Diseases [3], in which he posed the question in his writings ‘Is [rheumatism] not in some degree hereditary?’. Today, it is well established that there are key genetic risk factors, such as the shared epitope human leukocyte antigen (HLA) alleles [4,5] and environmental risk factors, such as smoking [6]. Within the past 2 years, genetic studies have produced a rapidly growing list of risk loci for RA and other autoimmune diseases, and investigators are struggling to understand how these loci augment our knowledge about human disease and identify individuals at risk. Currently, known risk alleles, however, account for only a fraction of the heritability of RA, and as genotyping technology evolves and case–control collections enlarge, the number of known risk variants will undoubtedly continue to expand.
Mouse models of arthritis have helped to elucidate the key genetic factors and highlight potential therapeutic interventions [7–10], but do not always correlate perfectly with human disease. Genetic studies can help us identify the key genes and pathways that are involved at the inception of human disease. As genetic variants are present at birth and precede disease, the convincing association of a specific variant with disease demonstrates that either the variant or a highly correlated nearby variant plays a key role in disease pathogenesis. Consequently, pathways that these genes are involved in may be targeted effectively by therapeutics in early disease and furthermore are potentially promising for curative therapy.
Main text
In the present review, I seek to review recent advances in RA genetics. First, I will review the available data demonstrating that RA is a heritable disease. Second, I will review the RA risk loci identified so far and the strategies used to discover them. Third, I will talk about some of the biological insights that might be gleaned from the current list of RA risk loci. Finally, I will discuss the application of these loci to predicting RA risk.
Rheumatoid arthritis is a genetic disease
Overwhelming evidence from familial studies has long implicated genetic factors in RA. Most notably, familial studies have demonstrated an increased prevalence of disease in first-degree relatives. Twin studies have shown an increased rate of disease in monozygous (identical) versus dizygotic twins.
First-degree relatives
One common approach to assess and quantify the possible role of genetic factors is to measure the concordance of disease between related individuals. A highly heritable disease will have an increased prevalence in individuals related to affected probands compared with the general population. For example, a proband with a rare and highly penetrant monogenic dominant disease will have 50% of their siblings affected by a rare disease compared with a less than 1% population prevalence. The increased incidence of RA among those individuals related to probands with disease is a key observation that suggests the heritability of disease. Of course, this observation could be explained by shared environmental factors as well; for example, family members living together close to heavy traffic might all be exposed to the same risk [11]. A key descriptive parameter is λR – the ratio of the prevalence of a disease among first-degree relatives (parents, children, and siblings) compared with the general population. For highly penetrant monogenic diseases, λR can be very high; for example, λR is approximately 500 for cystic fibrosis. Complex diseases have more modest λR values; for example, it is approximately 15 for type I diabetes.
RA is commonly cited as having an λR as 2–17 [12], though more careful estimates are often more modest. The earliest evidence of familial segregation was made in 1950 by the Empire Rheumatism Council [13]. They noted that fathers, mothers, and sibling relatives of affected individuals were each about twice as likely as age-matched and sex-matched controls. This finding was reproduced in many additional studies in the same time-frame as reviewed by Lawrence [14]. Several large studies that have carefully ascertained controls to match or adjust for age and sex have shown that the relative risk of RA for first-degree relatives is also about 2–4 [15–17]. Jones et al. [16] looked at 134 RA probands from the Norfolk Arthritis Registry compared with age-matched and sex-matched controls. When they compared the prevalence of RA among 418 first-degree proband relatives compared with control relatives, they noted a 2.2-increased relative risk; however, this increase was not statistically significant. Del Junco et al. [15] examined 1631 first-degree relatives of 78 RA probands in Rochester, Minnesota and noted a significant increased rate of 1.7 compared with age-adjusted and sex-adjusted local rates. A recent large study using the Multigeneration Register in Sweden used diagnostic codes from hospital discharges to demonstrate an increased relative rate of disease in siblings of affected individuals to be 4.6 and in children to be 3.0 [17]. These values are likely dependent on the severity of the disease, serologic status of patients, age, and sex of the proband [14,18]. Additionally, hospital-based cohorts seem to suggest greater heritability than community-based ones.
Twin studies
Twin studies offer a powerful strategy to estimate the genetic aspects of disease. In the past, twin studies have been used to estimate the heritability of disease. Specifically, an increased rate of disease among monozygotic twins compared with dizygotic twins offers evidence that inherited factors predispose to disease risk, potentially above and beyond common environmental factors. To date, the most compelling analysis of twin studies suggests RA is approximately 65% heritable [19] based on data from a Finnish [20] and British cohort [21]. These estimates are based on a total of only 23 concordant monozygotic twins and 10 concordant dizygotic twins, and estimates of heritability are, therefore, potentially sensitive to modest biases. For example, a smaller Danish RA twin study demonstrated no heritability for RA whatsoever [22]. On the other hand, previous studies to estimate twin concordance had demonstrated higher degrees of heritability [14]. Possibly, diagnostic criteria and study designs have changed over time, and these more recent concordance rates are more modest.
In a recent extension of the British twin study, investigators genotyped twins for shared epitope alleles and for anticyclic citrullinated peptide (anti-CCP) antibody [23••]. They found that both seropositive and seronegative RAs were equally heritable with approximately 65% heritability for both diseases as previously noted. But, shared epitope alleles account for only 2.4% of the heritability of seronegative RA, but 18% of seropositive RA.
Expanding list of known rheumatoid arthritis risk variants with modern genetic approaches
Over the last year, a rapidly growing list of RA risk loci has emerged in the scientific literature as case–control sample collections grow and genotyping technologies develop (see Table 1, Fig. 1) [4,24,25••,26,27,28••,29••, 30,31,32••,33,34,35••,36••,37,38,39••,40,41]. Different genetic strategies have been effectively applied to identify risk loci in RA. These strategies can be broadly divided into unbiased genome-wide investigations and targeted investigations to examine specific alleles or genes for which there is increased prior probability of association based on previous functional or genetic information.
Table 1.
Known rheumatoid arthritis single nucleotide polymorphism associations
| SNP | Locus | Candidate gene | OR | Allele frequency | References |
|---|---|---|---|---|---|
| rs3890745 | 1p36.2 | TNFSF14 | 0.920 | 0.320 | Raychaudhuri 2008 [39••] |
| rs2240340a | 1p36.13 | PADI4 | 1.40 | 0.373 | Suzuki 2003 [40] |
| rs2476601 | 1p13.2 | PTPN22 | 1.750 | 0.100 | Begovich 2004 [33] |
| rs11586238 | 1p13.1 | CD2, IGSF2, CD58 | 1.120 | 0.227 | Raychaudhuri 2009 [36••] |
| rs7528684a | 1q23.1 | FCLR3 | 1.2 | 0.35 | Kochi 2005 [41] |
| rs12746613 | 1q23.2 | FCGR2A | 1.100 | 0.124 | Raychaudhuri 2009 [36••] |
| rs3766379a | 1q23.3 | CD244 | 1.31 | 0.53 | Suzuki 2008 [35••] |
| rs10919563 | 1q31.3 | PTPRC | 0.900 | 0.132 | Raychaudhuri 2009 [36••] |
| rs13031237 | 2p16.1 | REL | 1.207 | 0.340 | Gregersen 2009 [28••] |
| rs10865035 | 2q11.2 | AFF3 | 1.140 | 0.460 | Barton 2009 [32••] |
| rs7574865 | 2q32.3 | STAT4 | 1.320 | 0.180 | Remmers 2007 [34] |
| rs1980422 | 2q33.2 | CD28 | 1.100 | 0.238 | Raychaudhuri 2009 [36••] |
| rs3087243 | 2q33.2 | CTLA4 | 1.136 | 0.560 | Plenge 2005 [30] |
| rs6822844 | 4q27 | IL2/IL21 | 1.389 | 0.710 | Zhernakova 2007 [31] |
| rs2395175 (and others) | 6p21.32 | MHC | 1.000 | 0.021 | Gregersen 1987 [4] |
| rs548234 | 6q21 | PRDM1 | 1.100 | 0.322 | Raychaudhuri 2009 [36••] |
| rs10499194 | 6q23.3 | TNFAIP3 (a) | 1.220 | 0.220 | Plenge 2007 [24] |
| rs6920220 | 6q23.3 | TNFAIP3 (b) | 1.333 | 0.610 | Thomson 2007 [37] |
| rs5029937 | 6q23.3 | TNFAIP3 (c) | 1.34 | 0.04 | Orozco 2009 [25••] |
| rs394581 | 6q25.3 | TAGAP | 0.930 | 0.286 | Raychaudhuri 2009 [36••] |
| rs2736340 | 8p23.1 | BLK | 1.122 | 0.243 | Gregersen 2009 [28••] |
| rs2812378 | 9p13.3 | CCL21 | 1.100 | 0.355 | Raychaudhuri 2008 [39••] |
| rs3761847 | 9q33.1 | TRAF1 | 1.100 | 0.440 | Plenge 2007 [26]; Kurreeman 2007 [27] |
| rs2104286 | 10p15.1 | IL2RA | 0.92 | 0.28 | Thomson 2007 [37]; Kurreeman 2009 [38] |
| rs4750316 | 10p15.1 | PRKCQ | 0.910 | 0.183 | Raychaudhuri 2008 [39••]; Barton 2008 [29••] |
| rs540386 | 11p12 | RAG1, TRAF6 | 0.920 | 0.144 | Raychaudhuri 2009 [36••] |
| rs1678542 | 12q13.3 | KIF5A | 0.890 | 0.351 | Barton 2008 [29••]; Raychaudhuri 2008 [39••] |
| rs4810485 | 20q13.12 | CD40 | 0.910 | 0.231 | Raychaudhuri 2008 [39••] |
| rs3218253 | 22q12.3 | IL2RB | 1.110 | 0.730 | Barton 2008 [29••] |
Table represents known rheumatoid arthritis risk variants. In the three columns, I list the representative SNP, the cytogenetic location, and the likely candidate gene. In the next two columns, I list the odds ratios and allele frequency in control populations. I list appropriate references in the final column. OR, odds ratio; RA, rheumatoid arthritis; SNP, single nucleotide polymorphism.
Risk allele associated in east Asian populations; remaining loci associated in European populations.
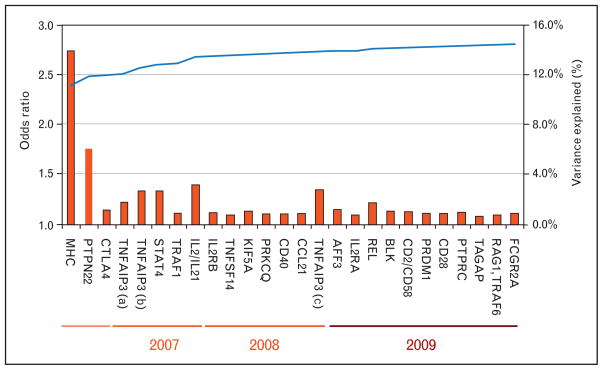
Figure 1. Variance explained by rheumatoid arthritis genetic associations.
I list each known rheumatoid arthritis (RA) risk variant along the bottom in the approximate order of their discovery from left to right. Each SNP is plotted along the Y-axis (left) with red bars the odds ratio of the risk allele. As time has progressed, case–control sample collections have increased in size and the ability to discover modest effect sizes has increased. On the Y-axis (right), I plot the increase in the percentage variance explained. A total of approximately 60% of the variance for RA liability is thought to be genetic. I have listed only variants shown to confer risk in European populations.
The single nucleotide polymorphism (SNP) has become the standard genetic marker to identify associated alleles [42–44]. Most confirmed SNP associations in RA are common SNPs – those SNPs in which the rarer nucleotide form is present in more than 5% of the population of chromosomes. Although these SNPs are associated with disease, in most cases the altered nucleotides do not necessarily cause disease themselves. But, they are likely to be correlated with common mutations within the region that affect a functional change in nearby genes and thereby cause disease. Methods to directly look for rare variation, such as de-novo mutations and rare structural variants, are ongoing and as of yet have not yielded confirmed RA risk variants, but have been successfully applied to other phenotypes.
Application of genome-wide scans
High throughput SNP genotyping technologies [45,46] have allowed investigators to genotype common variation across 100 000–1 000 000 SNPs efficiently. These SNPs typically capture up to 65–70% of common variation across the human genome [47,48] and can be used to do an unbiased investigation for novel risk loci for a phenotype of interest. Genome-wide association studies (GWASs) combined this technology with large case–control collections to allow investigators to rapidly identify associated alleles. Additionally, genome-wide genotyping offers investigators additional analytical advantages such as checking for duplicated or related individuals, imputation across the genome to determine genotype of ungenotyped SNPs, and application of strategies to check and correct for case–control stratification.
One key caveat for such studies is that as they assess variation across a large number of loci, very high levels of significance are necessary to convincingly rule out the possibility that an association is a false positive. Today, investigators typically require risk alleles to demonstrate significant association at about P value less than 5 × 10−8 before they accept it as truly associated. To achieve this level of stringency, effective genome-wide studies have required large-scale international collaborations between investigators worldwide.
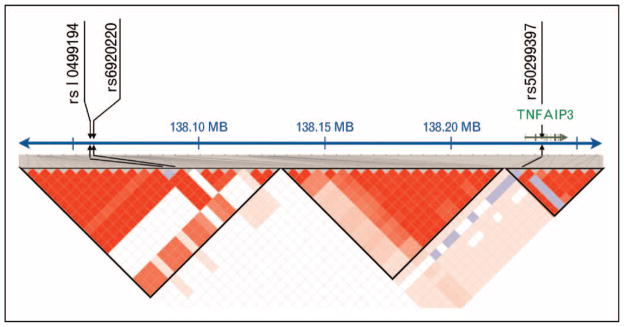
One of the first effective applications of the GWAS study in RA was to discover the association at TNFAIP3. Plenge et al. [24] identified an associated SNP (rs10499194) in that region using a collection of Boston-based cases and controls. A separate and independent TNFAIP3-associated SNP (rs6920220) was identified by Thomson et al. [37] in a follow-up study of the Wellcome Trust Case Control Consortium (WTCCC) RA GWAS study [49]. Subsequent investigation by the Orozco et al. [25••] identified a third independent SNP association in the same region in a fine mapping study. The TNFAIP3 locus highlights the potential complexity that can be present at a given locus involving multiple alleles marked by SNPs with independent effects (see Fig. 2). Other discoveries by the GWAS approach include an association at TRAF1 [26,27], REL, and BLK [28••].
Figure 2. TNFAIP3 locus.
Currently, there are three known associated single nucleotide polymorphism (SNP) loci in the TNFAIP3 locus. As indicated here, there are two risk SNPs far upstream of the gene in linkage disequilibrium with each other, but conferring independent risk of disease. A third risk SNP is located within the second intron of the gene and is not in linkage disequilibrium with the other two risk SNPs.
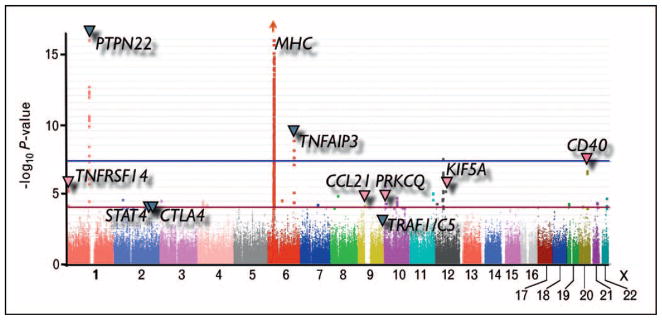
Another powerful aspect of genome-wide SNP data is that it can be used to conduct genome-wide meta-analyses across multiple studies, even if the same SNPs have not been genotyped in all studies. Powerful statistical techniques can be used to estimate or ‘impute’ missing genotypes prior to conducting meta-analyses [50,51•]. We conducted the most recent large-scale meta-analysis of three GWA scans [39••], including scans from the Epidemiological Investigation in RA (EIRA) consortium, the WTCCC, and The North American RA Consortium (NARAC; see Fig. 3). As the different studies used different genotyping platforms, we used imputation strategies to estimate SNP genotypes that were missing in individual studies. That study in conjunction with follow-up genotyping in independent cases and controls identified evidence of association at six separate loci. Of those, five associations at CCL21, KIF5A, CD40, PRKCQ, and TNFRSF14 have been independently described [29••,52•]. Imputation and meta-analysis techniques have become a standard tool to combine results of multiple GWA studies and have now been widely applied to many phenotypes, including Crohn’s disease [53], height [54], and diabetes [55].
Figure 3. A meta-analysis of three genome-wide association studies.
Here I present data from a meta-analysis of three separate genome-wide data sets. Each point represents a single nucleotide polymorphism (SNP). Along the X-axis, I plot the genomic location of an individual SNP. The height of each point indicates the strength of association for that SNP. The dark blue arrows indicate some known associations with rheumatoid arthritis (RA) at the time of this study. The pink arrows indicate SNPs that demonstrated nominal evidence of association in this study (P <0.0001) that were subsequently replicated in an independent collection of cases and controls.
Application of candidate single nucleotide polymorphism studies
One effective approach to discovering risk alleles in RA has been candidate SNP approach. In this approach, investigators have selected specific SNPs to investigate in RA, based on their known function, association with other autoimmune disease, or prior nominal evidence of association in linkage or case–control data.
Known risk alleles in type I diabetes have served as valuable candidates for investigation in RA. Investigators have tested multiple type I diabetes risk alleles for association with RA. Some of theses alleles have now been convincingly validated in RA, including CTLA4 [30], 4q27 [31], and AFF3 [32••]. Similarly, Zhernakova et al. [56] demonstrated preliminary evidence of association at SH2B3 by genotyping a known risk allele in celiac disease in RA samples [57]. These alleles highlight a very important theme in autoimmunity, which is the common SNP loci which seem to predispose to autoimmune diseases in general [56]. This theme was apparent even with the earliest discovery of a risk variant outside the major histocompatability complex (MHC) region in the PTPN22 gene with its association with RA, type I diabetes, lupus, and other autoimmune diseases [33,58].
At least two known RA associations have been driven by following up on regions with nominal evidence of association. Remmers et al. [34] identified a promising candidate gene in a known RA linkage peak – subsequent follow-up genotyping in a large set of independent cases and controls demonstrated association at the STAT4 locus. More recently, Suzuki et al. [35••] identified nominal evidence of association in a linkage disequilibrium block containing multiple signaling lymphocyte activation molecule (or SLAM) genes and mapped the signal to the rs3766379 SNP in Japanese patients. This SNP turned out to be a functional SNP that affected CD244 gene expression.
Pathway-based approaches can also be used to discover novel variants. The central assumption to these approaches is that disease-associated SNPs should implicate a common pathway. We described one novel approach that leverages both genome-wide data and information about gene function. In order to identify whether SNP associations implicated a set of genes that are functionally interrelated, we devised the GRAIL (gene relationships across implicated loci) algorithm [59•]. GRAIL applies computational text mining to PubMed abstracts to assess whether genes across multiple loci are functionally connected, and therefore, are likely to represent a common pathway. We used this approach to help identify novel RA risk loci. Based on the aforementioned meta-analysis, we identified 22 SNPs with nominal evidence of association (P <0.001), which were also near genes that were connected to genes already associated with RA. We genotyped these SNPs in independent samples and identified seven that replicated including associations at CD2/CD58, CD28, PRDM1, TAGAP, PTPRC, TRAF6/RAG1, and FCGR2A [36••].
Future challenges for rheumatoid arthritis genetics
As highlighted above, recent progress has identified a large number of RA risk alleles. Growing international collaboration, improving genotyping technologies, and enlarging patient sample collections will further enhance these discoveries. However, specific challenges remain to clearly understand how these variants cause disease and how these discoveries can be used to enhance patient care.
The major histocompatability complex
The largest contributor of the genetic variation has been attributed to HLA-DRB1 risk alleles in the MHC region discovered in the 1970s [5] and subsequently organized into the shared epitope alleles by Gregersen et al. [4]. Estimates of the contribution of the shared epitope alleles to the total genetic variability of RA have ranged from 18 to 37% [23••,60]. But, outside the shared epitope alleles within the HLA-DRB1 gene locus, additional risk alleles may exist within the MHC, but these alleles remain to be pinpointed precisely [61••,62••]. The MHC region is a highly complex region with extended linkage disequilibrium and complex structural features that reduce the effectiveness of standard SNP-based genotyping strategies and analytical approaches. In particular, converting SNP genotypes into HLA alleles is difficult. Although SNP GWA data are available for a large number of samples worldwide, in most cases HLA genotyping is expensive and unavailable. In the coming years, one of the challenges will be to use genotyped SNPs in GWAS to estimate HLA alleles and to then identify additional risk alleles within the MHC itself. Recent advances to use a panel of approximately 100 SNP genotypes across the MHC to estimate HLA genotypes could have a tremendous impact in this area and help investigators to clarify the genetics of the MHC and its impact on RA [63•,64].
Structural variants
Recently, new array-based and sequencing technologies have allowed investigators to examine the genome for structural variants [65–67], such as regions that are deleted and duplicated. Case–control studies have demonstrated association of structural variants with multiple diseases, including autoimmune diseases. In one striking example, McCarroll et al. [68] examined a strongly associated SNP to Crohn’s disease and recognized that it correlated perfectly with the presence of a deletion in the promoter region of the IRGM gene and therefore affected gene expression. Another compelling example is the β-defensin gene cluster; duplications of β-defensin have been shown to be associated with greater risk of psoriasis [69]. Rare variants can also now be detected with current technologies, and the role of rare or single-event deletions and duplications has been demonstrated in neuoropsychiatric diseases [70,71]. No convincing examples of common or rare structural variants conferring disease risk have been recognized for RA yet. However, there is mounting evidence in the literature that a microdeletion in the CCR5 gene, the CCR5Δ32 polymorphism, protects individuals from disease; this variant has been demonstrated to play a role in HIV disease progression [72]. There has also been published evidence suggesting that duplications of the CCL3L1 gene may increase RA risk [73].
Implicating pathways to rheumatoid arthritis risk
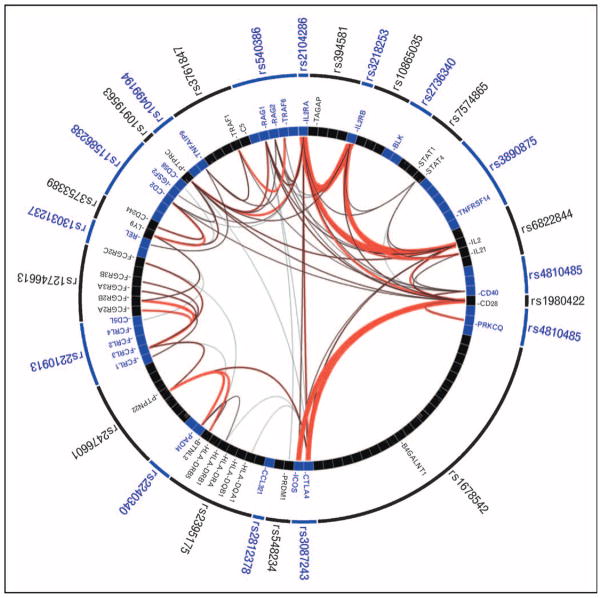
One of the key goals of genetics is to identify biological pathways and processes that predispose to risk. In Fig. 4, I have used GRAIL to demonstrate the compelling functional connections across genes near RA-associated SNPs; these connections strongly suggest that common pathways are present across multiple RA risk loci.
Figure 4. Gene relationships across rheumatoid arthritis-associated loci.
Here I list each of the known rheumatoid arthritis (RA)-associated single nucleotide polymorphisms (SNPs) along the outer ring; the internal ring represents the genes near each SNP with a box. I illustrate the literature-based functional connectivity between these genes with lines drawn between them as assessed by the gene relationships across implicated loci (GRAIL) algorithm. The more red and thicker the lines are, the stronger the connectivity between the genes is. RA SNPs implicate a small number of highly connected genes – those genes are indicated by labeled boxes.
For example, risk alleles highlight genes involved in T-cell activation by antigen presenting cells (class II MHC region, PTPN22, STAT4, CD28, and CTLA4). More recent discoveries have demonstrated the role of the CD40 signaling pathway and downstream activation of nuclear factor-κB (NF-κB) signaling pathway (CD40, TRAF1, TRAF6, TNFSF14, TNFAIP3). Recently, Gregersen et al. [28••] conducted an unbiased GWAS to definitively identify an associated risk allele at c-REL, one of the five NF-κB family proteins.
Recent associations have also now clearly implicated the IL2 signaling pathway, a critical cytokine involved in T-cell activation and proliferation. Follow-up genotyping of nominally associated SNPs (P <10−5) in the WTCCC RA GWAS demonstrated evidence of association at IL2RA that was subsequently confirmed by an independent group [37,38]. Follow-up genotyping of nominally associated SNPs (P <10−4) demonstrated evidence of association at IL2RB that was subsequently confirmed by an independent group [29••,38]. Finally, Zhernakova et al. [31] demonstrated association of a SNP implicating the IL2/IL21 locus, which was subsequently confirmed in an independent study by Barton et al. [32••]. Functional connections across the genes within these loci are clearly highlighted in Fig. 4.
Seronegative disease
The majority of the discoveries presented here have been identified with predominantly seropositive (anti-CCP or rheumatoid factor positive) samples. Many studies explicitly exclude seronegative samples to assure diagnostic certainty and homogeneity. The differences in the genetics of anti-CCP-positive and anti-CCP-negative RA have now been demonstrated most strikingly in the role that the shared epitope alleles play in multiple studies [23••,74], whereas in seropositive disease, shared epitope alleles are the strongest risk factor; they seem to play a much more modest role if any at all in seronegative disease. More recently, Ding et al. [62••] conducted a GWA study and examined the association of MHC SNPs in seronegative cases and observed no significant association of any SNP. However, outside the MHC, additional studies are necessary to demonstrate similarities and differences between the risk loci for seronegative and seropositive disease.
Predicting disease risk
One of the goals of genetic studies is to be able to predict individual risk for patients. For such predictive strategies to be clinically applicable, they will need to achieve a high degree of specificity.
Based on twin studies, it is unlikely that full ascertainment of the genome alone will result in highly accurate clinical risk prediction in RA. Concordance rates among dizygotic twins offer a sense for the upper-bound for the predictive power of a genetic test. As monozygotic twins have identical genotypes, a hypothetical predictive method that uses only genetic information will assign the same prediction to both twins. For monozygotic twin pairs with at least one affected twin, RA has a maximum of 15% concordance. Any method that is 100% sensitive when applied to probands will accurately classify them as affected. But, the same sensitive method when applied to the related twins will also classify all of them as affected, and the positive predictive value will be at most 15%. On the other hand, a 100% specific method will accurately classify at least 85% of the related twins as unaffected; but the same method when applied to the affected probands will only be at most 15% sensitive. A specific and clinically applicable method that uses only genetic information can, therefore, identify at most 15% of affected patients in advance of clinical symptoms.
Certainly, the addition of additional factors such as epigenetic information, biomarkers, clinical predictors, and environmental factors will improve predictive power and may result in a more useful approach.
However, current SNP associations and MHC alleles in aggregate can offer insight into risk of RA for individuals within a population, assuming representative ethnic background. Karlson et al. [75•] used published RA risk SNP alleles and their odds ratios along with MHC risk alleles to demonstrate that they could be used to stratify patients into seven genetic risk categories; they showed that the highest risk category was at three-fold the risk of the population baseline and six-fold the lowest risk category.
Conclusion
In the last year, investigators have identified multiple additional novel RA risk loci; but these loci likely represent early steps in understanding RA genetics. Much additional work remains to understand the complexity of each of these loci, the functional role that each locus plays in the pathogenesis of RA and of autoimmune disease in general, and to discover the complete list of risk loci. Ultimately, these risk loci will continue to provide key insights into the pathogenesis of RA.
Acknowledgments
S.R. is supported by an NIH Career Development Award (1K08AR055688).
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 000–000).
- 1.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 2.Isenberg D. Oxford textbook of rheumatology. 3. Oxford: New York: Oxford University Press; 2004. [Google Scholar]
- 3.Heberden W. Commentaries on the history and cure of diseases. 3. London: Printed for T. Payne; 1806. [Google Scholar]
- 4.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 5.Stastny P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N Engl J Med. 1978;298:869–871. doi: 10.1056/NEJM197804202981602. [DOI] [PubMed] [Google Scholar]
- 6.Stolt P, Bengtsson C, Nordmark B, et al. Quantification of the influence of cigarette smoking on rheumatoid arthritis: results from a population based case–control study, using incident cases. Ann Rheum Dis. 2003;62:835–841. doi: 10.1136/ard.62.9.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Remmers EF, Longman RE, Du Y, et al. A genome scan localizes five non-MHC loci controlling collagen-induced arthritis in rats. Nat Genet. 1996;14:82–85. doi: 10.1038/ng0996-82. [DOI] [PubMed] [Google Scholar]
- 8.Wandstrat A, Wakeland E. The genetics of complex autoimmune diseases: non-MHC susceptibility genes. Nat Immunol. 2001;2:802–809. doi: 10.1038/ni0901-802. [DOI] [PubMed] [Google Scholar]
- 9.Keffer J, Probert L, Cazlaris H, et al. Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J. 1991;10:4025–4031. doi: 10.1002/j.1460-2075.1991.tb04978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams RO, Feldmann M, Maini RN. Antitumor necrosis factor ameliorates joint disease in murine collagen-induced arthritis. Proc Natl Acad Sci U S A. 1992;89:9784–9788. doi: 10.1073/pnas.89.20.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart JE, Laden F, Puett RC, et al. Exposure to traffic pollution and increased risk of rheumatoid arthritis. Environ Health Perspect. 2009;117:1065–1069. doi: 10.1289/ehp.0800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seldin MF, Amos CI, Ward R, Gregersen PK. The genetics revolution and the assault on rheumatoid arthritis. Arthritis Rheum. 1999;42:1071–1079. doi: 10.1002/1529-0131(199906)42:6<1071::AID-ANR1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Lewis-Faning E. Report on an enquiry into the aetiological factors associated with rheumatoid arthritis. Ann Rheum Dis. 1950;9:1–94. [Google Scholar]
- 14.Lawrence JS. Heberden Oration, 1969. Rheumatoid arthritis: nature or nurture? Ann Rheum Dis. 1970;29:357–379. doi: 10.1136/ard.29.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.del Junco D, Luthra HS, Annegers JF, et al. The familial aggregation of rheumatoid arthritis and its relationship to the HLA-DR4 association. Am J Epidemiol. 1984;119:813–829. doi: 10.1093/oxfordjournals.aje.a113802. [DOI] [PubMed] [Google Scholar]
- 16.Jones MA, Silman AJ, Whiting S, et al. Occurrence of rheumatoid arthritis is not increased in the first degree relatives of a population based inception cohort of inflammatory polyarthritis. Ann Rheum Dis. 1996;55:89–93. doi: 10.1136/ard.55.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemminki K, Li X, Sundquist J, Sundquist K. Familial associations of rheumatoid arthritis with autoimmune diseases and related conditions. Arthritis Rheum. 2009;60:661–668. doi: 10.1002/art.24328. [DOI] [PubMed] [Google Scholar]
- 18.Lynn AH, Kwoh CK, Venglish CM, et al. Genetic epidemiology of rheumatoid arthritis. Am J Hum Genet. 1995;57:150–159. [PMC free article] [PubMed] [Google Scholar]
- 19.MacGregor AJ, Snieder H, Rigby AS, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 20.Aho K, Koskenvuo M, Tuominen J, Kaprio J. Occurrence of rheumatoid arthritis in a nationwide series of twins. J Rheumatol. 1986;13:899–902. [PubMed] [Google Scholar]
- 21.Silman AJ, MacGregor AJ, Thomson W, et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br J Rheumatol. 1993;32:903–907. doi: 10.1093/rheumatology/32.10.903. [DOI] [PubMed] [Google Scholar]
- 22.Svendsen AJ, Holm NV, Kyvik K, et al. Relative importance of genetic effects in rheumatoid arthritis: historical cohort study of Danish nationwide twin population. BMJ. 2002;324:264–266. [PMC free article] [PubMed] [Google Scholar]
- 23••.van der Woude D, Houwing-Duistermaat JJ, Toes RE, et al. Quantitative heritability of anticitrullinated protein antibody-positive and anticitrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum. 2009;60:916–923. doi: 10.1002/art.24385. In this study, investigators revisited a previously published twin study and checked samples for CCP status and genotype samples. They quantified the heritability of seropositive and seronegative RA and demonstrated that in both cases, RA is approximately 65% heritable. They also demonstrated that shared epitope HLA alleles contribute substantially only to the genetic variance of seropositive RA. [DOI] [PubMed] [Google Scholar]
- 24.Plenge RM, Cotsapas C, Davies L, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Orozco G, Hinks A, Eyre S, et al. Combined effects of three independent SNPs greatly increase the risk estimate for RA at 6q23. Hum Mol Genet. 2009;18:2693–2699. doi: 10.1093/hmg/ddp193. The authors conducted a fine-mapping study of the 6q23/TNFAIP3 locus in 3962 RA patients and 3531 controls. In addition to known associations at rs13207033 (a perfect proxy for rs10499194) and rs6920220, the authors demonstrated independent association at another variant at rs5029937, located in the intron 2 of TNFAIP3. This paper highlights the complexity of the disease association of this locus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plenge RM, Seielstad M, Padyukov L, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis: a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurreeman FA, Padyukov L, Marques RB, et al. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med. 2007;4:e278. doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Gregersen PK, Amos CI, Lee AT, et al. REL, encoding a member of the NF-κB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41:820–823. doi: 10.1038/ng.395. Investigators conducted a GWAS of 2418 RA cases and 4504 controls and identified an association at c-Rel, one of the five NF-κB family members. The association was reproduced in 2604 cases and 2882 controls. Its association in addition to CD40, TRAF1, TNFAIP3 and PRKCQ the key role of NF-κB signaling in RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Barton A, Thomson W, Ke X, et al. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet. 2008;40:1156–1159. doi: 10.1038/ng.218. This study examined 49 SNPs demonstrating nominal evidence (P <0.0001) of association in a GWA study conducted by the WTCCC. SNPs were genotyped in 4000 RA cases and 11 000 controls to identify SNP associations at KIF5A and PRKCQ along with that in the study by Raychaudhuri et al. 2008; it also noted an association at IL2RB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plenge RM, Padyukov L, Remmers EF, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77:1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhernakova A, Alizadeh BZ, Bevova M, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81:1284–1288. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Barton A, Eyre S, Ke X, et al. Identification of AF4/FMR2 family, member 3 (AFF3) as a novel rheumatoid arthritis susceptibility locus and confirmation of two further pan-autoimmune susceptibility genes. Hum Mol Genet. 2009;18:2518–2522. doi: 10.1093/hmg/ddp177. Here, investigators tested 18 SNPs, known to be associated with type I diabetes in 6819 RA cases and 12 650 controls. They demonstrated novel association at AFF3 with RA. They reproduced previously observed associations at CTLA4 and 4q27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Begovich AB, Carlton VE, Honigberg LA, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Remmers EF, Plenge RM, Lee AT, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Suzuki A, Yamada R, Kochi Y, et al. Functional SNPs in CD244 increase the risk of rheumatoid arthritis in a Japanese population. Nat Genet. 2008;40:1224–1229. doi: 10.1038/ng.205. These investigators identified a linkage disequilibrium block associated with RA in the chromosome 1q region containing multiple SLAM family genes. In follow-up genotyping, they demonstrated peak association at two functional SNPs (rs3766379 and rs6682654) in CD244 in two independent rheumatoid arthritis cohorts from Japan. They further demonstrated that RA-susceptible alleles increased the expression in luciferase and allele-specific transcript quantification assays of CD244. [DOI] [PubMed] [Google Scholar]
- 36••.Raychaudhuri S, Thomson BP, Remmers EF, et al. Genetic variants at CD28, PRDM1, and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. In this study, we examined 179 independent loci with P value less than 0.001 in a previously published meta-analysis of RA GWAS (Raychaudhuri et al. 2008). We used GRAIL to objectively score these 179 loci for functional relationships to genes in 16 established RA disease loci. We identified and genotyped 22 representative SNPs in an independent set of 20 000 cases and controls; of those seven replicated convincingly and are now considered novel RA risk loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson W, Barton A, Ke X, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurreeman FA, Daha NA, Chang M, et al. Association of IL2RA and IL2RB with rheumatoid arthritis: a replication study in a Dutch population. Ann Rheum Dis. 2009;68:1789–1790. doi: 10.1136/ard.2008.106393. [DOI] [PubMed] [Google Scholar]
- 39••.Raychaudhuri S, Remmers EF, Lee AT, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. This study presents a large-scale meta-analysis of three genome-wide association studies in RA, consisting of 15 800 cases and controls. This study first proposed and replicated RA risk loci at CD40, CCL21, PRKCQ, KIF5A, and TNFRSF14 in an additional independent 9700 samples. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki A, Yamada R, Chang X, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 41.Kochi Y, Yamada R, Suzuki A, et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet. 2005;37:478–485. doi: 10.1038/ng1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halushka MK, Fan JB, Bentley K, et al. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- 43.Cargill M, Altshuler D, Ireland J, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 44.Wang DG, Fan JB, Siao CJ, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 45.Matsuzaki H, Loi H, Dong S, et al. Parallel genotyping of over 10 000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res. 2004;14:414–425. doi: 10.1101/gr.2014904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen R, Fan JB, Campbell D, et al. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 47.Bhangale TR, Rieder MJ, Nickerson DA. Estimating coverage and power for genetic association studies using near-complete variation data. Nat Genet. 2008;40:841–843. doi: 10.1038/ng.180. [DOI] [PubMed] [Google Scholar]
- 48.de Bakker PI, Yelensky R, Pe’er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 49.The Wellcome Trust. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchini J, Howie B, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 51•.de Bakker PI, Ferreira MA, Jia X, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. A practical guide about using GWAS data to impute missing SNP data, and conducting meta-analyses across multiple GWASs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Orozco G, Eyre S, Hinks A, et al. Association of CD40 with rheumatoid arthritis confirmed in a large UK case-control study. Ann Rheum Dis. 2009 doi: 10.1136/ard.2009.109579. In this study, investigators sought to confirm associations at CD40, CCL21, CDK6, and CD244 reported by Raychaudhuri et al. 2008 and Suzuki et al. 2008. SNPs were genotyped in a UK cohort comprising 3962 UK RA cases and 3531 healthy controls. They found evidence of association at CD40 and CCL21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lettre G, Jackson AU, Gieger C, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 57.Coenen MJ, Trynka G, Heskamp S, et al. Common and different genetic background for rheumatoid arthritis and coeliac disease. Hum Mol Genet. 2009;18:4195–4203. doi: 10.1093/hmg/ddp365. [DOI] [PubMed] [Google Scholar]
- 58.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 59•.Raychaudhuri S, Plenge RM, Rossin EJ, Ng ACY. Ng ACY; International Schizophrenia Consortium, Purcell SM, Sklar P, Scolnick EM, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. In: Purcell SM, Sklar P, Scolnick EM, et al., editors. PLOS Genet. Vol. 5. 2009. p. e1000534. This paper presents an online tool to leverage functional information contained in the published literature to examine genes near SNP associations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deighton CM, Walker DJ, Griffiths ID, Roberts DF. The contribution of HLA to rheumatoid arthritis. Clin Genet. 1989;36:178–182. doi: 10.1111/j.1399-0004.1989.tb03185.x. [DOI] [PubMed] [Google Scholar]
- 61••.Lee HS, Lee AT, Criswell LA, et al. Several regions in the major histocompatibility complex confer risk for anti-CCP-antibody positive rheumatoid arthritis, independent of the DRB1 locus. Mol Med. 2008;14:293–300. doi: 10.2119/2007-00123.Lee. Here, investigators attempted to identify risk loci independent of the class II HLA-DRB1 locus, elsewhere in the MHC. They genotyped SNPs within the MHC for 372 cases with 372 matched 1:1 by DRB1 genotype. This analysis suggested the presence of at least two additional regions of association with RA in the class I region, independent of DRB1 genotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62••.Ding B, Padyukov L, Lundstrom E, et al. Different patterns of associations with anticitrullinated protein antibody-positive and anticitrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum. 2009;60:30–38. doi: 10.1002/art.24135. The authors examined the role of the MHC region in anti-CCP-positive and anti-CCP-negative RA cases separately. When they examined 640 anti-CCP-negative cases compared with controls, they observed no significant SNP associations within the MHC region. In contrast, when they examined a total of 1255 anti-CCP-positive cases and 1719 controls, they observed independent associations at the known DRB1 risk loci and identified additional independent associations with SNPs near HLA-DPB1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. The authors present a statistical strategy that uses approximately 100 SNP markers within the MHC to predict HLA genotypes at key class I and II loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Bakker PI, McVean G, Sabeti PC, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 66.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alkan C, Kidd JM, Marques-Bonet T, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hollox EJ, Huffmeier U, Zeeuwen PL, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prahalad S. Negative association between the chemokine receptor CCR5-delta32 polymorphism and rheumatoid arthritis: a meta-analysis. Genes Immun. 2006;7:264–268. doi: 10.1038/sj.gene.6364298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKinney C, Merriman ME, Chapman PT, et al. Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 2008;67:409–413. doi: 10.1136/ard.2007.075028. [DOI] [PubMed] [Google Scholar]
- 74.van der Helmvan Mil AH, Verpoort KN, Breedveld FC, et al. The HLA-DRB1 shared epitope alleles are primarily a risk factor for anticyclic citrullinated peptide antibodies and are not an independent risk factor for development of rheumatoid arthritis. Arthritis Rheum. 2006;54:1117–1121. doi: 10.1002/art.21739. [DOI] [PubMed] [Google Scholar]
- 75•.Karlson EW, Chibnik LB, Kraft P, et al. Cumulative association of twenty-two genetic variants with seropositive rheumatoid arthritis risk. Ann Rheum Dis. in press. The authors genotyped 14 validated RA risk SNPs and eight HLA alleles in two populations. They created a weighted genetic risk score (GRS), in which the weight for each risk allele is the log of the published odds ratio. They demonstrated that the weighted GRS significantly stratifies individuals for RA risk beyond clinical risk factors alone. [Google Scholar]