Abstract
Inflammation is common to many disorders and responsible for tissue and organ damage. In many disorders, the associated peripheral cytokine milieu is dilute and difficult to measure, necessitating development of more sensitive and informative biomarkers for mechanistic studies, earlier diagnosis, and monitoring therapeutic interventions. Previously, we have shown that plasma of recent-onset (RO) Type 1 diabetes patients induces a disease-specific proinflammatory transcriptional profile in fresh peripheral blood mononuclear cells (PBMC) compared with that of healthy controls (HC). To eliminate assay variance introduced through the use of multiple donors or multiple draws of the same person over time, we evaluated human leukemia cell lines as potential surrogates for fresh PBMC. We 1) tested seven different cell lines in their power to differentiate RO from HC plasma and 2) compared the similarity of the signatures generated across the seven cell lines to that obtained with fresh PBMC. While each cell line tested exhibited a distinct transcriptional response when cultured with RO or HC plasma, the expression profile induced in any single cell line shared little identity with that of the other cell lines or fresh PBMC. In terms of regulated biological pathways, the transcriptional response of each cell line shared varying degrees of functional identity with fresh PBMC. These results indicate that use of human leukemia cell lines as surrogates for fresh PBMC has potential in detecting perturbations to the peripheral cytokine milieu. However, the response of each is distinct, possessing varying degrees of functional relatedness to that observed with PBMC.
Keywords: gene expression analysis, NCI60 leukemia cell lines, cytokines
genomic technologies including expression arrays offer unprecedented opportunities to advance our diagnostic capabilities in human disease. The circulatory and lymphatic systems that flow through all parts of the body are the basis for blood being the most frequently sampled human tissue. It is within the field of oncology where, arguably, microarrays have made the largest clinical impact. In this venue they have been found sufficiently accurate to diagnose and classify acute lymphoblastic leukemia (ALL) subtypes as well as guide the selection of the most appropriate treatment modality (40, 51, 62). This success has been facilitated by the fact that ALL is, at least initially, a clonal disease, and relevant sample (peripheral blood) is readily available. Microarrays have made a lesser impact in most other clinical situations where it is often not possible to readily biopsy the most relevant tissue.
Two general array-based strategies have been successfully applied to human diseases that utilize peripheral blood sampling. The more common approach has been to directly profile peripheral blood mononuclear cells (PBMC) or purified immunocyte subsets of cases and controls (46). This approach relies on the fact that lymphocytes migrate to and from sites of injury/infection/inflammation as well as circulate through lymphoid organs and requires that a sufficient percentage of peripheral cells have transcriptionally responded in a disease-specific manner to enable detection. The less common approach has been to use subject serum or plasma to induce gene expression in healthy, unrelated third-party PBMC (32, 45, 60). In this application, the cells are used as reporters that sensitively respond to disease-associated factors present in the serum/plasma.
Previously, we reported that plasma of recent-onset (RO) Type 1 diabetes (T1D) patients induce a unique proinflammatory gene expression profile in fresh unrelated PBMC relative to that induced by plasma collected from healthy controls (HC) (60). This signature included innate immunity genes and genes regulated by IL-1. These studies utilized samples collected during the first 6 mo postonset, a period termed the “honeymoon,” during which there remains residual endogenous insulin production as well as ongoing autoimmunity toward the remaining pancreatic β-cells. The RO signature was distinct from that induced by plasma of unrelated HC, long-standing T1D patients, or patients possessing other inflammatory diseases (45, 60). Presently, the detection of auto-antibodies toward β-cell antigens is the best prognostic indicator of T1D risk. Importantly, in the retrospective analysis of a limited number of longitudinal samples, the RO transcriptional signature was evident as early as five years prior to T1D onset, before detection of auto-antibodies (60). Thus, our studies support the hypothesis that the peripheral cytokine milieu associated with autoimmunity toward the pancreatic β-cells is sufficient to induce a unique T1D-specific transcriptional profile in healthy responder cells; this signature reflects active autoimmunity, is disease specific, may improve disease prediction beyond islet auto-antibodies, and is potentially mechanistically informative (60).
Applying this strategy to precious sample repositories as well as any future clinical application in T1D or other disease will require definition and control of the assay's variables. Among these is the influence of the responder cells. Our previous studies utilized fresh PMBC isolated from >15 healthy blood donors (60). Despite the potential heterogeneity in the responsiveness of PBMC isolated from different donors, we found subject plasma within a given subject cohort to induce remarkably homogeneous profiles. However, we anticipate that eliminating or minimizing the heterogeneity in the responder cell used in assays conducted over time should result in greater sensitivity to detect early disease and better specificity to distinguish related inflammatory states. Furthermore, use of a single, widely available responder cell type would serve to standardize the approach, making results generated over time and across multiple laboratories more comparable than those of fresh PBMC collected from multiple donors or even multiple draws of the same individual.
While they differ from their native counterparts, cell lines derived from human tissues and tumors have been extensively used by laboratories for decades as models to study essentially all aspects of human biology (12, 21, 35, 38, 43, 44, 52). The National Cancer Institute's Developmental Therapeutics Program has carried out extensive studies of 60 cancer cell lines (the NCI60) derived from a variety of tissues (50) (http://dtp.nci.nih.gov). Based upon their myeloid and lymphoid origins, in this report, we evaluate the transcriptional responses of seven NCI60 cell lines as potential surrogates for fresh PBMC in our transcriptional bioassay.
MATERIALS AND METHODS
Subjects and subject characterization.
Caucasian RO T1D patients (n = 20) were recruited through Children's Hospital of Wisconsin. Diabetes was defined according to World Health Organization criteria and included blood glucose levels of >200 mg/dl with symptoms confirmed by a physician (1). Samples of normoglycemic RO T1D patients were collected after stabilization on exogenous insulin (2–7 mo after diagnosis). Healthy Caucasian control (n = 20) recruitment criteria included fasting blood glucose of <100 mg/dl, no familial history of any autoimmune/autoinflammatory disorder, <39 yr of age, and negativity for islet auto-antibodies at the 99th percentile (61). All study subjects were free of known infection at the time of sample collection.
All peripheral blood samples were drawn by trained phlebotomists at Children's Hospital of Wisconsin. Peripheral blood was aseptically collected in acid citrate dextrose solution A (RO T1D and HC subjects) or K+EDTA (healthy blood donors for responder PBMC isolation) anticoagulant, and components were immediately separated by Ficoll-Histopaque density gradient centrifugation. Plasma was stored at −80°C until use. Auto-antibody titers for glutamic acid decarboxylase (GAD), protein tyrosine phosphatase-2 (IA2), and insulin (IAA) were determined as previously described (61). Genotyping of both HLA-DQA1 and HLA-DQB1 loci in all subjects was performed by direct sequencing of the second exon. HLA-DQB1 genotypes were determined with the SeCore DQB1 sequencing kit in accordance with the manufacturer's instructions (Invitrogen Life Technologies, Brown Deer, WI) and HLA-DQA1-DQB1 haplotypes were inferred using reported European Caucasian haplotype frequencies (34). The study was approved by the Institutional Review Board (IRB) of the Children's Hospital of Wisconsin (IRB 01-15), and informed consent was obtained from parents/legal guardians. Subject characteristics are shown in Table 1.
Table 1.
Subject characteristics
| Antibody Status* |
HLA Genotypes |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample Identifier | Sex | HgA1c, % | GAD | IA2 | IAA† | Age at Diagnosis, yr | Sample Relative to Onset | DQA1 | DQB1 |
| Recent Onset T1DM | |||||||||
| RO1 | male | 8.8 | 0.0500 | 0.3562 | 0.0772 | 9.75 | +2.3 mo | 0102/0501 | 0201/0502 |
| RO2 | female | 7.3 | 0.0120 | 0.9537 | 2.4592 | 6.24 | +5.9 mo | 0301/0301 | 0302/0302 |
| RO3 | female | 8.0 | 0.0373 | 0.3809 | 1.0658 | 8.99 | +4.2 mo | 0501/0401 | 0201/0402 |
| RO4** | female | 7.1 | 0.1644 | 0.0014 | 0.1225 | 8.44 | +3.8 mo | 0501/0501 | 0201/0201 |
| RO5 | female | 7.0 | 0.0660 | 0.2404 | 0.6100 | 7.06 | +2.2 mo | 0501/0301 | 0201/0302 |
| RO6 | female | 7.2 | 0.3078 | 0.0165 | 3.2237 | 5.70 | +3.8 mo | 0103/0201 | 0202/0603 |
| RO7 | male | 5.9 | −0.0004 | −0.0046 | 7.2202 | 5.76 | +6.0 mo | 0501/0401 | 0201/0402 |
| RO8 | female | 7.2 | 0.0022 | 0.5050 | 0.6193 | 5.21 | +4.2 mo | 0501/0301 | 0201/0302 |
| RO9 | female | 6.5 | 0.0736 | 0.4490 | 0.0397 | 7.65 | +4.3 mo | 0301/0102 | 0302/0604 |
| RO10 | female | 8.8 | 0.6031 | 0.7622 | 4.5672 | 5.39 | +5.5 mo | 0101/0501 | 0201/0501 |
| RO11** | female | 9.8 | 0.6068 | 0.7542 | 0.6435 | 9.75 | +0.9 mo | 0301/0501 | 0201/0302 |
| RO12 | male | 7.5 | 1.3191 | 0.5670 | 3.6172 | 6.82 | +4.4 mo | 0301/0501 | 0201/0302 |
| RO13 | male | 7.7 | 0.0327 | 0.5428 | 5.2753 | 5.42 | +4.2 mo | 0301/0501 | 0302/0201 |
| RO14** | female | 5.5 | 0.1312 | 0.1777 | 1.9395 | 9.38 | +3.2 mo | 0302/0501 | 0303/0301 |
| RO15 | male | 7.7 | 0.0615 | −0.0003 | 0.1669 | 7.45 | +3 mo | 0302/0501 | 0301/0201 |
| RO16 | female | 6.9 | 0.7122 | 0.8799 | 0.4855 | 7.62 | +2.9 mo | 0201/0302 | 0202/0301 |
| RO17 | male | 7.0 | 1.1043 | 0.0005 | 0.6271 | 7.87 | +3.8 mo | 0501/0501 | 0201/0201 |
| RO18 | female | 7.6 | 0.0760 | 0.7319 | 2.3014 | 5.14 | +3.6 mo | 0201/0301 | 0202/0302 |
| RO19** | male | 7.9 | 0.1275 | 0.0036 | 4.9503 | 6.21 | +6.0 mo | 0101/0501 | 0501/0201 |
| RO20 | female | 8.8 | 0.7234 | 0.0073 | 0.2875 | 7.76 | +4.7 mo | 0102/0501 | 0502/0201 |
| Healthy Controls | |||||||||
| HC1 | male | N/A | 0.0046 | −0.0003 | −0.0035 | 18.71‡ | 0102/0401 | 0402/0502 | |
| HC2** | female | N/A | 0.0155 | −0.0023 | 0.0035 | 28.67‡ | 0501/0501 | 0201/0201 | |
| HC3** | female | N/A | 0.0150 | −0.0022 | 0.0043 | 27.73‡ | 0102/0101 | 0501/0602 | |
| HC4 | female | N/A | 0.0166 | −0.0003 | 0.0137 | 19.07‡ | 0102/0501 | 0201/0602 | |
| HC5 | female | N/A | 0.0065 | −0.0002 | 0.0096 | 24.20‡ | 0302/0103 | 0301/0603 | |
| HC6 | female | N/A | 0.0144 | −0.0008 | 0.0144 | 30.96‡ | 0102/0501 | 0604/0201 | |
| HC7 | male | N/A | −0.0063 | −0.0001 | 0.0067 | 28.09‡ | 0302/0501 | 0301/0301 | |
| HC8 | male | N/A | −0.0044 | −0.0014 | 0.0071 | 20.07‡ | 0103/0501 | 0301/0601 | |
| HC9 | male | N/A | −0.0098 | −0.0043 | 0.0000 | 23.01‡ | 0101/0102 | 0501/0604 | |
| HC10 | female | N/A | no results | 23.87‡ | 0501/0501 | 0201/0301 | |||
| HC11 | male | N/A | 0.0109 | −0.0013 | 0.0097 | 28.16‡ | 0101/0104 | 0501/0503 | |
| HC12 | female | N/A | 0.0163 | −0.0021 | 0.0067 | 25.04‡ | 0102/0301 | 0602/0302 | |
| HC13 | male | N/A | 0.0126 | −0.0003 | 0.0022 | 25.49‡ | 0101/0501 | 0501/0201 | |
| HC14** | female | N/A | 0.0166 | 0.0004 | 0.1812 | 21.3‡ | 0201/0501 | 0202/0201 | |
| HC15 | female | N/A | −0.0023 | −0.0001 | 0.0055 | 26.23‡ | 0201/0501 | 0202/0301 | |
| HC16 | female | N/A | 0.0018 | −0.0012 | −0.0068 | 25.47‡ | 0101/0101 | 0501/0501 | |
| HC17** | female | N/A | 0.0088 | −0.0010 | 0.0088 | 23.3‡ | 0102/0201 | 0602/0303 | |
| HC18 | male | N/A | −0.0027 | −0.0025 | −0.0081 | 25.22‡ | 0104/0201 | 0503/0202 | |
| HC19 | male | N/A | −0.0003 | −0.0011 | −0.0224 | 30.76‡ | 0102/0102 | 0502/0604 | |
| HC20 | female | N/A | −0.0017 | −0.0020 | 0.0054 | 23.45‡ | 0103/0501 | 0603/0201 | |
Threshold cutoffs for GAD, IA2, and IAA were 0.07, 0.007, and 0.05 respectively. Samples exceeding cutoffs shown in boldface;
Insulin autoantibody;
age at sample collection;
among the 4 samples assayed independently. RO, recent onset; HC, healthy control; N/A, not available.
PBMC and cell line cultures.
Seven myeloid/lymphoid cell lines (K562, chronic myelogenous leukemia; IM9, B-lymphoblastic leukemia; HL-60, acute myeloid leukemia; KG-1, erythroleukemia; CCRF-CEM, T-acute lymphoblastic leukemia; THP-1, acute monocytic leukemia; U-937, myelomonocytic lymphoma) were purchased from the American Tissue Culture Collection (Manassas, VA). These were selected on the basis of their original cell type, properties, and characteristics (Table 2). Cells were grown in complete media per the provider's product information sheets with 100 U/ml penicillin and 100 μg/ml streptomycin. The induction of gene expression was accomplished by culturing each of the seven cell lines for 6 h at 37°C in 5% CO2 with pooled HC plasma or pooled RO plasma (n = 20 individual plasma/pool). Cultures were prepared in a Costar 24-well plate (Corning, Corning, NY) using 500,000 cells/well in 400 μl of RPMI 1640 medium plus 100 μl (20%) plasma. When inducing gene expression in the cell lines with the RO and HC plasma pools, three independent replicates were generated for each experimental condition (each cell line was cultured in triplicate with the RO or HC plasma pool). When inducing gene expression in fresh PBMC, we generated three independent replicates with both the RO and HC plasma pools. When culturing fresh PBMC with individual plasma samples, we independently analyzed plasma of four RO and four HC subjects (members of the respective pools) as previously described (60). After culture, the content of each well (independent replicate) was centrifuged, and the pellet lysed/resuspended by vortexing in 1 ml of TRIzol (Invitrogen).
Table 2.
Cell line characteristics
| Patient Characteristics |
||||||||
|---|---|---|---|---|---|---|---|---|
| Cell Line | Morphology: Cell Type | Organ | Antigen Expression | Disease | Age, yr | Sex | Race | Other Characteristics |
| K562 | lymphoblast: B lymphocyte | bone marrow | CD7 | chronic myelogenous leukemia (CML) in terminal blast crisis | 53 | female | Caucasian | erythroleukemia cell line; lacks MHC complex; highly undifferentiated and are of the granulocytic series; nonadherent and rounded cells; inducible DC properties |
| CCRF-CEM | lymphoblast: T lymphocyte | blood | CD3, CD4, CD5, CD7 | acute lymphoblastic leukemia | 4 | female | Caucasian | polymorph cells, big nuclei; formation of microvilli |
| IM9 | lymphoblast: B lymphocyte | blood | CD11a, CD19, CD20, CD38-, CD49e | multiple myeloma | data not available | female | Caucasian | EBV transformed |
| HL-60 | myeloblast: promyelocyte | blood | acute promyelocytic leukemia | 36 | female | Caucasian | cellular products: TNF-a, after stimulation with PMA; inducible DC properties | |
| KG-1 | myeloblast: myelo-monocyte | bone marrow | HLA A30, A31, B35, Cw4, CD34, CD86, CD13, CD33 | erythroleukemia that developed into acute myelogenous leukemia | 59 | male | Caucasian | develops DC phenotype in response to cytokines or PMA; expresses TNFR1 and TNFR2 |
| U-937 | monocyte; myeloid lineage; myelo-monocyte | Fas | histiocytic lymphoma | 37 | male | Caucasian | cellular products: lysozyme; beta 2 microglobulin, secretes TNF-a, after stim. with PMA; constitutively produces IL-1; receptors: complement (C3) | |
| THP-1 | monoblast: monocyte | blood | HLA A2, A9, B5, DRw1, DRw2 | acute monocytic leukemia | 1 | male | Caucasian | expresses Fc and C3b; lack surface and cytoplasmic immunoglobulins; produce IL-1; are phagocytic; large, round, single cells; develops DC phenotype |
RNA extractions and GeneChip analysis.
Total RNA was extracted for each replicate; cRNA was synthesized, labeled, fragmented, and independently hybridized to the array in accordance to the Affymetrix GeneChip expression analysis technical manual (Affymetrix, Santa Clara, CA). The GeneChip human genome U133 plus 2.0 array was selected for these studies for its comprehensive coverage. In terms of controlling technical factors, 1) all cell lines were grown within a 1 mo period of time and challenged with aliquots of the same plasma pools; 2) RNA labeling and array hybridizations were conducted within a 1 mo period where array and labeling reagent lot numbers were controlled; and 3) RO and HC cultures for each cell line were analyzed in a paired fashion. After hybridization, arrays were washed and stained with Affymetrix fluidics protocol FS450_0001 and scanned with a 7G Affymetrix GeneChip Scanner. Image data were analyzed with Affymetrix GeneChip Microarray Suite software and normalized with Robust Multichip Analysis (http://www.bioconductor.org/) to determine signal log ratios. The statistical significance of differential gene expression and false discovery rates (FDR) were derived through a Rank Product algorithm available as an analysis package in R (25).
Pathway enrichment analysis was performed with DAVID (Database for Annotation, Visualization, and Integrated Discovery) (13, 26), which conducts an overrepresentation analyses of the functional gene categories detected relative to the total functional gene categories assayed by the array using the Gene Ontology (GO) project databases as references. Identification of genes belonging to regulated pathways common to the responses in PBMC, and the various cell lines also relied on ToppGene (Transcriptome Ontology Pathway PubMed based prioritization of Genes; Refs. 10, 11), which utilizes human gene annotations, mouse phenotype data, and literature cocitations for human complex disease candidate gene identification and prioritization. Hierarchical clustering was conducted with Genesis (58) and principal component analysis (PCA) was performed with Partek Genomics Suite (http://www.partek.com/). The data discussed in this publication have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) (19) and are accessible through GEO Series accession number GSE24147.
RESULTS
Our objectives in this study were to determine 1) whether plasma of RO and HC induced distinct transcriptional programs in the seven selected cell lines and 2) whether the gene expression signature induced by RO versus HC plasma in any of the evaluated cell lines shared identity to that observed when using freshly isolated PBMC as the responder cell.
An obstacle to studying human disease, in particular pediatric diseases, is sample availability and volume. This study, being one of assay development, necessitated that the transcriptional responses of each cell line be assessed with the same RO and HC plasmas while not depleting banked samples. Thus RO and HC plasma pools were created, each from the small contribution of 20 subjects (Table 1). To ensure induced transcription was not related to elevated blood glucose levels, only plasma of RO subjects possessing normal blood glucose levels and histories of good glycemic control (HbA1c : mean ± SD : 7.5% ± 1.01) were used in generating the pools.
Plasma pools recapitulate transcriptional patterns induced by individual RO and HC samples.
To assess their validity, fresh PBMC of a healthy male Caucasian were cultured with the RO and HC plasma pools (in triplicate) to induce transcription as previously described (60). These PBMC were also cultured with individual plasma samples of four RO and four HC (members of the RO and HC pools each analyzed a single time) as well as autologous (self) plasma. For either the pooled or individual conditions, the transcriptional responses of healthy PBMCs to HC plasma or RO plasma were normalized with those induced by autologous plasma. The objective of this study was to assess the transcriptional response of cell lines to RO and HC plasma, specifically, in their ability to capture the proinflammatory signature we previously observed when using fresh PBMC as responder cells. This signature consisted of 233 regulated probe sets (192 unique UniGenes) that exhibited a log2 ratio > 0.58 (1.5-fold) and significance thresholds of P < 0.01 (Student's t-test) and FDR < 0.01 between the RO and HC groups (60). Therefore, this subset of genes was used to assess the correlativeness of the transcription induced by the pooled plasma to that induced by 1) the four individual RO and HC plasmas cultured with the same PBMC of a single donor, as well as 2) the 16 RO and 16 HC plasma individually assayed on a total of seven different PBMC donors [i.e., the four RO and HC examined here as well as the 12 RO and 12 HC examined previously (60)]. When we examined the autologous normalized RO intensities, Pearson's correlation coefficients of 0.84 and 0.89 were respectively observed between the RO pool versus the four individual RO plasma assayed in parallel and the 16 RO plasma assayed in total. When we examined the autologous normalized HC intensities, Pearson's correlation coefficients of 0.90 and 0.75 were respectively observed between the HC pool versus the four individual HC plasma assayed in parallel and the 16 HC plasma assayed in total. Thus, the RO and HC pools induced similar transcriptional responses to that of the individual plasma in the healthy PBMC drawn for this study, supporting the use of the pools for testing the response of the cell lines.
Relationship of baseline expression patterns to known histological origins and phenotypic characteristics.
The RO and HC plasma pools were then used to induce transcription in seven cell lines (triplicate RO and HC cultures, three independent replicates per condition, six arrays total/cell line). Each of the seven cell lines possesses distinct features: K562, CCRF-CEM, and IM9 are of lymphoid origin, while HL-60, KG-1, U-937, and THP-1 are of myeloid origin. The cell lines also differ based on sex of the source patient, neoplastic disease from which they were derived, cell surface antigen expression, and other features (Table 2). To determine whether the response of each cell line was related to one or more of its phenotypic variables, the gene expression profile induced by RO or HC plasma was compared across cell lines. Specifically, for each cell line, a ratio was calculated between the mean normalized intensity value of a gene across the triplicate samples and the mean expression of that gene across all cell lines (7 cell lines × 3 replicates per cell line = 21 data points). Next, the dataset was filtered to include only those transcripts that exhibited differential expression (|log2 ratio| > 0.5) between the two conditions in at least two of the seven cell lines. Hierarchical clustering (not shown) did not reveal any relationship among the induced signatures based on common histological, phenotypic, or patient characteristics in response to either HC or RO plasma.
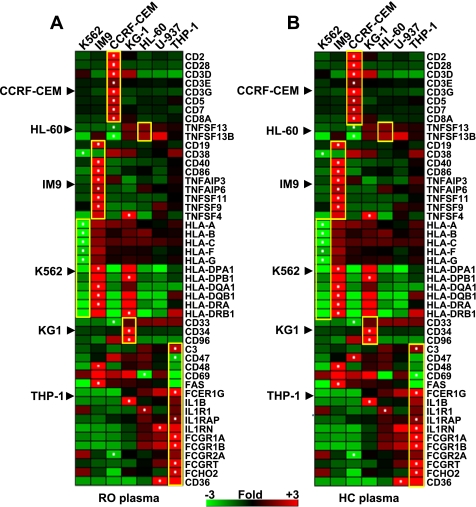
Next, from these two datasets of baseline expression ratios (in response to HC or RO plasma), a set of 49 genes was identified based upon previously known gene or cell surface antigen expression in one or more of the cell lines. In response to both RO plasma (Fig. 1A) and HC plasma (Fig. 1B), the observed gene expression trends strongly correlated with those reported in previous studies. Moreover, this response was similar with either RO or HC plasma, indicating that transcriptional response of this subset of genes reflected the nature of the individual cell lines. Specifically, CCRF-CEM has been reported to express CD3, CD4, CD5, and CD7 (6) consistent with its observed overexpression of these genes relative to the other cell lines in the present study. As reported (47), we also observed increased expression of CD19, CD40, and CD86 and downregulation of CD38 in the IM9 cell line. Several HLA class I and class II genes were highly underexpressed in K562, consistent with its known lack of MHC expression (33). Paralleling the known positivity of the KG-1 cell line for CD33, CD34, and CD96 (8, 35, 57), we observed high expression of these genes relative to the other cell lines. In general, for this limited subset of genes, the exhibited expression trends observed here parallel the published literature and are consistent with their known cellular characteristics.
Fig. 1.
Expression trends in a set of genes specific to each cell line. The gene expression profile induced by recent-onset (RO, A) and healthy control (HC, B) plasma was used to compare the cell lines directly. A baseline expression ratio was calculated for each gene across all cell lines (mean normalized intensity value across the triplicate samples per cell line to the mean expression of that gene across all cell lines). One-way hierarchical clustering (genes only) on baseline expression ratios was carried out on a set of 49 genes selected based upon previously known gene or cell surface antigen expression in one or more of the cell lines. A block of genes bound by a yellow box show the genes specific to a cell line (shown on the left of the heat map). A white dot in a cell denotes the cell line in which maximum differential expression was observed for that gene across the 7 cell lines. The scale denotes fold of change relative to the mean normalized intensity value across all conditions.
Cell lines display expression signatures that can distinguish RO from HC plasma.
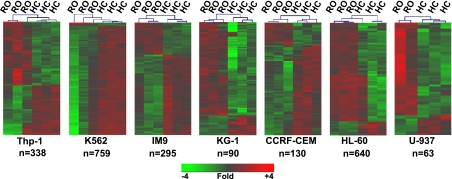
We examined the gene expression signature induced after culturing cells with RO versus HC plasma (the RO:HC ratio) for each of the seven cell lines. Gene lists were defined as those probe sets exhibiting a |log2 ratio| > 0.5, (mean absolute 1.4-fold change between RO and HC inductions) and an FDR < 10%. The number of significantly regulated genes varied from a low of 63 genes in U-937 to a high of 759 in K562. Consistent with our observations with fresh PBMC (60), each cell line exhibited a distinct response when cultured with either RO or HC plasma (Fig. 2).
Fig. 2.
Each of the 7 cell lines display distinct expression signatures distinguishing the response of RO plasma and HC plasma. For each cell line, lists of differentially expressed genes were defined as probe sets exhibiting a |log2 ratio| > 0.5, (mean absolute 1.4-fold change between RO and HC inductions) and a rank product false discovery rate < 10%. Two-way hierarchical clustering was conducted using these gene lists differentially regulated in each cell line. The lineage of each cell line and the number of genes meeting the cutoff for differential expression are indicated below each cluster picture. For the U-937 cell line, 1 of the 3 RO samples was removed as it was flagged as an outlier during the initial QC analysis. The scale denotes fold of change relative to the intensity value across all replicates of each cell line.
Differential gene expression induced in cell lines exposed to RO and HC plasma is distinct from those observed in fresh PBMC.
To further assess the suitability of each cell line as a possible surrogate for fresh PBMC in detecting the inflammatory state associated with T1D, we examined the differential gene expression induced upon culture with RO versus HC plasma (the RO:HC ratio) for its concordance and correlation with those regulated when using fresh PBMC responder cells. For each condition (the 16 RO and 16 HC plasmas individually assayed on a total of seven different PBMC donors; the four individual RO and HC plasmas cultured with the fresh PBMC drawn from a single donor for this study; the pooled RO and HC plasma cultured with the fresh PBMC drawn from a single donor for this study; and the pooled RO and HC plasma cultured with each of the seven cell lines) genes meeting the thresholds of |log2 ratio| > 0.5 and FDR <10% were identified and compared. Table 3 lists the number of commonly identified genes and the mean Pearson's correlation coefficients between signatures generated under each of the possible pairings of the various conditions.
Table 3.
Concordance and mean correlation between RO:HC signatures generated in fresh PBMC vs. cell lines
| 16RO:16HC Multi-PBMC | RO:HC Pools Single PBMC | 4RO:4HC Single PBMC | Thp1 | K562 | IM9 | CCRF-CEM | HL-60 | KG1 | U937 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 16RO:16HC multi-PBMC | 496 (100)a | |||||||||
| 496 (100)b | ||||||||||
| 1.00c | ||||||||||
| RO:HC pools single PBMC | 112 (22.6) | 765 (100) | ||||||||
| 99 (88.4) | 765 (100) | |||||||||
| 0.35 | 1.00 | |||||||||
| 4RO:4HC single PBMC | 177 (36.7) | 118 (15.4) | 653 (100) | |||||||
| 168 (94.9) | 107 (90.7) | 653 (100) | ||||||||
| 0.76 | 0.46 | 1.00 | ||||||||
| Thp1 | 16 (3.2) | 28 (3.7) | 19 (2.9) | 338 (100) | ||||||
| 6 (37.5) | 8 (28.6) | 7 (36.8) | 338 (100) | |||||||
| −0.12 | −0.12 | −0.10 | 1.00 | |||||||
| K562 | 17 (3.4) | 86 (11.2) | 13 (2.0) | 36 (10.7) | 759 (100) | |||||
| 8 (47.1) | 1 (1.2) | 8 (61.5) | 34 (94.4) | 759 (100) | ||||||
| −0.01 | −0.16 | −0.02 | 0.35 | 1.00 | ||||||
| IM9 | 16 (3.2) | 17 (2.2) | 6 (0.9) | 11 (3.3) | 19 (2.5) | 295 (100) | ||||
| 15 (93.8) | 4 (23.5) | 0 (0) | 5 (45.5) | 11 (57.9) | 295 (100) | |||||
| 0.20 | −0.09 | −0.16 | −0.14 | 0.00 | 1.00 | |||||
| CCRF-CEM | 2 (0.4) | 5 (0.7) | 3 (0.5) | 10 (3.0) | 7 (0.9) | 32 (10.8) | 130 (100) | |||
| 2 (100) | 0 (0) | 2 (66.7) | 8 (80.0) | 5 (71.4) | 32 (100) | 130 (100) | ||||
| 0.20 | −0.10 | 0.01 | 0.32 | −0.10 | 0.44 | 1.00 | ||||
| HL60 | 25 | 124 (16.2) | 27 (4.1) | 35 (10.4) | 145 (19.1) | 22 (7.5) | 12 (9.2) | 640 (100) | ||
| 5.0 (36.0) | 120 (96.8) | 12 (44.4) | 5 (14.3) | 2 (1.4) | 8 (36.4) | 9 (75.0) | 640 (100) | |||
| −0.02 | 0.27 | 0.02 | −0.21 | −0.49 | 0.05 | 0.20 | 1.00 | |||
| KG1 | 7 (1.4) | 10 (1.3) | 8 (1.2 | 4 (1.2) | 4 (0.5) | 2 (0.7) | 2 (1.5) | 5 (0.8) | 90 (100) | |
| 4 (57.1) | 10 (100) | 4 (50.0) | 2 (50.0) | 0 (0) | 0 (0) | 0 (0) | 5 (100) | 90 (100) | ||
| −0.03 | 0.24 | 0.11 | −0.16 | −0.33 | −0.26 | −0.09 | 0.54 | 1.00 | ||
| U937 | 2 (0.4) | 7 (0.9) | 2 (0.3) | 5 (1.5) | 24 (3.2) | 2 (0.7) | 0 (0) | 23 (3.6) | 1 (1.1) | 63 (100) |
| 0 (0) | 7 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 23 (100) | 1 (100) | 63 (100) | |
| −0.07 | 0.23 | −0.02 | −0.29 | −0.54 | −0.15 | −0.03 | 0.55 | 0.49 | 1.00 |
Each cell within the table provides the statistics pertaining to the 2-way comparison between the intersecting (top vs. side) conditions.
Total number of genes (percentage) commonly regulated between both conditions (RO:HC |log2 ratio| >0.5, false discovery rate < 0.10).
total number of genes (percentage) commonly regulated under both conditions that are directionally concordant.
mean Pearson's correlation coefficient between the 2 gene lists compared. PBMC, peripheral blood mononuclear cell.
When comparing all conditions to the signature induced by the independently analyzed 16 RO and 16 HC plasmas, we find that signatures generated with fresh PBMC identified more commonly regulated genes and exhibited higher correlations (Table 3, first data column). Clearly, when the RO:HC ratios are examined, limitations of the pooling strategy are evident, as the signature of the RO and HC pools on fresh cells exhibited the lowest correlativeness among the conditions using fresh PBMC. The signatures generated on the cell lines exhibited little overlap and were even less correlative.
When comparing all the conditions to the signature induced by the RO and HC pools in fresh PBMC, we again observed the highest correlations with the other conditions where fresh PBMC were used (Table 3, second data column). The highest number of commonly regulated genes induced with the plasma pools was observed with HL-60 (120/765), which also showed the best correlation coefficient among the cell lines (0.27). While HL-60 exhibited the second highest number of differentially expressed genes among the cell lines (n = 640, Fig. 2), a relationship between the number of genes differentially regulated by RO versus HC plasma in a cell line and its correlativeness with assays conducted on fresh PBMC cannot be drawn, as K562 regulated the highest number of genes (n = 759, Fig. 2) yet shared little identity with signatures generated in fresh PBMC. Among the seven cell lines themselves, little identity was observed between the signatures generated in terms of the number of commonly regulated genes meeting the fold of change and statistical cutoffs. However, it must be noted that identity exists in the RO:HC signatures of U-937, KG-1, and HL-60, as reflected by the Pearson's correlation coefficients among these cell lines. This may be related to their common myeloid lineage.
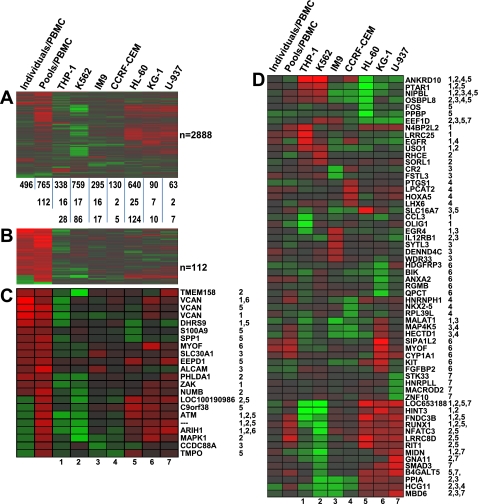
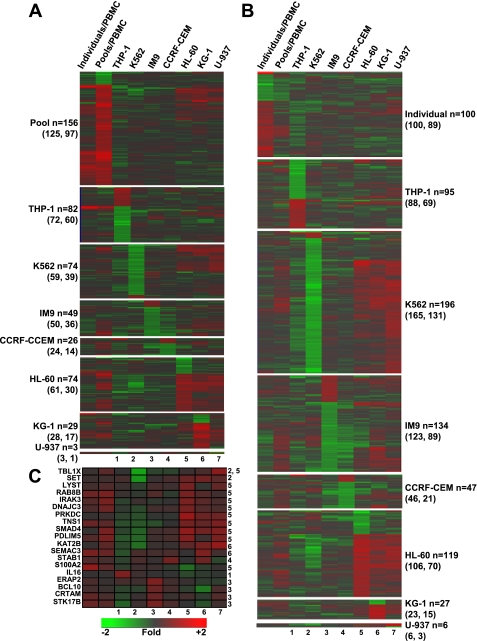
One-way hierarchical clustering was conducted using the union of the differentially expressed genes identified when 1) the 16 RO and 16 HC plasmas were individually assayed on seven different PBMC donors, 2) the pooled RO and HC plasma were assayed on PBMC of a single donor, and 3) the pooled RO and HC plasma were cultured with each of the seven cell lines (n = 2,888 genes) to identify groups of genes that may be similarly regulated across the various conditions (Fig. 3A). This analysis shows that genes commonly regulated by the pooled and individual RO and HC plasmas in fresh PBMC, in general, are not regulated by the cell lines in the same manner. The analysis did reveal a group of genes that are upregulated in fresh PBMC as well as HL-60, KG-1, and U-937 as a function of being challenged with the pooled samples. This analysis also identified genes commonly regulated in fresh PBMC by both individual and pooled plasma, which likely represent the genes most reliably regulated by T1D plasma (those grouped at the top of Fig. 3A). When we focus only on the 112 genes commonly regulated in fresh PBMC by both individual and pooled plasma meeting the statistical and fold-change thresholds (Fig. 3B), it is clear that for this subset of genes the cell line signatures are highly distinct. From this subset of 112 genes, we identified only 22 genes (probe sets) that were significantly expressed in at least one of the seven cell lines (Fig. 3C). Even among this select subset, no significant directional concordance across all cell lines was observed. Finally, the most highly regulated genes (n = 10/cell line, in terms of fold of change and meeting the statistical threshold) within each cell line were extracted and subjected to one-way hierarchical clustering (Fig. 3D). Again, none of the top differentially regulated genes showed any significant similarity to the expression trends observed when using fresh PBMC as responders.
Fig. 3.
Comparison of expression signatures between cell lines and the fresh peripheral blood mononuclear cells (PBMC). A: 1-way hierarchical clustering (genes only) was conducted on a gene set encompassing the union of the differentially expressed (DE) genes (RO vs. HC) identified among the 7 cell lines and fresh PBMC (2,888 unique genes). The number of DE genes in each experimental condition is indicated below the heat map. The number of genes in the signature of each cell line that overlap with the 496 or 765 gene signatures observed in fresh PBMC challenged independently or with plasma pools are indicated below the heat map. B: 1-way hierarchical clustering (genes only) on the 112 genes commonly regulated in fresh PBMC challenged independently or with plasma pools. C: 1-way hierarchical clustering (genes only) on a set of 22 genes (among the 112 genes illustrated in B) that were differentially expressed in fresh PBMC as well as in at least 1 of the 7 cell lines. Each of the 7 experimental conditions in this heat map is given a number (1 through 7) as indicated below the heat map. The numbers next to each gene symbol indicate the cell line/s in which that gene was differentially expressed. D: genes with the top 10 ranks within each cell line (5 most upregulated and 5 most downregulated) were extracted, merged, duplicates removed (n = 61), and subjected to 1-way hierarchical clustering along with the expression trend in fresh PBMC. The numbers next to each gene symbol indicate the cell line/s in which that gene was differentially expressed.
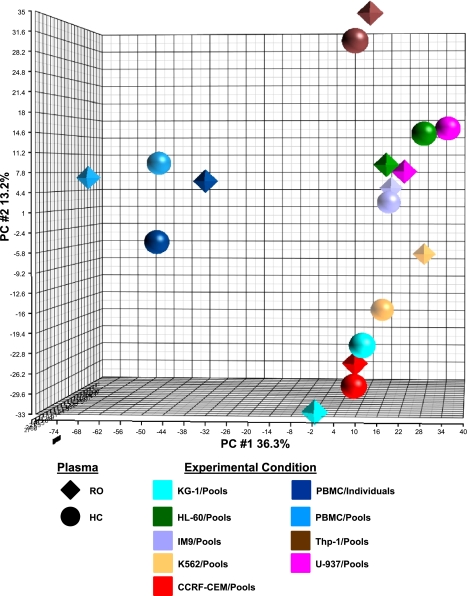
To visualize the relatedness of the signatures generated among the seven cell lines as well as their similarity to the signature generated with fresh PBMC, we carried out PCA using the union of the differentially expressed genes identified among the seven cell lines and fresh PBMC (n = 2,888 genes, Fig. 4). The PCA plot reveals that all the cell lines exhibit distinct separation among themselves as well as from the fresh PBMC signature.
Fig. 4.
Principal component analysis (PCA) of the signatures induced in cell lines and fresh PBMC. The union of the significantly regulated genes observed in the 7 cell lines (2,888 genes) was used for PCA, adding the expression values in fresh PBMC for those genes. Shown is the PCA plot, where each point represents the position of a cell line or fresh PBMC, in the 3D space. The ROs are represented by tetrahedrons and the HCs are represented by spheres. Responder cell type is reflected by color.
Comparison of highly enriched pathways across cell lines and fresh PBMC.
For each of the signatures induced in the seven cell lines and fresh PBMC, pathway analysis was conducted using the DAVID pathway enrichment tool to identify significantly enriched GO Biological Processes and GO Molecular Functions (P < 0.01). Each of the seven lists of enriched pathway terms was compared with that of fresh PBMC challenged with RO and HC plasma independently or with the pools, with the objective of identifying commonly regulated pathways. With the exception of seven terms commonly identified (among a total of 188) when either fresh PBMC and HL-60 cells were challenged with the pooled RO and HC plasma, there were no pathway terms in common between the enriched pathways in fresh PBMC and any of the seven cell lines (data not shown).
To further explore potential commonalities in the responses of PBMC and the various cell lines to RO T1D and HC plasma, we employed ToppGene (10, 11). ToppGene prioritizes genes based on functional similarity to a training gene list using gene expression, protein domains and interactions, transcription factor binding site and miRNA, ontologies, human disease and mouse phenotype, drug-gene associations, and literature cocitation. The analysis was conducted using training lists consisting of either 1) the 233 regulated probe sets (|log2 ratio| > 0.58, P < 0.01, FDR < 0.01) originally identified when PBMC were cultured with individual RO and HC plasma (60) or 2) the 765 regulated probe sets (|log2 ratio| > 0.5, FDR < 0.10) identified in this study when PBMC were cultured with pooled RO and HC plasma. When we compared the cell line signatures to that of the PBMC/individual plasma training set (Fig. 5A), the number of significantly identified (P < 0.01) functionally related genes ranged from a low of three (U-937) to a high of 82 (THP-1). When we compared the cell line signatures to that of the PBMC/pooled plasma training set (Fig. 5B), the number of significantly identified (P < 0.01) functionally related genes ranged from a low of six (U-937) to a high of 196 (K562). While prioritized genes identified within the K652 signature were largely directionally discordant relative to that observed in PBMC, selected genes related to immunological activation, signal transduction, and transcriptional regulation identified in the signatures of the other cells lines showed varying degrees of directional concordance (Fig. 5C).
Fig. 5.
Functional analysis of cell line signatures. Candidate gene prioritization with ToppGene was conducted for each cell line using the significantly regulated probe sets defined in Table 3. Analyses were conducted with training lists consisting of either the 233 regulated probe sets [|log2 ratio| > 0.58, P < 0.01, false discovery rate (FDR) < 0.01] identified when PBMC were cultured with individual RO and HC plasma (60) (A) or the 765 regulated probe sets (|log2 ratio| > 0.5, FDR < 0.10) (B) identified when PBMC were cultured with pooled RO and HC plasma. The number of significantly identified (P < 0.01) functionally related candidate probe sets are indicated. In parentheses are indicated the total number of genes identified and the total number of unique genes identified. C: a selection of annotated genes related to immunological activation, signal transduction, or transcriptional regulation. Each of the 7 experimental conditions in this heat map is given a number (1 through 7) as indicated below the heat map. The numbers next to each gene symbol indicate the cell line/s in which that gene was differentially expressed. The scale denotes fold of change relative to the intensity value across all replicates of each cell line.
DISCUSSION
In T1D, the insulin-producing pancreatic β-cells and associated lymph nodes are the most relevant tissues, and aside from the laparoscopic pancreatic biopsies being conducted in Japan (28–30), in general, these tissues are inaccessible. In terms of peripheral blood-based biomarkers, clinical onset of T1D is preceded by the development of auto-antibodies toward islet antigens (4, 9, 60). While the measurement of antibodies is certainly useful, the humoral immune response is thought to be secondary, appearing late during prediabetes (2, 14, 55). Moreover, the observation of T1D development in severe hereditary B-cell deficiency suggests that auto-antibodies may not be required for either the initiation or the progression of T1D (39) at least in a subset of patients. Thus, development of more sensitive and informative biomarkers is a high priority. Treatment of most disorders has a higher likelihood of success when specific intervention can be made early in the disease process, and since T1D patients have lost an estimated 80% of β-cell mass at the time of diagnosis, reliable, disease-specific, preonset biomarkers are highly desirable. Moreover, as we move toward individualized medicine, better tools are needed to measure a patient's response to targeted therapies.
The two blood-based array strategies have both been applied to T1D (31, 60). Kaizer et al. (31) reported the direct expression profiling of PBMC isolated from healthy controls versus T1D patients at diagnosis as well as at 1 and 4 mo postdiagnosis. These studies revealed, in patient PBMC collected at diagnosis, an expression signature that included IL1β, similar to our studies that employed the alternative serum-based approach (60); however, it was reported to resolve by 4 mo postdiagnosis despite the fact that pancreatic β-cell destruction is ongoing during this time, prompting the authors to hypothesize that the at-onset signature in PBMC may be in part a direct or indirect consequence of hyperglycemia. We too have found it difficult to detect differences between the transcriptomes of PBMC collected from RO and HC during the months following onset after glycemic control has been established (60).
We elected to investigate the plasma-induced transcription approach because 1) T1D pathogenesis involves a cytokine-based arm that kills pancreatic β-cells (22, 23, 54); 2) elevated peripheral IL-1 (α/β), IL-6, IL-8, IL-18, TNF-α, CXCL10, and IFN-γ levels have been reported in RO T1D cohorts (15, 20, 27, 41, 42, 48); and 3) although commercial ELISA kits offer sensitivities as low as pg/ml levels, the peripheral cytokine concentrations associated with T1D are generally too dilute for prognostic measurement by current technologies. Furthermore, in studying systemic onset juvenile idiopathic arthritis (SOJIA), Pascual et al. (45) observed that the gene expression differences between PBMC incubated with SOJIA vs. control sera were more robust than those observed between the direct gene expression profiling of case and control PBMC. Thus we reasoned that plasma-induced transcriptional profiling may be more sensitive in detecting the inflammatory state associated with T1D.
Collectively, our studies of T1D in humans as well as T1D in the BioBreeding rat model indicate that transcriptional signatures reflective of active autoimmunity are induced using plasma collected prior to T1D onset (32, 60). These signatures appear to be mechanistically informative and show promise as a measure during therapeutic intervention. In our previous studies of human T1D, we used healthy, unrelated third-party fresh PBMC as responder cells. Prior to analyzing larger clinical cohorts we aim to limit the potential heterogeneity introduced by cells drawn from multiple blood donors. Here we have investigated the potential use of human myeloid and lymphoid cell lines as surrogates for fresh PBMC. Such lines are well established as powerful tools for the investigation of gene function, the dissection of signal transduction pathways, and the analysis of drug effects (16–18). While they are known to differ from both normal and cancerous tissue, we reasoned they may serve as a reproducible, reliable and widely available source of responder cells, provided 1) their transcriptional response was found distinct when cultured with RO vs. HC plasma and 2) the response mimicked, at least in part, that observed when using fresh PBMC.
Our results show that although there was no clustering among the induced signatures based on common phenotypic or lineage characteristics, expression trends in a set of key genes specific to each cell line paralleled the trends reported in the literature (Fig. 1). Furthermore, within each cell line, there was sufficient power among the differentially expressed genes to distinguish the responses to RO plasma and HC plasma, although the number and identity of those genes varied across cell lines (Fig. 2). Furthermore, at the gene level, the transcriptional programs induced in the cell lines by RO vs. HC plasma displayed low correlation and concordance with that induced in fresh PBMC (Table 3). The basis for this can be attributed, but not limited, to the facts that cell lines 1) are derived from hematological malignancies, 2) are not terminally differentiated, 3) have been shaped by the in vitro conditions under which they have been propagated, and 4) have been passed countless times over many years and thus have probably deviated significantly from normal blood cells (i.e., altered receptor repertoire and signal transduction pathways). However, using ToppGene, we observed functional relatedness between the genes regulated by RO T1D plasma in PBMC and the various cell lines.
This study has a number of limitations. First, the benchmark for the response of the cell lines were assays conducted using a fresh heterogeneous PBMC population consisting of a monocyte/lymphocyte fraction isolated from whole blood by density gradient centrifugation. Therefore, this fresh responder population was mixed, consisting of T cells, B cells, and monocytes. We are presently investigating the response of PBMC subpopulations in an effort to better characterize their individual contribution to the response observed in this bioassay. A second limitation is that this study used pooled RO and HC plasma instead of individual plasmas. Pooling of human samples, which can exhibit high heterogeneity compared with in-bred animal models or in vitro culture systems, is not ideal, especially for array studies that capture comprehensive data sets. However, our banked pediatric T1D samples represent a considerable recruitment effort and are limited in supply such that their extensive use in assay development cannot be justified. Therefore we used a small volume of plasma from numerous individuals to create two pools versus exhausting multiple individual samples and introducing intersubject heterogeneity in the examination of the cell lines. When using fresh PBMC as responder cells, we found the signatures induced by the plasma pools less correlative than those induced by individual plasmas; we attribute this result to outlying samples influencing the mean response observed with the pools. A third limitation is the age difference between the RO (7.2 ± 1.5 yr) and HC (24.9 ± 3.5 yr) subjects used in these studies. Age has been observed to influence peripheral cytokine/chemokine levels in both the elderly and the very young (5, 7, 24, 37, 53). In our previous report (60), the “signature-positive” RO cases (5.5 ± 1.3 yr) were also younger than the “signature-negative” unrelated healthy controls (18.1 ± 4.3 yr). However, in that study, we also examined healthy age-matched at risk siblings of probands that possessed one or more islet auto-antibodies, where we observed a mixture of signature-positive and signature-negative subjects, supporting that the inflammatory signature was not age related. Furthermore, we examined longitudinal samples collected from progressors to T1D, where we observed a crescendo in the inflammatory signature as subjects approached onset, spanning the gap in age between the cross-sectionally studied cases and controls, again associating the proinflammatory signature induced in healthy PBMCs by RO plasma with disease not age. However, it is possible that the cell lines examined here express different receptors or utilize different signal transduction pathways allowing them to recognize ligands to which fresh healthy PBMC do not respond, some of which may be found in different concentrations in younger versus older subject plasma. Thus it cannot be excluded that some of the differential gene expression induced by RO and HC plasma in the various cell lines may be age related.
Our ongoing efforts include the evaluation of other lymphoblastoid cell lines (49) and viably frozen mixed PBMC populations that are now commercially available. These are collected from highly characterized blood donors by aphaeresis, thus billions of cells are harvested during a single draw and are rapidly processed for high viability. Cryopreserved PBMC have been found to be suitable in assessment of immunological markers in children with T1D (3), and studies have reported that such PBMC maintain functionality and cytokine production after cryopreservation (36, 56, 59).
Overall our results thus far suggest that fresh PBMC as well as myeloid and lymphoid cell lines have exquisite sensitivity to sense their environment and transcriptionally respond in a specific manner. In this sense, all seven of the cell lines could distinguish RO and HC plasma and thus have the capacity to serve as a biosensor. However, relative to a fresh, mixed PBMC population, the transcriptional response generated from each of the cell lines is distinct, each possessing varying degrees of functional relatedness.
GRANTS
This work was supported by National Institutes of Health Grants R01AI-078713 (M. J. Hessner) and R01DK-080100 (X. Wang), Juvenile Diabetes Research Foundation International Grant 1-2008-1026 (M. J. Hessner), and The Children's Hospital of Wisconsin Foundation.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
The authors acknowledge Dr. Soumitra Ghosh and the physicians, nurses, and staff of Children's Hospital of Wisconsin and The Max McGee National Research Center for Juvenile Diabetes who assisted in subject recruitment and sample collection/processing. The authors sincerely thank the recruited patients and families that have participated in this study.
REFERENCES
- 1. Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 15: 539–553, 1998 [DOI] [PubMed] [Google Scholar]
- 2. Atkinson MA, Eisenbarth GS. Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet 358: 221–229, 2001 [DOI] [PubMed] [Google Scholar]
- 3. Axelsson S, Faresjo M, Hedman M, Ludvigsson J, Casas R. Cryopreserved peripheral blood mononuclear cells are suitable for the assessment of immunological markers in type 1 diabetic children. Cryobiology 57: 201–208, 2008 [DOI] [PubMed] [Google Scholar]
- 4. Baekkeskov S, Aanstoot HJ, Christgau S, Reetz A, Solimena M, Cascalho M, Folli F, Richter-Olesen H, De Camilli P. Identification of the 64K autoantigen in insulin-dependent diabetes as the GABA-synthesizing enzyme glutamic acid decarboxylase. Nature 347: 151–156, 1990 [DOI] [PubMed] [Google Scholar]
- 5. Beharka AA, Meydani M, Wu D, Leka LS, Meydani A, Meydani SN. Interleukin-6 production does not increase with age. J Gerontol A Biol Sci Med Sci 56: B81–B88, 2001 [DOI] [PubMed] [Google Scholar]
- 6. Belov L, de la Vega O, dos Remedios CG, Mulligan SP, Christopherson RI. Immunophenotyping of leukemias using a cluster of differentiation antibody microarray. Cancer Res 61: 4483–4489, 2001 [PubMed] [Google Scholar]
- 7. Berdat PA, Wehrle TJ, Kung A, Achermann F, Sutter M, Carrel TP, Nydegger UE. Age-specific analysis of normal cytokine levels in healthy infants. Clin Chem Lab Med 41: 1335–1339, 2003 [DOI] [PubMed] [Google Scholar]
- 8. Berges C, Naujokat C, Tinapp S, Wieczorek H, Hoh A, Sadeghi M, Opelz G, Daniel V. A cell line model for the differentiation of human dendritic cells. Biochem Biophys Res Commun 333: 896–907, 2005 [DOI] [PubMed] [Google Scholar]
- 9. Bingley PJ, Bonifacio E, Williams AJ, Genovese S, Bottazzo GF, Gale EA. Prediction of IDDM in the general population: strategies based on combinations of autoantibody markers. Diabetes 46: 1701–1710, 1997 [DOI] [PubMed] [Google Scholar]
- 10. Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–311, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics 8: 392, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins SJ, Ruscetti FW, Gallagher RE, Gallo RC. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc Natl Acad Sci USA 75: 2458–2462, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: P3, 2003 [PubMed] [Google Scholar]
- 14. Devendra D, Liu E, Eisenbarth GS. Type 1 diabetes: recent developments. BMJ 328: 750–754, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dogan Y, Akarsu S, Ustundag B, Yilmaz E, Gurgoze MK. Serum IL-1beta, IL-2, and IL-6 in insulin-dependent diabetic children. Med Inf (Lond) 2006: 59206, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drexler HG, Matsuo AY, MacLeod RA. Continuous hematopoietic cell lines as model systems for leukemia-lymphoma research. Leuk Res 24: 881–911, 2000 [DOI] [PubMed] [Google Scholar]
- 17. Drexler HG, Matsuo Y. Malignant hematopoietic cell lines: in vitro models for the study of multiple myeloma and plasma cell leukemia. Leuk Res 24: 681–703, 2000 [DOI] [PubMed] [Google Scholar]
- 18. Drexler HG, Matsuo Y. Malignant hematopoietic cell lines: in vitro models for the study of natural killer cell leukemia-lymphoma. Leukemia 14: 777–782, 2000 [DOI] [PubMed] [Google Scholar]
- 19. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res 30: 207–210, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Erbagci AB, Tarakcioglu M, Coskun Y, Sivasli E, Sibel Namiduru E. Mediators of inflammation in children with type I diabetes mellitus: cytokines in type I diabetic children. Clin Biochem 34: 645–650, 2001 [DOI] [PubMed] [Google Scholar]
- 21. Fahey JL, Buell DN, Sox HC. Proliferation and differentiation of lymphoid cells: studies with human lymphoid cell lines and immunoglobulin synthesis. Ann NY Acad Sci 190: 221–234, 1971 [DOI] [PubMed] [Google Scholar]
- 22. Green EA, Flavell RA. The temporal importance of TNFalpha expression in the development of diabetes. Immunity 12: 459–469, 2000 [DOI] [PubMed] [Google Scholar]
- 23. Guerder S, Picarella DE, Linsley PS, Flavell RA. Costimulator B7–1 confers antigen-presenting-cell function to parenchymal tissue and in conjunction with tumor necrosis factor alpha leads to autoimmunity in transgenic mice. Proc Natl Acad Sci USA 91: 5138–5142, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hasegawa Y, Sawada M, Ozaki N, Inagaki T, Suzumura A. Increased soluble tumor necrosis factor receptor levels in the serum of elderly people. Gerontology 46: 185–188, 2000 [DOI] [PubMed] [Google Scholar]
- 25. Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22: 2825–2827, 2006 [DOI] [PubMed] [Google Scholar]
- 26. Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol 4: R70, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hussain MJ, Peakman M, Gallati H, Lo SS, Hawa M, Viberti GC, Watkins PJ, Leslie RD, Vergani D. Elevated serum levels of macrophage-derived cytokines precede and accompany the onset of IDDM. Diabetologia 39: 60–69, 1996 [DOI] [PubMed] [Google Scholar]
- 28. Imagawa A, Hanafusa T, Itoh N, Miyagawa J, Nakajima H, Namba M, Kuwajima M, Tamura S, Kawata S, Matsuzawa Y, Harlan DM. Islet-infiltrating t lymphocytes in insulin-dependent diabetic patients express CD80 (B7–1) and CD86 (B7–2). J Autoimmun 9: 391–396, 1996 [DOI] [PubMed] [Google Scholar]
- 29. Imagawa A, Hanafusa T, Itoh N, Waguri M, Yamamoto K, Miyagawa J, Moriwaki M, Yamagata K, Iwahashi H, Sada M, Tsuji T, Tamura S, Kawata S, Kuwajima M, Nakajima H, Namba M, Matsuzawa Y. Immunological abnormalities in islets at diagnosis paralleled further deterioration of glycaemic control in patients with recent-onset Type I (insulin-dependent) diabetes mellitus. Diabetologia 42: 574–578, 1999 [DOI] [PubMed] [Google Scholar]
- 30. Imagawa A, Hanafusa T, Tamura S, Moriwaki M, Itoh N, Yamamoto K, Iwahashi H, Yamagata K, Waguri M, Nanmo T, Uno S, Nakajima H, Namba M, Kawata S, Miyagawa JI, Matsuzawa Y. Pancreatic biopsy as a procedure for detecting in situ autoimmune phenomena in type 1 diabetes: close correlation between serological markers and histological evidence of cellular autoimmunity. Diabetes 50: 1269–1273, 2001 [DOI] [PubMed] [Google Scholar]
- 31. Kaizer EC, Glaser CL, Chaussabel D, Banchereau J, Pascual V, White PC. Gene expression in peripheral blood mononuclear cells from children with diabetes. J Clin Endocrinol Metab 92: 3705–3711, 2007 [DOI] [PubMed] [Google Scholar]
- 32. Kaldunski M, Jia S, Geoffrey R, Basken J, Prosser S, Kansra S, Mordes JP, Lernmark A, Wang X, Hessner MJ. Identification of a serum-induced transcriptional signature associated with type 1 diabetes in the BioBreeding rat. Diabetes 59: 2375–2385, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klein E, Ben-Bassat H, Neumann H, Ralph P, Zeuthen J, Polliack A, Vanky F. Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int J Cancer 18: 421–431, 1976 [DOI] [PubMed] [Google Scholar]
- 34. Klitz W, Maiers M, Spellman S, Baxter-Lowe LA, Schmeckpeper B, Williams TM, Fernandez-Vina M. New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans. Tissue Antigens 62: 296–307, 2003 [DOI] [PubMed] [Google Scholar]
- 35. Koeffler HP, Golde DW. Acute myelogenous leukemia: a human cell line responsive to colony-stimulating activity. Science 200: 1153–1154, 1978 [DOI] [PubMed] [Google Scholar]
- 36. Kreher CR, Dittrich MT, Guerkov R, Boehm BO, Tary-Lehmann M. CD4+ and CD8+ cells in cryopreserved human PBMC maintain full functionality in cytokine ELISPOT assays. J Immunol Methods 278: 79–93, 2003 [DOI] [PubMed] [Google Scholar]
- 37. Kubo M, Cinader B. IL-3 production as a function of age and its correlation with splenomegaly: age versus disease-related change. Immunol Lett 24: 133–136, 1990 [DOI] [PubMed] [Google Scholar]
- 38. Lozzio BB, Lozzio CB. Properties and usefulness of the original K-562 human myelogenous leukemia cell line. Leuk Res 3: 363–370, 1979 [DOI] [PubMed] [Google Scholar]
- 39. Martin S, Wolf-Eichbaum D, Duinkerken G, Scherbaum WA, Kolb H, Noordzij JG, Roep BO. Development of type 1 diabetes despite severe hereditary B-lymphocyte deficiency. N Engl J Med 345: 1036–1040, 2001 [DOI] [PubMed] [Google Scholar]
- 40. Moos PJ, Raetz EA, Carlson MA, Szabo A, Smith FE, Willman C, Wei Q, Hunger SP, Carroll WL. Identification of gene expression profiles that segregate patients with childhood leukemia. Clin Cancer Res 8: 3118–3130, 2002 [PubMed] [Google Scholar]
- 41. Nicoletti F, Conget I, Di Marco R, Speciale AM, Morinigo R, Bendtzen K, Gomis R. Serum levels of the interferon-gamma-inducing cytokine interleukin-18 are increased in individuals at high risk of developing type I diabetes. Diabetologia 44: 309–311, 2001 [DOI] [PubMed] [Google Scholar]
- 42. Nicoletti F, Conget I, Di Mauro M, Di Marco R, Mazzarino MC, Bendtzen K, Messina A, Gomis R. Serum concentrations of the interferon-gamma-inducible chemokine IP-10/CXCL10 are augmented in both newly diagnosed Type I diabetes mellitus patients and subjects at risk of developing the disease. Diabetologia 45: 1107–1110, 2002 [DOI] [PubMed] [Google Scholar]
- 43. Olsson I, Gullberg U, Ivhed I, Nilsson K. Induction of differentiation of the human histiocytic lymphoma cell line U-937 by 1 alpha,25-dihydroxycholecalciferol. Cancer Res 43: 5862–5867, 1983 [PubMed] [Google Scholar]
- 44. Ortaldo JR, Oldham RK, Cannon GC, Herberman RB. Specificity of natural cytotoxic reactivity of normal human lymphocytes against a myeloid leukemia cell line. J Natl Cancer Inst 59: 77–82, 1977 [DOI] [PubMed] [Google Scholar]
- 45. Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J Exp Med 201: 1479–1486, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pascual V, Chaussabel D, Banchereau J. A genomic approach to human autoimmune diseases. Ann Rev Immunol 28: 535–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pellat-Deceunynk C, Amiot M, Bataille R, Van Riet I, Van Camp B, Omede P, Boccadoro M. Human myeloma cell lines as a tool for studying the biology of multiple myeloma: a reappraisal 18 years after. Blood 86: 4001–4002, 1995 [PubMed] [Google Scholar]
- 48. Perez F, Oyarzun A, Carrasco E, Angel B, Albala C, Santos JL. [Plasma levels of interleukin-1beta, interleukin-2 and interleukin-4 in recently diagnosed type 1 diabetic children and their association with beta-pancreatic autoantibodies]. Rev Med Chil 132: 413–420, 2004 [DOI] [PubMed] [Google Scholar]
- 49. Reiss CS, Hemler ME, Englehard VH, Mier JW, Strominger JL, Burakoff SJ. Development and characterization of allospecific long-term human cytolytic T-cell lines. Proc Natl Acad Sci USA 77: 5432–5436, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet 24: 227–235, 2000 [DOI] [PubMed] [Google Scholar]
- 51. Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, Liu HC, Mahfouz R, Raimondi SC, Lenny N, Patel A, Downing JR. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood 102: 2951–2959, 2003 [DOI] [PubMed] [Google Scholar]
- 52. Sandstrom PA, Buttke TM. Autocrine production of extracellular catalase prevents apoptosis of the human CEM T-cell line in serum-free medium. Proc Natl Acad Sci USA 90: 4708–4712, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schultz C, Rott C, Temming P, Schlenke P, Moller JC, Bucsky P. Enhanced interleukin-6 and interleukin-8 synthesis in term and preterm infants. Pediatr Res 51: 317–322, 2002 [DOI] [PubMed] [Google Scholar]
- 54. Shimabukuro M, Koyama K, Lee Y, Unger RH. Leptin- or troglitazone-induced lipopenia protects islets from interleukin 1beta cytotoxicity. J Clin Invest 100: 1750–1754, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Skyler JS. Prediction and prevention of type 1 diabetes: progress, problems, and prospects. Clin Pharmacol Ther 81: 768–771, 2007 [DOI] [PubMed] [Google Scholar]
- 56. Sobota V, Bubenik J, Indrova M, Vlk V, Jakoubkova J. Use of cryopreserved lymphocytes for assessment of the immunological effects of interferon therapy in renal cell carcinoma patients. J Immunol Methods 203: 1–10, 1997 [DOI] [PubMed] [Google Scholar]
- 57. St Louis DC, Woodcock JB, Franzoso G, Blair PJ, Carlson LM, Murillo M, Wells MR, Williams AJ, Smoot DS, Kaushal S, Grimes JL, Harlan DM, Chute JP, June CH, Siebenlist U, Lee KP. Evidence for distinct intracellular signaling pathways in CD34+ progenitor to dendritic cell differentiation from a human cell line model. J Immunol 162: 3237–3248, 1999 [PubMed] [Google Scholar]
- 58. Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208, 2002 [DOI] [PubMed] [Google Scholar]
- 59. Wang SY, Hsu ML, Tzeng CH, Hsu HC, Ho CK. The influence of cryopreservation on cytokine production by human T lymphocytes. Cryobiology 37: 22–29, 1998 [DOI] [PubMed] [Google Scholar]
- 60. Wang X, Jia S, Geoffrey R, Alemzadeh R, Ghosh S, Hessner MJ. Identification of a molecular signature in human type 1 diabetes mellitus using serum and functional genomics. J Immunol 180: 1929–1937, 2008 [DOI] [PubMed] [Google Scholar]
- 61. Woo W, LaGasse JM, Zhou Z, Patel R, Palmer JP, Campus H, Hagopian WA. A novel high-throughput method for accurate, rapid, and economical measurement of multiple type 1 diabetes autoantibodies. J Immunol Methods 244: 91–103, 2000 [DOI] [PubMed] [Google Scholar]
- 62. Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui CH, Evans WE, Naeve C, Wong L, Downing JR. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1: 133–143, 2002 [DOI] [PubMed] [Google Scholar]