Abstract
Primary sclerosing cholangitis (PSC) is a chronic cholestatic liver disease characterized by inflammation and fibrosis of the bile ducts. Both environmental and genetic factors contribute to its pathogenesis. To further clarify its genetic background, we investigated susceptibility loci recently identified for ulcerative colitis (UC) in a large cohort of 1186PSC patients and 1748 controls. Single nucleotide polymorphisms (SNPs) tagging 13 UC susceptibility loci were initially genotyped in 854 PSC patients and 1491 controls from the Benelux (331 cases, 735 controls), Germany (265 cases, 368 controls) and Scandinavia (258 cases, 388 controls). Subsequently, a joint analysis was performed with an independent second Scandinavian cohort (332 cases, 257 controls). SNPs at chromosomes2p16 (p value 4.12×10−4), 4q27 (p value 4.10×10−5) and 9q34 (p value 8.41×10−4) were associated with PSC in the joint analysis after correcting for multiple testing. In PSC patients without inflammatory bowel disease(IBD), SNPs at 4q27and9q34 were nominally associated (p<0.05). We applied additional in silico analyses to identify likely candidate genes at PSC susceptibility loci. To identify non-random, evidence-based links we used GRAIL analysis showing interconnectivity between genes in six out of in total nine PSC-associated regions. Expression quantitative trait analysis from 1469 Dutch and UK individuals demonstrated that five out of nine SNPs had an effect on cis-gene expression. These analyses prioritized IL2, CARD9 and REL as novel candidates.
Conclusion
We have identified three UC susceptibility loci to be associated with PSC, harboring the putative candidate genes REL, IL2 and CARD9. These results add to the scarce knowledge on the genetic background of PSC and imply an important role for both innate and adaptive immunological factors.
Keywords: chronic liver disease, auto-immunity, inflammation, complex disease, genetics
Primary sclerosing cholangitis (PSC) is a chronic cholestatic liver disease characterized by inflammation and fibrosis of both intrahepatic and extrahepatic bile ducts.(1) Currently there is no curative therapy available and, in most cases, PSC ultimately leads to liver cirrhosis and liver failure requiring liver transplantation.(2, 3) Population-based studies from the USA and Norway show an incidence of 0.9 and 1.3 per 100,000/year and prevalence of 14.2 and 8.5 per 100,000, respectively.(4, 5) About 50 to 80 percent of the patients with PSC also have inflammatory bowel disease (IBD), compatible with a diagnosis of ulcerative colitis (UC) in approximately 80 percent of these cases and of Crohn’s disease (CD) or unclassified IBD in the remaining 20 percent.(6)
PSC is a complex disease, with both environmental and genetic factors likely to determine its development and course. Siblings of PSC patients are 9 to 39 times more likely to develop PSC than the overall population. They also have an eight times increased risk of developing UC, suggesting a shared genetic component between both diseases.(7) Very little is known about the pathogenesis of PSC, but its high concordance with other immune-mediated diseases and its known genetic associations within the HLA complex suggest that PSC is an immune-mediated disease.(8)
Genome-wide association scans and candidate gene studies have identified more than 30 risk loci for UC in the past decade. Eighteen of these loci reached the threshold of genome-wide significance.(9–16) Identifying these loci has led to a better understanding of the biological pathways involved and implicatedTh1 and Th17 responses and the epithelial barrier in UC pathogenesis.(17) However, little is known about the genetic architecture of PSC. Since the discovery of associations between the HLA variants DR3 and HLA-B8 and PSC in 1982,(18) many studies have confirmed genetic associations between variants in the HLA complex and PSC.(19) Recently, two genome-wide association scans identified five PSC risk loci outside the HLA complex including 2q13, 2q35, 3p21, 10p15 and 13q31.(20, 21) Of these loci,2q35 and3p21are also associated with UC.(10–12) The causal genes for these regions still need to be determined, but the locus on chromosome 2q35 harbors a compelling candidate gene (the bile acid receptor TGR5).(22)
BCL2L11 is an interesting candidate for the association at 2q13because of its role in maintaining immunological tolerance, and IL2RA is of interest in PSC at 10p15 since IL2ra −/− mice spontaneously develop intestinal and biliary inflammation.(21)
Our aim was to further clarify the genetic background of PSC. Because of the clinical overlap between UC and PSC, we first investigated the association of 13recently identified UC susceptibility loci in a cohort of 1186PSC patients and 1748healthy controls. We further analyzed whether associations were limited to PSC patients with concomitant IBD or to PSC patients without IBD. Finally, we used eQTL mapping and Gene Relationships Across Implicated Loci (GRAIL) analysis (23) to propose likely causal genes from the nine known PSC-associated loci, including the three new ones.
Methods
Subjects
We included 1186 primary sclerosing cholangitis (PSC) patients and 1748healthy controls in our cohorts. The initial three cohorts comprised 331 Benelux, 265 German and 258 Scandinavian patients, and735 Benelux, 368 German and 388Scandinavian healthy individuals. A second Scandinavian cohort included in the joint analysis consisted of 332 patients and 257 controls. Benelux patients were recruited at the Academic Medical Center in Amsterdam and the University Medical Center Groningen, the Netherlands, and at the University Hospital Leuven, Belgium. The German patients were recruited through the Northern German biobank popgen (http://www.popgen.de), and contain patients from the University Medical Center Hamburg-Eppendorf, the Hannover Medical School and the Christian-Albrechts-University Hospital Kiel. The PSC patients in the Scandinavian cohorts were recruited at the Rikshospitalet, Oslo University Hospital, Oslo, Norway, the Huddinge University Hospital, Stockholm, Sweden and the Sahlgrenska University Hospital, Gothenburg, Sweden. A description of the clinical characteristics of the PSC cases is provided in Supplementary table 1.
Healthy controls from the Benelux were blood donors recruited from donor centers in Groningen, the Netherlands, and from healthy volunteers recruited via the University Hospital Leuven, Belgium. DNA from the German controls was obtained from blood donors through the Northern German biobank popgen. The Scandinavian controls were randomly selected from the Norwegian Bone Marrow Donor Registry. Although blood bank donors and bone marrow donors are a selected group compared to population controls, they are regularly and confidently being used in genetic association studies.(24)
The diagnosis of primary sclerosing cholangitis was based on standard clinical, biochemical, histological and cholangiographic criteria.(25) The diagnosis of IBD was based on accepted clinical, radiologic, endoscopic and histopathologic criteria.(26) Phenotype information on IBD status was available for 984cases, of which 739cases had concurrent IBD and 245cases did not. Written informed consent was obtained from all participants. Recruitment of the participants was approved by the ethics committees at each of the recruiting hospitals.
Genotyping and SNP selection
At the start of the study all known UC loci identified through GWAS and large candidate gene studies were considered. In total we included 13 single nucleotide polymorphisms (SNPs) tagging 13 recently identified UC susceptibilityloci(14–16, 27, 28) (Table 1). Fifteen other UC susceptibility variants had been tested before as part of a GWAS in PSC in the Scandinavian PSC population and were therefore not included in the current study.(20)
Table 1.
Allelic Association Analysis of 13 UC Susceptibility Loci in Three PSC Cohorts and a Combined Analysis
| Chr | SNP | Gene of interest | Alleles (min/maj) |
Benelux (309 cases, 700 controls) |
Germany (262 cases, 361 controls) |
Scandinavia (252 cases, 384 controls) |
Combined cohorts (823 cases, 1445 controls) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF (Case/ctrl) |
p-value | MAF (Case/ctrl) |
p-value | MAF (Case/ctrl) |
p-value | OR | 95% CI | CMH-test p-value |
BD p-value | ||||
| 4 | rs6822844 | IL2/21 | T/G | 0.12/0.15 | 5.39×10−2 | 0.090/0.13 | 1.95×10−2 | 0.15/0.19 | 8.66×10−2 | 0.73 | 0.61–0.88 | 6.87×10−4 | 0.74 |
| 9 | rs4077515 | CARD9 | T/C | 0.49/0.43 | 7.32×10−3 | 0.47/0.42 | 0.11 | 0.48/0.46 | 0.37 | 1.21 | 1.07–1.37 | 2.18×10−3 | 0.58 |
| 2 | rs6706689 | PUS10/REL | A/G | 0.35/0.40 | 4.29×10−2 | 0.34/0.38 | 0.15 | 0.36/0.41 | 8.77×10−2 | 0.82 | 0.73–0.93 | 2.62×10−3 | 0.98 |
| 20 | rs6017342 | HNF4A | A/C | 0.45/0.48 | 0.18 | 0.42/0.47 | 6.90×10−2 | 0.47/0.48 | 0.90 | 0.89 | 0.79–1.00 | 5.51×10−2 | 0.48 |
| 22 | rs5771069 | IL17REL | A/G | 0.48/0.49 | 0.53 | 0.49/0.49 | 0.98 | 0.44/0.50 | 4.98×10−2 | 0.91 | 0.81–1.03 | 0.15 | 0.34 |
| 16 | rs4781011 | CIITA | T/G | 0.25/0.27 | 0.26 | 0.28/0.27 | 0.72 | 0.23/0.26 | 0.37 | 0.93 | 0.81–1.07 | 0.32 | 0.55 |
| 17 | rs2305480 | ORMDL3 region | T/C | 0.45/0.45 | 0.90 | 0.48/0.44 | 0.22 | 0.49/0.51 | 0.55 | 1.06 | 0.94–1.20 | 0.37 | 0.60 |
| 16 | rs1728785 | CDH1 | A/C | 0.21/0.22 | 0.57 | 0.20/0.23 | 0.25 | 0.20/0.20 | 0.80 | 0.94 | 0.81–1.09 | 0.39 | 0.62 |
| 7 | rs7809799 | SMURF1-KPNA7 | G/A | 0.034/0.033 | 0.89 | 0.039/0.053 | 0.24 | 0.048/0.020 | 4.74×10−3 | 1.14 | 0.82–1.57 | 0.44 | 1.35×10−2 |
| 1 | rs10800309 | FCGR2A/FCGR2C | A/G | 0.36/0.37 | 0.90 | 0.40/0.39 | 0.88 | 0.35/0.32 | 0.24 | 1.04 | 0.92–1.18 | 0.53 | 0.60 |
| 5 | rs4957048 | Cep72/SLC9A3 | T/C | 0.24/0.23 | 0.46 | 0.19/0.23 | 0.11 | 0.19/0.18 | 0.79 | 0.98 | 0.85–1.14 | 0.81 | 0.21 |
| 9 | rs10758669 | JAK2 | C/A | 0.35/0.35 | 0.93 | 0.33/0.37 | 0.23 | 0.37/0.35 | 0.37 | 0.99 | 0.87–1.12 | 0.82 | 0.33 |
| 7 | rs6949033 | LAMB1 | A/G | 0.39/0.41 | 0.50 | 0.43/0.38 | 6.34×10−2 | 0.39/0.42 | 0.27 | 1.00 | 0.88–1.13 | 0.98 | 7.78×10−2 |
p-values in bold indicate association. Bonferroni corrected threshold for significance: p=0.004. MAF, Minor allele frequency, CMH-test, Cochrane-Mantel-Haenszel test, BD, Breslow-Day test, OR, odds ratio, CI, confidence interval. The third column depicts the genes of interest as described by previous studies.
Genotyping of the initial three cohorts (Benelux, German, and Scandinavian I) was performed using TaqMan technology from Applied Biosystems(Foster City, California, USA) according to the manufacturer’s recommendations. The patient and control DNA samples were processed in 384-wells plates, with each plate containing 16 genotyping controls (four duplicates of four Centre d’Etude du Polymorphisme Humain (CEPH) DNA). The results from the genotyping assays were analyzed using SDS 2.3 software compatible with the TaqMan system. Genotype data for the Scandinavian replication panel (Scandinavian II) were extracted from a previous PSC GWAS. The patients and controlsin this study were genotyped with the Affymetrix® Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA). Details on quality control and imputation of additional genotypes (using MACH software version 1.0.16) are described previously.(21, 29) Individuals overlapping with the Taqman genotyped Scandinavian study panel were excluded from the GWAS data (n=5 controls).
For the directly genotyped SNPs genotyping cluster plots were manually inspected to ensure high genotyping quality while for the imputed SNPs good imputation quality was required (r2>0.3).
Allelic association analysis
Statistical analysis of the genotyping data from the initial three cohorts genotyped with Taqman assays was performed using PLINK software version 1.06.(30) Quality controls excluded samples with a low genotyping success rate (missing one or more SNPs) and SNPs with a genotyping success rate below 95%. SNPs showing deviation from Hardy-Weinberg equilibrium (HWE) in the controls (P < 0.004) would be discarded from further analysis. All 13 SNPs were in HWE, therefore no SNPs were discarded. To correct for multiple testing forthe13 SNPs, we determined a significance threshold for P at 0.05/13 → P <0.004.
Differences in allele frequencies between cases and controls were analyzed in the three individual cohorts with a χ2 test. Association analysis for the combined German, Benelux and Scandinavian I cohort was performed with a Cochrane-Mantel-Haenszel (CMH) test. A Breslow-Day test for heterogeneity of odds ratios was also performed for the combined cohort.
Association analysis for the Scandinavian II cohort was performed as follows: To take account of uncertainty from the imputation, allele dosages were used for all calculations except for Hardy-Weinberg equilibrium, which was evaluated using the best-guess genotypes in PLINK v1.06.(30) For one SNP(rs6017342) the controls showed deviation from HWE, therefore this SNP was discarded for further analysis. The SNPs were tested for association using binary logistic regression as implemented in the R statistical package version 2.11.1.
The combined analysis of the Taqman genotyped data and the GWAS data was conducted using weighted Z-scores.(31) To take account of unequal numbers of cases and controls in the study panels, the effective sample size was used for the weighting of the Z-scores.(32) To check for heterogeneity of the associations between the four different cohorts a Breslow-Day test was performed in R statistical package. Regarding the Scandinavian II cohort, 2×2 tables with allele frequencies in cases/controls were recalculated using allele frequencies based on the allele dosages from the GWAS.
In silico Analyses
GRAIL analysis
To obtain insight into the functional relation of the PSC risk loci, we performed a Gene Relationship Across Implicated Loci (GRAIL) pathway analysis (http://www.broadinstitute.org/mpg/grail). GRAIL is a statistical tool that uses text mining of PubMed abstracts to annotate candidate genes in loci associated with disease risk.(23) We included all six regions that showed an association in previous studies,(20, 21) in combination with the three new loci we identified. Specifically, GRAIL evaluated each gene in a PSC-associated locus for non-random correlation with genes in the other eight loci, via word-usage in PubMed abstracts related to the gene. We used HG17 and December 2006 PubMed datasets, default settings for SNP rsNumber submission, and all nine PSC loci as query and seed. To assess the accuracy of the statistical significance of the set of connections, we conducted simulations in which we selected 1000 sets of nine SNPs. Since PSC associated SNPs were in genomic regions implicating 100 genes, we selected SNP sets using rejection sampling that implicated 100 genes ± 5%. Each of these 1000 sets were also scored with GRAIL.
Expression quantitative trait locus analysis
We used genetical genomics data of 1469 peripheral blood DNA and RNA (PAXgene) samples from Dutch and UK individuals, described in detail by Dubois et al.(33) to perform an expression quantitative traitlocus (eQTL) analysis. All samples had been genotyped using either an Illumina Hap370 or 610-Quad platform. Imputation for ungenotyped SNPs was performed using IMPUTE software (https://mathgen.stats.ox.ac.uk/impute/impute.html). RNA from these individuals was hybridized either to an Illumina HumanRef-8 v2 array (229 samples, Ref-8 v2, Gene Expression Omnibus (GEO) accession GSE20332) or to an Illumina HumanHT-12 array (1,240 samples, HT-12, GEO accession GSE20142), and raw probe intensity extracted using BeadStudio. The Ref-8 v2 samples were jointly quantile normalized and log2 transformed, as were the HT-12 samples. Expression variation due to batch and technical effects were removed by using principal component analysis (PCA) and removal of 50 PCAs. Subsequent analyses were conducted separately for both datasets, up to the eventual eQTL mapping, which used a meta-analysis framework combining eQTL results from both arrays. We applied a window of 500kb around each SNP (250kb on each side).
To prevent spurious associations due to outliers, a non-parametric Spearman’s rank correlation analysis was performed. When a particular probe-SNP pair was present in both the HT-12 and Ref-8 v2datasets, an overall joint P value was calculated using a weighted Z-method (square root of the dataset’s sample number). To correct for multiple testing, we controlled the false-discovery rate (FDR). The distribution of observed P values was used to calculate the FDR, by permuting expression phenotypes relative to genotypes 25 times within the HT-12 and Ref-8 v2dataset. cis-eQTLs were considered statistically significant with a Spearman P < 0.0028, corresponding to a 5% FDR. Finally, we removed any probes from the analysis that contained a known SNP (1000Genomes CEU(Utah residents with European ancestry) SNP data, April 2009 release) to prevent false-positive associations, assuming the “probe SNP” caused differential hybridization. Six regions that showed an association in previous studies, along with the three loci identified in our study, were included in the eQTL analysis.
Results
Initially, we genotyped 13 SNPs in 854 PSC patients and 1491 controls. After applying quality control measures, 823 cases (96,4%) and 1445 controls (96,9%) were available for further analysis. Next, a joint analysis was performed with an independent Scandinavian cohort consisting of 332 cases and 257 controls. Thus, in total1155PSC patients and 1702 controls were analyzed. Both in the initial analysis as in the joint analysis we did not observe any heterogeneity of the ORs between the cohorts(Breslow Day test p>0.004)
Allelic association analysis of UC susceptibility loci in PSC
Combined analysis of the initial three cohorts revealed three loci associated with PSC. The strongest association was observed for SNP rs6822844 at locus 4q27 with a P-value of 6.87×10−4 (ORT allele 0.73; 95% CI 0.61–0.88). SNP rs4077515 at 9q34 was associated with a p-value in the combined analysis of 2.18×10−3 (ORT allele 1.21; 95% CI 1.07–1.37) and rs6706689 at 2p16 showed a P-value of 2.62×10−3 (ORA allele 0.82; 95% CI 0.73–0.93). All the results are shown in Table 1. Joint analysis with an independent Scandinavian cohort of 332 cases and 257 controls further strengthened the association signal(p=4.10×10−5 for rs6822844, p= 8.41×10−4 for rs4077515 and p= 4.12×10−4 for rs6706689, Table 2). All currently identified allelic association signals for PSC are in the same direction as the original identified associations for ulcerative colitis.
Table 2.
Joint analysis of the initial three PSC cohorts with an additional Scandinavian cohort.
| Chr. | SNP | Gene | Combined cohorts Benelux, German, Scandinavian I (823 cases, 1445 controls) | Scandinavian II (332 cases, 257 controls) | Meta-analysis (1155 cases, 1702 controls) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| z-score | CMH p-value | MAF (cases/controls) | z-score | p-value | Weighted z-score | p-value | BD p-value | |||
| 4 | rs6822844 | IL2/21 | −3.39 | 6.87×10-4 | 0.16/0.22 | −2.37 | 1.8×10-2 | −4.10 | 4.10×10-5 | 0.96 |
| 9 | rs4077515 | CARD9 | 3.06 | 2.18×10-3 | 0.48/0.45 | 1.34 | 0,18 | 3.34 | 8.41×10-4 | 0.90 |
| 2 | rs6706689 | PUS10/REL | −3.01 | 2.62×10-3 | 0.39/0.45 | −1.87 | 6.08×10-2 | −3.53 | 4.12×10-4 | 1.00 |
| 20 | rs6017342* | HNF4A | −1.92 | 5.51×510-2 | - | - | - | - | - | - |
| 22 | rs5771069 | IL17REL | −1.45 | 0.15 | 0.48/0.54 | −2.09 | 3.67×10-2 | −2.24 | 2.48×10-2 | 0.66 |
| 16 | rs4781011 | CIITA | −1.00 | 0.32 | 0.27/0.26 | 0.47 | 0.64 | −0.68 | 0.50 | 0.82 |
| 17 | rs2305480 | ORMDL3 region | 0.90 | 0.37 | 0.47/0.47 | 0.14 | 0.89 | 0.87 | 0.39 | 0.83 |
| 16 | rs1728785 | CDH1 | −0.85 | 0.39 | 0.16/0.18 | −1.04 | 0.30 | −1.23 | 0.22 | 0.90 |
| 7 | rs7809799 | SMURF1-KPNA7 | 0.78 | 0.44 | 0.038/0.027 | 0.98 | 0.33 | 1.14 | 0.26 | 0.26 |
| 1 | rs10800309 | FCGR2A/FCGR2C | 0.63 | 0.53 | 0.32/0.34 | −0,49 | 0.63 | 0.34 | 0.74 | 0.87 |
| 5 | rs4957048 | Cep72/SLC9A3 | −0.25 | 0.81 | 0.19/0.20 | −0.14 | 0.89 | −0.29 | 0.78 | 0.66 |
| 9 | rs10758669 | JAK2 | −0.23 | 0.82 | 0.37/0.32 | 2.11 | 3.48×10-2 | 0.75 | 0.45 | 0.42 |
| 7 | rs6949033 | LAMB1 | −0.02 | 0.98 | 0.41/0.43 | −0.61 | 0.54 | −0.30 | 0.77 | 0.43 |
Joint analysis between the combined Cochrane-Mantel-Haenszel analysis of Benelux, German and Scandinavian I cohorts by Taqman assays, and an additional independent Scandinavian II cohort of which genotype data was extracted from a previous PSC GWAS. The joint analysis of the Taqman genotyped data and the GWAS data was conducted using weighted Z-scores.
In the Scandinavavian II cohort, the rs6017342 SNP showed deviation from Hardy-Weinberg equilibrium (HWE) in the controls (p< 0.004).
Therefore this SNP was discarded for the joint analysis.
Association with PSC in patients with or without IBD
Of the 984PSC cases for which IBD phenotype information was available, there were 739cases with concurrent IBD and 245 cases without it. After quality control, 732PSC cases with IBD and 241PSC cases without IBD were available for further analysis. Two of the three SNPs associated in the entire PSC cohort were also nominally associated in PSC patients without IBD: rs6822844 at 4q27 (P = 1.54×102) and rs4077515 at chromosome 9q34 (P =1.62×10−2), but they were not statistically significant after correcting for multiple testing.(Table 3)
Table 3.
Allelic association analysis in PSC cases with IBD and without IBD
| Chr | SNP | Gene | PSC patients with IBD (739 cases, 1445 controls) | PSC patients without IBD (n=241 cases, 1445 controls) | ||
|---|---|---|---|---|---|---|
| z-score | p-value | z-score | p-value | |||
| 4 | rs6822844 | IL2/21 | −3.42 | 6.21×10-4 | −2.42 | 1.54×10-2 |
| 9 | rs4077515 | CARD9 | 2.90 | 3.72×10-3 | 2.40 | 1.62×10-2 |
| 2 | rs6706689 | PUS10/REL | −3.50 | 4.73×10-4 | −0.89 | 0.37 |
| 20 | rs6017342* | HNF4A | −1.70 | 8.87×10-2 | −0.51 | 0.61 |
| 22 | rs5771069 | IL17REL | −1.45 | 0.15 | −2.12 | 3.40×10-2 |
| 16 | rs4781011 | CIITA | −0.26 | 0.80 | 1.45 | 0.15 |
| 17 | rs2305480 | ORMDL3 region | 0.59 | 0.55 | 1.33 | 0.18 |
| 16 | rs1728785 | CDH1 | −1.19 | 0.23 | 0.084 | 0.93 |
| 7 | rs7809799 | SMURF1-KPNA7 | 1.39 | 0.16 | 0.039 | 0.97 |
| 1 | rs10800309 | FCGR2A/FCGR2C | 0.19 | 0.85 | −0.28 | 0.78 |
| 5 | rs4957048 | Cep72/SLC9A3 | −0.022 | 0.98 | −0.28 | 0.78 |
| 9 | rs10758669 | JAK2 | 0.44 | 0.66 | 1.03 | 0.30 |
| 7 | rs6949033 | LAMB1 | 0.056 | 0.96 | 1.18 | 0.24 |
In PSC patients with known IBD status, subgroup analyses were performed in PSC patients with IBD and PSC patients without IBD.
In the Scandinavavian II cohort, the rs6017342 SNP showed deviation from Hardy-Weinberg equilibrium (HWE) in the controls (p< 0.004). Therefore this SNP was discarded for the joint analysis. The shown z-score and p-value for this SNP is from the CMH-test of the initial three cohorts (Benelux, German, Scandinavian I).
In silico GRAIL and eQTL analysis
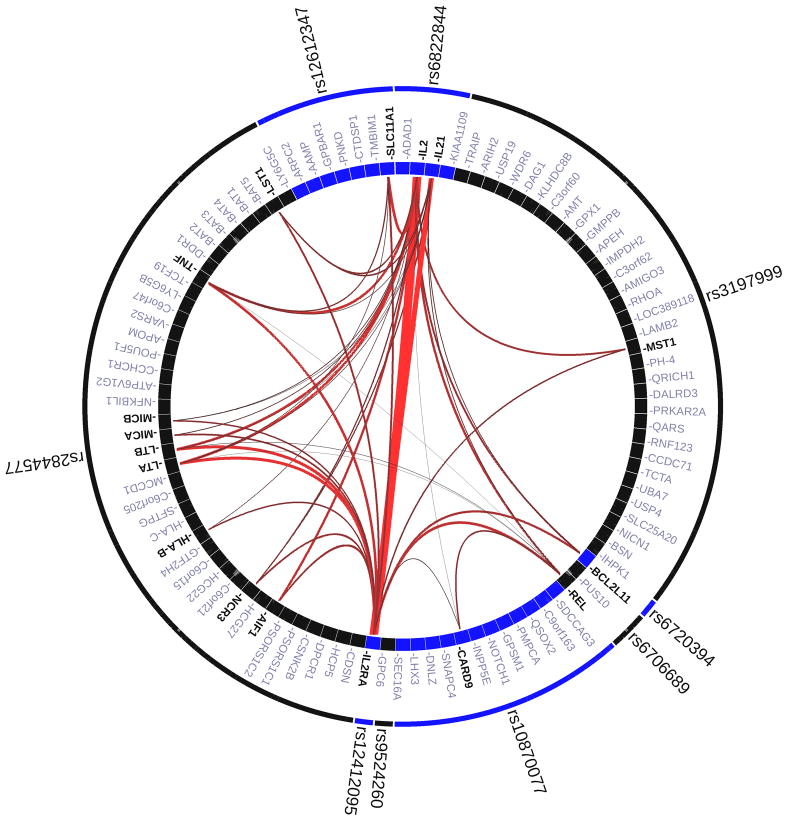
For both the GRAIL and eQTL analyses, we took the three new risk loci (rs6822844, rs4077515 and rs6706689) and added the six previously reported loci, including the HLA locus (rs6720394, rs12612347, rs3197999, rs3134792, rs10905718 and rs9524260). GRAIL shows significant interconnectivity between genes at six out of the nine PSC associated regions. Results of the GRAIL analysis are shown in Figure 1.
Figure1. Gene Relationships Across Implicated Loci (GRAIL) pathway analysis.
Links between genes at nine Primary Sclerosing Cholangitis associated loci. Specifically, all of the nine PSC-associated SNPs are in LD intervals containing, or within 50kb of at least one known gene. In total there are 100 genes implicated by proximity to these nine SNPs. Each observed PSC-association was scored with GRAIL, which takes all genes mapping within PSC-associated intervals and evaluates for each whether it is non-randomly linked to the other genes, via word-usage in PubMed abstracts. The lines between genes represent individually significant connections that contribute to the positive signal, with the thickness of the lines being inversely proportional to the probability that a literature-based connection would be seen by chance.
The mean number of P <0.05 hits in a simulated list was 0.291, with a range in the 1000 sets from zero to five. The likelihood of observing six hits with P <0.05 is therefore less than 0.1%.
Five out of the nine SNPs had a significant effect (Spearman P <0.0028; FDR 0.05) on expression of one or more nearby genes. The results of both the GRAIL and eQTL analyses are summarized in Table 4. Detailed results of the eQTL analysis for each locus are given in the online supplementary materials (Supp. Table 2 and Supp. Figs 1A-P).
Table 4.
Overview of PSC Loci and Implicated Candidate Genes
| Chr | SNPdb ID | Candidate gene | GRAIL | Cis eQTL |
|---|---|---|---|---|
| 2p16 | rs6706689 | PUS10/REL | REL | AHSA2 |
| 2q13 | rs6720394 | BCL2L11 | BCL2L11 | - |
| 2q35 | rs12612347 | GPBAR1 | SLC11A1 |
AAMP SLC11A1 IL8RA IL8RB ARPC2 |
| 3p21 | rs3197999 | MST1 | - |
UBA7 MST1///APEH RNF123///AMIGO3///GMPPB |
| 4q27 | rs6822844 | IL2/21 |
IL2 IL21 |
- |
| 6p21 | rs3134792* | HLA-B |
LTA LTB TNF AIF1 |
HCP5 |
| 9q34 | rs4077515* | CARD9 | - |
CARD9 INPP5E SDCCAG3 INPP5E///SEC16A SNAPC4 |
| 10p15 | rs10905718* | IL2RA | IL2RA | - |
| 13q31 | rs9524260 | GPC6 | - | - |
Overview of PSC loci identified in the current study and previous genetic studies in PSC.(20,21)Candidate gene: positional candidate gene or gene of interest annotated in previous studies, GRAIL: most likely candidates based on GRAIL analysis; Cis eQTL: cis-expression of one or more genes.
These SNPs were not in Hapmap r21, so perfect proxies (r2 = 1) were used in the GRAIL analysis: rs2844577 for rs3134792, rs12412095 for rs10905718, and rs10870077 for rs4077515
Discussion
In a large, northern European cohort consisting of 1186PSC cases and 1748controls we identified three UC susceptibility loci on chromosomes2p16, 4q27 and 9q34 to be associated with PSC. In silico analysis using GRAIL and eQTL analyses prioritized REL, IL2, IL21 and CARD9 as putative candidate genes from these associated loci. These results add to the scarce knowledge about the genetic background of PSC and candidate genes residing at these and previously identified loci highlight an important role for both the innate and adaptive immunological factors in disease pathogenesis.
On the 4q27 locus SNP rs62822844 resides in a non-coding region upstream of IL21 and downstream of IL2. The region shows extensive linkage disequilibrium (LD) and harbours four genes: KIAA1109, TENR, IL2 and IL21. Due to the LD structure, defining the causal variant by conventional genetic testing is difficult. From a functional perspective and from secondary interconnectivity analysis IL2 and IL21 are the most compelling candidate genes. Furthermore IL2RA, encoding the alpha subunit of the IL2 receptor, was found to be associated with PSC in a recent genome-wide analysis, suggesting an important role for the IL2 pathway in disease pathogenesis.(21) The IL2-IL21 region appears to be a general risk locus for many immune mediated diseases since it is associated with multiple diseases such as celiac disease, UC, type 1 diabetes, systemic lupus erythematosus, rheumatoid arthritis and psoriasis.(27, 34–38) IL2 serves as a T-cell growth factor, augments natural killer (NK) cell cytolytic activity and promotes immunoglobulin production by B cells. IL2 is also involved in the development of regulatory T-cells playinga role in T-cell tolerance. (39) The other possible candidate gene from the 4q27 locus is IL-21 which has a broad function, including promotion of differentiation of B cells to plasma cells, driving the expansion of CD8+ T cells, and acting as a pro-apoptotic factor for NK cells and incompletely activated B cells.(39)
The 9q34 locus constistutes an extended haplotype block spanning 120 kb including multiple genes (among others CARD9, GPSM1, SNAPC4, SDCCAG3, PMPCA, INPP5E, and KIAA0310). Although a number of genes located in this LD block are attractive candidates for association with PSC, previous reports on IBD prioritized CARD9 as the most likely candidate gene.(37) Furthermore, the associated SNP rs4077515 exerts a very strong effect on the expression of CARD9 (Cis-eQTL p-value 5.81×10−125) in our current analysis. In addition, CARD9 is interesting from a functional point of view as the encoded protein of CARD9 plays an essential role in stimulating innate immune signaling by intracellular and extracellular pathogens. After binding of fungi to the Dectin-1 receptor or bacteria to the Toll-like receptor (TLR), CARD9 stimulates T cells to differentiate into type 17 helper T cells (TH17 cells) and also promotes the pro-inflammatory cytokine production.(40, 41) TH17 cells are increasingly recognized as being involved in host defense and inducing autoimmunity and tissue inflammation.(42, 43) On chromosome 2p16 the REL gene is a possible candidate for the association with rs6706689. REL is a key mediator in NF-κB inflammatory signaling. Besides UC, this locus is also associated with celiac disease(44) and rheumatoid arthritis.(45)
From the previously identified PSC loci, TGR5, the G protein-coupled bile acid receptor 1 (GPBAR1) is a compelling candidate gene at chromosome 2q35 because of its function in bile duct homeostasis and inflammation; recent re-sequencing and functional studies have provided insight into the function of TGR5. Due to the strong linkage disequilibrium (LD), other genes at the 2q35 locus cannot be excluded as PSC-associated genes(21) and our analyses showed that SLC11A1 (NRAMP1) might also be a candidate gene. SLC11A1 encodes a multi-pass membrane protein and is involved in iron metabolism and host resistance to certain pathogens.(46, 47) and the locus is also implicated in other immune-related diseases such as rheumatoid arthritis.(48)
As PSC is strongly associated with IBD, and in particular with UC, it is difficult to identify loci specifically associated with PSC (Supp. Table 3). In the literature it is debated whether IBD in PSC patients is indeed UC or Crohn’s Disease (CD) or whether it represents a distinct entity of IBD. Disease in patients with the PSC-IBD phenotype is, for instance, more frequently characterized by rectal sparing and backwash ileitis compared to chronic UC patients.(49) Furthermore, previous candidate gene studies investigating IBD genes in PSC did not yield any associations with PSC.(50) It is compelling that two out of three loci were associated in the subgroup of PSC patients without IBD despite the low power of this subgroup analysis. These associations do not pass stringent correction for multiple testing, but the fact that the association is in the same direction as for the whole cohort suggests association irrespective of concomitant IBD. However, it is important to realize that the group of PSC patients without IBD is difficult to define precisely since the IBD in PSC can present either before the onset of PSC or up to several years after the PSC diagnosis,(1) and even sometimes after liver transplantation.(51)
Since only five non-HLA genetic risk loci for PSC had been identified before, this study provides a major addition to the current knowledge on the genetic background of PSC. The PSC susceptibility loci we identified in our study were not found in the two recently published GWAS.(20, 21) A probable reason for this is that the number of samples in the discovery sets for both GWAS were small (285 and 715 cases, respectively) and due to the general design of GWAS studies, many true positive findings will be discarded and not followed up due to stringent thresholds for multiple testing, hereby ignoring large amounts of data. In the current study, the associations reported do not reach genome wide significant levels but are robust to correction for multiple testing using a conservative Bonferroni approach. Although the current cohort is large, not reaching genome wide significance might be due to the lack of power to identify genes with smaller effect sizes than the previously reported associations.(20, 21) Since PSC is a relatively rare condition, large-scale international collaborations will be needed to identify additional genetic loci with even smaller effect sizes.
In conclusion, we have identified three UC susceptibility loci to be associated with PSC. Likely candidate genes at these and other PSC susceptibility loci highlight an important role for the innate immune system (REL, CARD9) and the adaptive immune system (IL2-IL2R pathway). Large international collaborations, using custom array-based technologies, have started fine-mapping studies aimed at identifying causal variants within the PSC-associated loci. These studies will hopefully lead to a better understanding of PSC pathogenesis and identify possible targets for therapy.
Supplementary Material
Acknowledgments
Financial Support
This study was supported by a clinical fellowship grant (90.700.281) from the Netherlands Organization for Scientific Research (NWO) to RKW. SR is supported by an NIH Career Development Award (1K08AR055688). The study was supported by The Norwegian PSC research center (http://www.rikshospitalet.no/nopsc), the German Ministry of Education and Research (BMBF): through the National Genome Research Network (NGFN) and the Integrated Research and Treatment Center Transplantation (reference number: 01EO0802). This study received infrastructure support through the Research Computing Services at the University of Oslo. L.H.v.d.B. and R.A.O acknowledge funding from the Amyotrophic Lateral Sclerosis Association. R.A.O acknowledges funding from the National Institute of Neurological Disorders and Stroke(NINDS)
We thank Jackie Senior for editing the manuscript. Dr. Benedicte A. Lie and the Norwegian Bone Marrow Donor Registry at Oslo University Hospital, Rikshospitalet are acknowledged for contributing the healthy Norwegian control population.
List of abbreviations
- PSC
primary sclerosing cholangitis
- IBD
inflammatory bowel disease
- UC
ulcerative colitis
- CD
Crohn’s disease
- GWAS
genome-wide association scan/study
- HLA
human leucocyte antigen
- BCL2L11
BCL2-like 11
- IL2RA
interleukin 2 receptor, alpha
- eQTL
expression quantitative trait
- GRAIL
Gene Relationships Across Implicated Loci
- SNP
single nucleotide polymorphism
- CEPH
Centre d’Etude du Polymorphisme Humain
- HWE
Hardy-Weinberg equilibrium
- CMH
Cochran-Mantel-Haenszel
- GEO
Gene Expression Omnibus
- PCA
principal component analysis
- FDR
false discovery rate
- OR
odds ratio
- IL2/21
interleukin 2/21
- CARD9
caspase recruitment domain-containing protein 9
- REL
v-rel reticuloendotheliosis viral oncogene homolog
- LD
linkage disequilibrium
- NK
natural killer
- TLR
toll-like receptor
- GPBAR1
G-protein coupled bile acid receptor 1
- SLC11A1
solute carrier family 11 member 1
- NRAMP1
natural resistance-associated macrophage protein 1
Footnotes
Authors’ disclosure
The authors have no financial, professional or personal conflicts of interest to declare.
References
- 1.Chapman RW, Arborgh BA, Rhodes JM, Summerfield JA, Dick R, Scheuer PJ, et al. Primary sclerosing cholangitis: A review of its clinical features, cholangiography, and hepatic histology. Gut. 1980;21:870–877. doi: 10.1136/gut.21.10.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broome U, Olsson R, Loof L, Bodemar G, Hultcrantz R, Danielsson A, et al. Natural history and prognostic factors in 305 swedish patients with primary sclerosing cholangitis. Gut. 1996;38:610–615. doi: 10.1136/gut.38.4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Card TR, Solaymani-Dodaran M, West J. Incidence and mortality of primary sclerosing cholangitis in the UK: A population-based cohort study. J Hepatol. 2008;48:939–944. doi: 10.1016/j.jhep.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Boberg KM, Aadland E, Jahnsen J, Raknerud N, Stiris M, Bell H. Incidence and prevalence of primary biliary cirrhosis, primary sclerosing cholangitis, and autoimmune hepatitis in a norwegian population. Scand J Gastroenterol. 1998;33:99–103. doi: 10.1080/00365529850166284. [DOI] [PubMed] [Google Scholar]
- 5.Bambha K, Kim WR, Talwalkar J, Torgerson H, Benson JT, Therneau TM, et al. Incidence, clinical spectrum, and outcomes of primary sclerosing cholangitis in a united states community. Gastroenterology. 2003;125:1364–1369. doi: 10.1016/j.gastro.2003.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Fausa O, Schrumpf E, Elgjo K. Relationship of inflammatory bowel disease and primary sclerosing cholangitis. Semin Liver Dis. 1991;11:31–39. doi: 10.1055/s-2008-1040420. [DOI] [PubMed] [Google Scholar]
- 7.Bergquist A, Montgomery SM, Bahmanyar S, Olsson R, Danielsson A, Lindgren S, et al. Increased risk of primary sclerosing cholangitis and ulcerative colitis in first-degree relatives of patients with primary sclerosing cholangitis. Clin Gastroenterol Hepatol. 2008;6:939–943. doi: 10.1016/j.cgh.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Chapman R, Cullen S. Etiopathogenesis of primary sclerosing cholangitis. World J Gastroenterol. 2008;14:3350–3359. doi: 10.3748/wjg.14.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, et al. Replication of signals from recent studies of crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713–715. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 11.Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 12.Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in crohn’s disease. Nat Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.UK IBD Genetics Consortium. Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franke A, Balschun T, Sina C, Ellinghaus D, Hasler R, Mayr G, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL) Nat Genet. 2010;42:292–294. doi: 10.1038/ng.553. [DOI] [PubMed] [Google Scholar]
- 17.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrumpf E, Fausa O, Forre O, Dobloug JH, Ritland S, Thorsby E. HLA antigens and immunoregulatory T cells in ulcerative colitis associated with hepatobiliary disease. Scand J Gastroenterol. 1982;17:187–191. doi: 10.3109/00365528209182038. [DOI] [PubMed] [Google Scholar]
- 19.Karlsen TH, Schrumpf E, Boberg KM. Genetic epidemiology of primary sclerosing cholangitis. World J Gastroenterol. 2007;13:5421–5431. doi: 10.3748/wjg.v13.i41.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlsen TH, Franke A, Melum E, Kaser A, Hov JR, Balschun T, et al. Genome-wide association analysis in primary sclerosing cholangitis. Gastroenterology. 2010;138:1102–1111. doi: 10.1053/j.gastro.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 21.Melum E, Franke A, Schramm C, Weismuller TJ, Gotthardt DN, Offner FA, et al. Genome-wide association analysis in primary sclerosing cholangitis identifies two non-HLA susceptibility loci. Nat Genet. 2011;43:17–19. doi: 10.1038/ng.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hov JR, Keitel V, Laerdahl JK, Spomer L, Ellinghaus E, ElSharawy A, et al. Mutational characterization of the bile acid receptor TGR5 in primary sclerosing cholangitis. PLoS One. 2010;5:e12403. doi: 10.1371/journal.pone.0012403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, et al. International Schizophrenia Consortium. Identifying relationships among genomic disease regions: Predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman R, Fevery J, Kalloo A, Nagorney DM, Boberg KM, Shneider B, et al. Diagnosis and management of primary sclerosing cholangitis. Hepatology. 2010;51:660–678. doi: 10.1002/hep.23294. [DOI] [PubMed] [Google Scholar]
- 26.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 27.Festen EA, Goyette P, Scott R, Annese V, Zhernakova A, Lian J, et al. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut. 2009;58:799–804. doi: 10.1136/gut.2008.166918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, Monsuur AJ, et al. Genetic analysis of innate immunity in crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202–1210. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 32.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawalha AH, Kaufman KM, Kelly JA, Adler AJ, Aberle T, Kilpatrick J, et al. Genetic association of interleukin-21 polymorphisms with systemic lupus erythematosus. Ann Rheum Dis. 2008;67:458–461. doi: 10.1136/ard.2007.075424. [DOI] [PubMed] [Google Scholar]
- 37.Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81:1284–1288. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Helms C, Liao W, Zaba LC, Duan S, Gardner J, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colonna M. All roads lead to CARD9. Nat Immunol. 2007;8:554–555. doi: 10.1038/ni0607-554. [DOI] [PubMed] [Google Scholar]
- 41.Underhill DM, Shimada T. A pair of 9s: It’s in the CARDs. Nat Immunol. 2007;8:122–124. doi: 10.1038/ni0207-122. [DOI] [PubMed] [Google Scholar]
- 42.LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, et al. Syk-and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 43.Tesmer LA, Lundy SK, Sarkar S, Fox DA. Th17 cells in human disease. Immunol Rev. 2008;223:87–113. doi: 10.1111/j.1600-065X.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trynka G, Zhernakova A, Romanos J, Franke L, Hunt KA, Turner G, et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut. 2009;58:1078–1083. doi: 10.1136/gut.2008.169052. [DOI] [PubMed] [Google Scholar]
- 45.Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, Kastner DL, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41:820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Variations in the NRAMP1 gene and susceptibility to tuberculosis in west africans. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 47.Abel L, Sanchez FO, Oberti J, Thuc NV, Hoa LV, Lap VD, et al. Susceptibility to leprosy is linked to the human NRAMP1 gene. J Infect Dis. 1998;177:133–145. doi: 10.1086/513830. [DOI] [PubMed] [Google Scholar]
- 48.Shaw MA, Clayton D, Atkinson SE, Williams H, Miller N, Sibthorpe D, et al. Linkage of rheumatoid arthritis to the candidate gene NRAMP1 on 2q35. J Med Genet. 1996;33:672–677. doi: 10.1136/jmg.33.8.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loftus EV, Jr, Harewood GC, Loftus CG, Tremaine WJ, Harmsen WS, Zinsmeister AR, et al. PSC-IBD: A unique form of inflammatory bowel disease associated with primary sclerosing cholangitis. Gut. 2005;54:91–96. doi: 10.1136/gut.2004.046615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karlsen TH, Hampe J, Wiencke K, Schrumpf E, Thorsby E, Lie BA, et al. Genetic polymorphisms associated with inflammatory bowel disease do not confer risk for primary sclerosing cholangitis. Am J Gastroenterol. 2007;102:115–121. doi: 10.1111/j.1572-0241.2006.00928.x. [DOI] [PubMed] [Google Scholar]
- 51.Riley TR, Schoen RE, Lee RG, Rakela J. A case series of transplant recipients who despite immunosuppression developed inflammatory bowel disease. Am J Gastroenterol. 1997;92:279–282. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.