Abstract
Raf kinase inhibitor protein (RKIP) interacts with a number of different proteins and regulates multiple signaling pathways. Here, we show that locostatin, a small molecule that covalently binds RKIP, not only disrupts interactions of RKIP with Raf-1 kinase, but also with G protein-coupled receptor kinase 2. In contrast, we found that locostatin does not disrupt binding of RKIP to two other proteins: inhibitor of κB kinase α and transforming growth factor β-activated kinase 1. These results thus imply that different proteins interact with different regions of RKIP. Locostatin’s mechanism of action involves modification of a nucleophilic residue on RKIP. We observed that after binding RKIP, part of locostatin is slowly hydrolyzed, leaving a smaller RKIP-butyrate adduct. We identified the residue alkylated by locostatin as His86, a highly conserved residue in RKIP’s ligand-binding pocket. Computational modeling of the binding of locostatin to RKIP suggested that the recognition interaction between small molecule and protein ensures that locostatin’s electrophilic site is poised to react with His86. Furthermore, binding of locostatin would sterically hinder binding of other ligands in the pocket. These data provide a basis for understanding how locostatin disrupts particular interactions of RKIP with RKIP-binding proteins and demonstrate its utility as a probe of specific RKIP interactions and functions.
Keywords: RKIP, locostatin, mechanism of action, binding, Raf-1, GRK2, IKKα, TAK1
I. INTRODUCTION
Locostatin is an inhibitor of cell migration1 and cell-substratum adhesion2 that covalently binds Raf kinase inhibitor protein (RKIP).3 Locostatin disrupts the interaction of RKIP with Raf-1 kinase.3 RKIP has a number of functions, including modulation of the activity of Raf-1 kinase,4,5 G protein-coupled receptor kinase 2 (GRK2),6 and proteins involved in nuclear factor κB activation, including inhibitor of κB kinase α (IKKα)7,8 and transforming growth factor β-activated kinase 1 (TAK1).7,8 In addition, RKIP binds a number of endogenous small-molecule ligands, most notably phosphatidylethanolamine, and therefore is also known as phosphatidylethanolamine-binding protein (PEBP).9,10 There is evidence that RKIP/PEBP can bind morphine,11,12 morphine glucuronides,13,14 nucleotides,15 and a pyrazolopyrimidine phosphodiesterase-5 inhibitor.16 RKIP has also been identified as a potential odorant-binding protein.17 However, the effects of these various small molecules on the function of RKIP are unclear and, at least in the case of phosphatidylethanoloamine and morphine glucuronides, these interactions are of low affinity and appear nonspecific.14
The question of how RKIP binds its protein ligands is still poorly defined. RKIP binds with micromolar Kd values to phosphopeptides corresponding to a part of the N-region of Raf-1 that contains the sequence 338-Ser-Ser-Tyr-Tyr-341, while the nonphosphorylated peptide binds with lower affinity; phosphorylation of Ser338, Ser339, and Tyr341 appear to modulate binding affinity.18 Recently, Brady and coworkers have found that O-phosphotyrosine (pTyr) binds snugly in RKIP’s highly conserved ligand-binding pocket in an X-ray cocrystal structure [Protein Data Bank (PDB) code 2QYQ; R. Leo Brady, personal communication]. This structure is the first molecular description of an interaction that provides a model for how a protein ligand could conceivably bind in the ligand-binding pocket. The ligand-binding pocket interacts not just with pTyr but also other anionic ligands, such as cacodylate,19 a phosphate mimic, and O-phosphoethanolamine,20 the head group of phosphatidylethanolamine. The base of the pocket is composed of residues capable of hydrogen bonding to anions; additionally, the pocket is lined with aromatic residues that make the walls of the pocket more hydrophobic and may engage in van der Waals contacts with the hydrophobic portions of amphiphilic ligands such as pTyr.18 Although specific protein-protein interactions are unlikely to involve RKIP’s ligand-binding pocket alone, the pocket may be part of a larger interface of interaction with protein ligands.21,22
In this report, we show that locostatin not only disrupts the interaction of RKIP with Raf-1 kinase but also with GRK2. In contrast, we found that locostatin does not affect association of RKIP with IKKa or TAK1, suggesting that different proteins may bind different parts of RKIP. Locostatin also caused a partial aggregation of RKIP. Locostatin binds RKIP with slow kinetics.3 Preincubation of RKIP with locostatin before adding Raf-1 or sufficient incubation time when RKIP, Raf-1, and locostatin were combined simultaneously were required to observe disruption of the interaction. After binding RKIP, the crotonyl group of locostatin was slowly hydrolyzed from the oxazolidinone moiety, as determined by mass spectrometry (MS), leaving an RKIP-butyrate adduct. We identified the specific residue modified by locostatin as His86 by tandem MS. His86 is located in RKIP’s highly conserved ligand-binding pocket. Computational modeling of the recognition interaction between locostatin and RKIP resulted in positioning of His86 near the β-carbon of the crotonyl group of locostatin, the electrophilic site on locostatin with which His86 would be expected to react. These data add considerably to our understanding of the molecular mechanism of action of locostatin in its binding to RKIP and disruption of specific protein-protein interactions.
II. MATERIALS AND METHODS
II.A. Association of RKIP With Its Binding Partners in the Presence or Absence of Locostatin
His-tagged human RKIP-1/PEBP-1 in the pET-15b vector was expressed in Escherichia coli strain BL21(DE3) and purified with Ni-NTA agarose, according to the manufacturer’s instructions (Novagen). Mammalian expression constructs for hemagglutinin (HA)-tagged human Raf-1 in pcDNA3, FLAG-tagged human IKKa in pCMV-FLAG, and FLAG-tagged human TAK1 in pCMV-FLAG were transfected separately into COS-7 cells with Lipofectamine 2000 (Invitrogen). HA-Raf-1 was activated by treating the cells with phorbol 12-myristate 13-acetate (100 nM) for 20 min before cell lysis. Human Raf-1 was also baculovirally expressed as a FLAG-Raf-1 fusion in Sf9 cells. In all cases, the lysis buffer consisted of 20 mM Tris, pH 7.4, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.1% (v/v) protease inhibitor cocktail (P8340, Sigma-Aldrich), and 0.1% Nonidet P40 Substitute (USB Corp.). Purified recombinant human GRK2 protein was purchased from Invitrogen. Two micrograms of mouse monoclonal anti-HA antibody (Santa Cruz Biotechnology) for HA-Raf-1 or mouse monoclonal anti-FLAG antibody (Santa Cruz Biotechnology) for FLAG-tagged Raf-1, IKKa, or TAK1 was added to the cell lysates (Sf9 cell lysates for FLAG-Raf-1 or COS-7 cell lysates for the other FLAG-tagged proteins) or, in the case of GRK2, mouse monoclonal anti-GRK2 antibody (Santa Cruz Biotechnology) was added to the purified GRK2 sample. The samples were incubated at 4°C for 2 h and immunoprecipitated with Dynabeads protein G (Invitrogen) in a binding buffer of 20 mM Tris, pH 7.4, 2 mM EDTA. RKIP (100 nM) was preincubated with locostatin (200 µM) or 0.2% (v/v) dimethyl sulfoxide (DMSO) carrier solvent alone at 37°C for 6 h, also in 20 mM Tris, pH 7.4, 2 mM EDTA, then added to the immunoprecipitates and incubated at room temperature for 1 h with rotation. Samples were pelleted by centrifugation at 2500 × g for 2 min and washed five times with the same buffer, with centrifugation between each wash. In experiments where RKIP was not preincubated with locostatin first, RKIP and locostatin or RKIP and DMSO alone were added to the Raf-1 sample at the same time, with a buffer of 10 mM HEPES, pH 7.4, 3 mM MgCl2, 10 mM KCl, and 150 mM NaCl for binding. Samples were centrifuged at 2500 × g for 2 min and washed five times, with centrifugation between each wash. In all cases, the pellets were resuspended in sodium dodecyl sulfate (SDS) sample buffer, boiled and then subjected to SDS-polyacrylamide gel electrophoresis (PAGE), then transferred to polyvinylidene fluoride (PVDF) membranes for Western blot analysis. The blots were probed for RKIP with a rabbit anti-RKIP antibody (Invitrogen), and chemiluminescent detection was accomplished with a mouse anti-rabbit IgG conjugated to horseradish peroxidase (Santa Cruz Biotechnology).
II.B. RKIP Aggregation
RKIP (20 µM) was mixed with locostatin (100 µM) or 0.1% (v/v) DMSO alone in a 0.6 mL microcentrifuge tube and incubated at 37°C for 24 h in a buffer of 20 mM Tris, pH 7.5, 2 mM EDTA, and 2 mM dithiothreitol (DTT). The sample was then centrifuged at 2500 × g for 5 min to pellet aggregated RKIP from the solution. The supernatant was transferred to a new 0.6 ml microcentrifuge tube. Supernatant and pellet samples were subjected to SDS-PAGE, followed by transfer to a PVDF membrane and Western blot analysis with an anti-RKIP antibody, as described above. Percentage of aggregated RKIP was determined with Quantity One software (Bio-Rad).
II.C. MS Analysis
Covalent association of locostatin with intact RKIP was examined by liquid chromatography (LC)–electrospray ionization (ESI)–MS. A mixture of RKIP (20 µM) and locostatin (100 µM) or 0.1% (v/v) DMSO alone was incubated at 37°C for 6 h in 20 mM Tris, pH 7.5, 2 mM EDTA, and 2 mM DTT. The mixture was centrifuged at 2500 × g for 2 min to separate the precipitate from the solution. Aggregated protein was resuspended in an anionic acid-labile surfactant (Progenta AALS I from Protea Biosciences) that is compatible with LC-MS. The RKIP-locostatin mixture was then injected onto a C18 column (Hypersil GOLD, 1.9 µm, 100 mm × 1.0 mm, Thermo Scientific) on a 10ADvp high-performance LC (HPLC) system (Shimadzu). Protein was eluted with a linear gradient of acetonitrile containing 0.1% (v/v) acetic acid from 2% (v/v) acetonitrile/water to 90% (v/v) acetonitrile/water over 20 min at a flow rate of 50 mL/min. The LC effluent was fed into the ESI source of a QStar Elite mass spectrometer (Applied Biosystems/MDS Sciex). Typical source conditions were IS 5500 V, GS1 20, DP 80 V, FP 280 V, and DP2 15 V. Data acquisition was controlled with Analyst software (AB SCIEX).
For tandem MS analysis to identify the specific residue modified by locostatin, RKIP (20 µM) was incubated with locostatin (200 µM) or 0.1% (v/v) DMSO alone for 24 h at 37°C in 20 mM Tris, pH 7.5, 2 mM EDTA, and 2 mM DTT. The aggregated RKIP-locostatin mixture was resuspended in 15 µL 50 mM ammonium bicarbonate with 10 mM DTT. The sample was then incubated at 95°C for 5 min, cooled to room temperature, and mixed with 3 µL 100 mM iodoacetamide in the dark and incubated at room temperature for 20 min. Modified trypsin (Promega) was added for a final trypsin:RKIP ratio of 1:50 (w/w) and then incubated at 37°C for 24 h. The reaction was terminated by adding formic acid. The fragments were subjected to LC-ESI-MS/MS, under the same conditions and with the same HPLC system, mass spectrometer, and control software as above.
II.D. Computational Methods
The X-ray crystal structure of human RKIP complexed with pTyr (PDB code 2QYQ) was utilized for the docking calculations. The structure was processed with the protein preparation toolkit of Maestro, version 9 (Schrödinger). The process includes separation of nonprotein atoms and addition and optimization of hydrogen atoms. The pTyr coordinates were removed to allow for the docking of locostatin. The ligand structure for locostatin was obtained from a density functional theory minimization at the B3LYP/6-31g* theory level with Gaussian 09.23 Electrostatic potential charges were obtained from this calculation and assigned to locostatin. These charges were later used in the docking calculation.
Docking was performed with Glide, version 5.0 (Schrödinger), allowing for ring conformational sampling and bond rotation. Ten different poses were selected from the top 20 ranked poses according to the Glide docking score. The criteria for selecting these 10 poses was to make sure to include different orientations of the locostatin’s phenyl group relative to the center of the protein cavity. Using these 10 structures, quantum mechanics (QM)/molecular mechanics (MM) optimizations were performed at the b3lyp/6-31g*/OPLS2001 theory level with locostatin at the QM level and the rest of the protein at the MM level, leaving the protein frozen during minimization. Another set of QM/MM single-point energy calculations were performed on the QM/MM optimized structures with the same theory level including Poisson-Boltzmann (PB) continuum solvation. All QM/MM calculations were performed with the program Qsite, version 5.5 (Schrödinger). At the end, all ten poses were reranked according to the QM/MM and QM/MM/PB energies, with the lowest-energy pose the one shown in this report.
III. RESULTS AND DISCUSSION
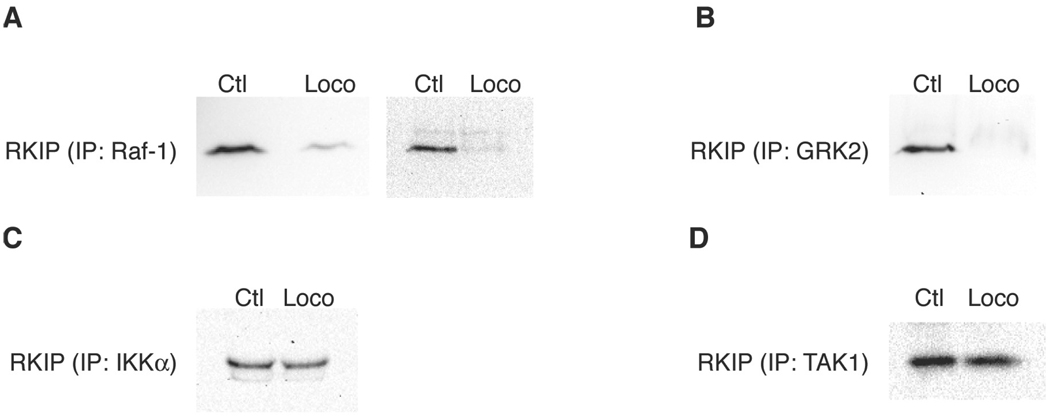
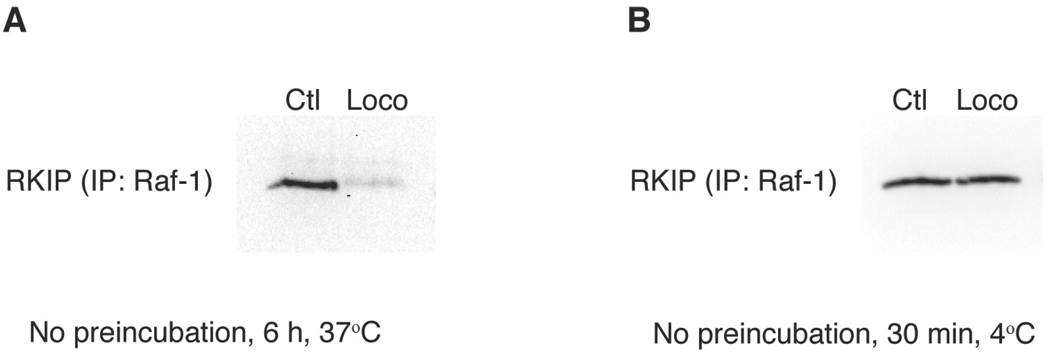
In addition to confirming that locostatin prevents association of RKIP with Raf-1 under conditions where RKIP is preincubated with locostatin (Fig. 1A and Ref. 3), we demonstrate in this report that locostatin added at the same time as RKIP to a Raf-1 sample results in disruption of binding of RKIP to Raf-1 when the sample is incubated at 37°C for 6 h, but not at 4°C for 30 min (Fig. 2). Locostatin also has been shown to disrupt the interaction of RKIP with Raf-1 in vivo.24
FIGURE 1.
Locostatin disrupts binding of RKIP with Raf-1 and GRK2 but not IKKa or TAK1. Purified recombinant human RKIP (100 nM) was preincubated with locostatin (200 µM; Loco) or DMSO alone (0.2%, corresponding to the concentration of DMSO carrier solvent in the experimental treatment) as control (Ctl) at 37°C for 6 h and then added to immunoprecipitated samples of the following recombinant human RKIP-binding proteins: (A) Raf-1 (shown are HA-tagged Raf-1 on the left and FLAG-tagged Raf-1 on the right); (B) GRK2; (C) IKKa; (D) TAK1. After allowing binding of RKIP to its binding proteins for 1 h at room temperature, the samples were centrifuged and washed five times. The pellets were then subjected to SDS-PAGE and Western blot analysis with an anti-RKIP antibody. Each Western blot is representative of at least three separate experiments.
FIGURE 2.
Locostatin disrupts binding of RKIP with Raf-1 without preincubation of RKIP with locostatin. RKIP (100 nM), immunoprecipitated FLAG-tagged Raf-1, and locostatin (200 µM; Loco) or DMSO alone (0.2%) as control (Ctl) were combined at the same time, then incubated for either (A) 6 h at 37°C or (B) 30 min at 4°C. After centrifugation and washing five times, the pellets were subjected to SDS-PAGE and Western blot analysis with an anti-RKIP antibody. Each Western blot is representative of at least three separate experiments.
Furthermore, Raf-1 is not the only RKIP-binding protein whose interaction with RKIP is disrupted by locostatin. We found that locostatin also prevents association of RKIP with GRK2 (Fig. 1B). Binding of GRK2 therefore appears to involve a region of RKIP that overlaps with the region to which Raf-1 binds. However, interaction of GRK2 with RKIP is enhanced by phosphorylation of Ser153 on RKIP, while binding of Raf-1 is reduced when Ser153 is phosphorylated.6 These data collectively imply that while both Raf-1 and GRK2 may partly interact in a similar manner within the same region of RKIP, such as in the ligand-binding pocket, additional divergent interactions exist. Raf-1’s association with RKIP is antagonized by phosphorylation of Ser153, perhaps sterically, while GRK2’s binding to RKIP is stabilized by the presence of a phosphorylated Ser153 residue.
In contrast to the binding of Raf-1 and GRK2, the interactions of RKIP with IKKα or TAK1 were not appreciably disrupted by locostatin (Figs. 1C and 1D). These results imply very different modes of binding to RKIP for these two sets of proteins. Raf-1 and GRK2 binding may at least partly, but critically, depend on interactions in the same region of RKIP, which is sterically blocked or conformationally altered by locostatin binding. IKKα and TAK1, on the other hand, either bind in another region entirely or, if they do bind in an overlapping manner with Raf-1 and GRK2, the overall interactions of IKKα and TAK1 are not as strongly destabilized by locostatin binding.
Another report suggested that locostatin does not disrupt the interaction of RKIP with Raf-1; however, in that study, locostatin, RKIP and Raf-1 were mixed together at the same time and the subsequent incubation was done at 4°C for only 30 min.25 These conditions are not sufficient to observe disruption of the RKIP-Raf-1 interaction (Fig. 2B). Locostatin alkylates RKIP with slow kinetics, i.e., the calculated second-order rate constant for the irreversible association at 25°C is 13.09 ± 1.36 M−1 s−1 (mean ± standard deviation).3 Therefore, binding of locostatin to RKIP requires either preincubation of RKIP with locostatin prior to addition of Raf-1 or sufficient incubation time when RKIP, Raf-1, and locostatin are simultaneously combined. Under conditions that reflect the kinetics of locostatin’s binding to RKIP and are not unduly stringent, locostatin abrogates the interaction of RKIP with Raf-1 and GRK2 (Figs. 1 and 2, and Ref. 3).
The binding of Raf-1 to RKIP would presumably have much faster association kinetics than the binding of locostatin to RKIP. Without preincubation of locostatin with RKIP, there are then two possibilities to explain how locostatin eventually displaces Raf-1 from RKIP. If the two binding events are mutually exclusive sterically, the binding of locostatin would have to occur when Raf-1 is in the off state under equilibrium conditions. If, however, locostatin can still bind to the RKIP-Raf-1 complex, this would suggest that the binding sites are distinct and that locostatin causes a conformational change resulting in dissociation of Raf-1. With either possibility, however, locostatin would prevent reassociation of Raf-1 or cause its dissociation over time, since locostatin, unlike Raf-1, binds irreversibly, having a thermodynamic, though not a kinetic, advantage in binding.
At high concentrations of RKIP and locostatin, we observed aggregation of RKIP, as previously reported.25 We determined the fraction of RKIP that aggregated when RKIP (20 µM) was incubated with locostatin (100 µM) at 37°C for 24 h. Under these conditions, 22.2% of the RKIP appeared as a pelletable aggregate (Table 1). The aggregate could be resolublized with SDS sample buffer for SDS-PAGE for Western blot analysis (Table 1) or with an anionic acid-labile surfactant (Progenta AALS I from Protea Biosciences).
TABLE I.
Binding of Locostatin to RKIP Causes Aggregation of RKIPa
| Sample | Mean (%) | Standard deviation (%) |
|---|---|---|
| RKIP with DMSO alone: supernatant (n = 3) | 100 | 0 |
| RKIP-locostatin: supernatant (n = 3) | 77.8 | 7.9 |
| RKIP-locostatin: precipitate (n = 3) | 22.2 | 7.9 |
His-tagged RKIP (20 µM) was incubated with locostatin (100 µM) or DMSO alone (0.1%) as control at 37°C for 24 h. The samples were centrifuged, and supernatants and pellets were separately collected and subjected to SDS-PAGE and Western blot analysis with an anti-RKIP antibody. The data represent band intensities for three independent Western blots.
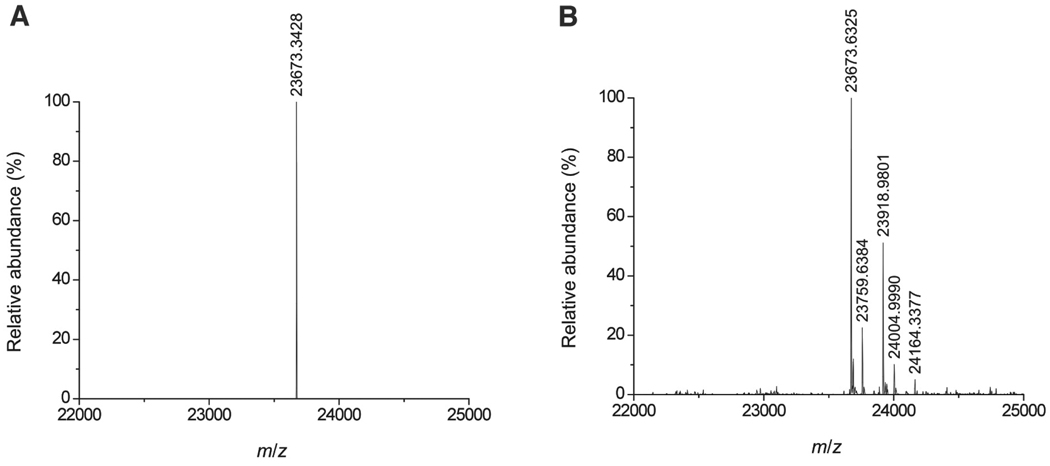
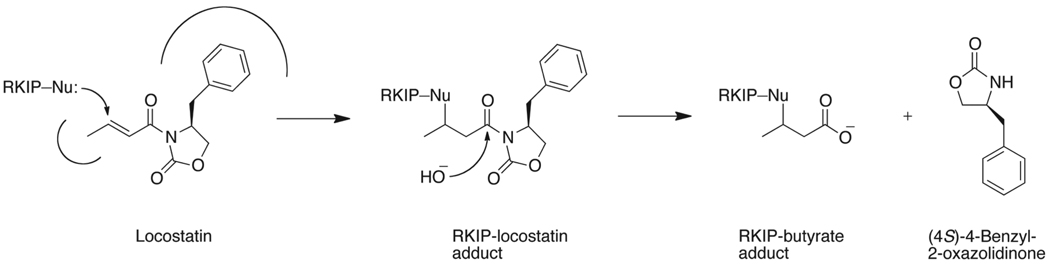
We sought to further define the mode of binding of locostatin to RKIP. We found by LC-MS that locostatin primarily alkylates one residue on RKIP, with a small degree of alkylation of a second (Fig. 3). The mass spectra showed peaks corresponding to products of modification of RKIP by locostatin and by a smaller entity, the latter corresponding in mass to a butyrate moiety (Fig. 3). The RKIP-locostatin signal became less intense and the RKIP-butyrate signal became more intense slowly over time (data not shown), suggesting conversion of the first species to the second. The MS data thus suggested that the bound locostatin is slowly converted to a bound butyrate moiety as a result of hydrolysis of the acyl group from the oxazolidinone (Fig. 4).
FIGURE 3.
Modification of RKIP by locostatin. RKIP (20 µM) was incubated with locostatin (100 µM) or DMSO alone (0.1%) as control at 37°C for 6 h and then subjected to LC-MS. Mass spectra are shown for (A) the control RKIP sample and (B) the locostatin-treated RKIP sample. In addition to the His-tagged RKIP molecular ion peak, the mass spectrum in (B) shows a prominent peak at an m/z of RKIP+246 (corresponding to an RKIP-locostatin adduct) and a weaker peak at RKIP+86 (corresponding to an RKIP-butyrate adduct), as well as still weaker peaks at RKIP+2×246 and RKIP+246+86 (corresponding to a small degree of alkylation of a secondary site).
FIGURE 4.
Mechanism of modification of RKIP by locostatin and hydrolysis of the bound locostatin. The scheme shows the putative mechanism of alkylation of RKIP by locostatin and hydrolysis of the RKIP-locostatin adduct to an RKIP-butyrate adduct, consistent with the MS data shown in Fig. 3.
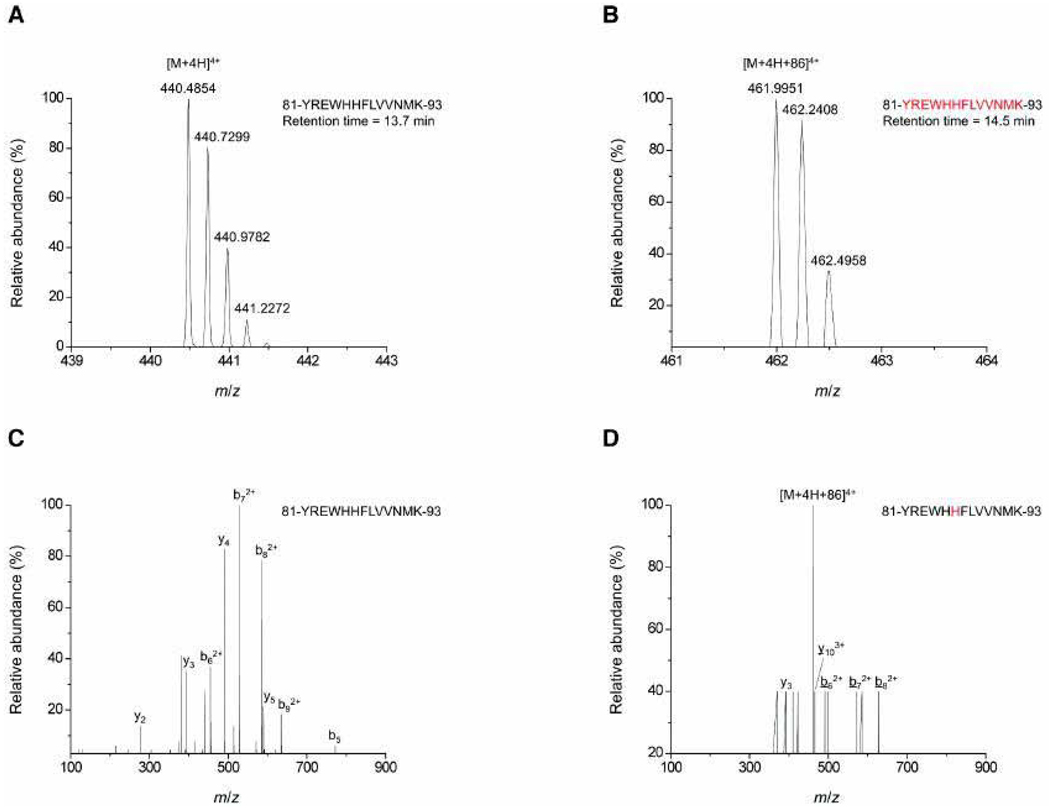
Next, we focused on identification of the primary site of alkylation of RKIP by locostatin. Prior work has shown that the residue alkylated by locostatin is not RKIP’s regulatory Ser153 residue,3,25 whose phosphorylation results in dissociation of RKIP from Raf-1 and association with GRK2.6 Therefore, in order to identify the main residue on RKIP alkylated by locostatin, we subjected RKIP treated with locostatin to LC-MS/MS. We identified His86 as the alkylated residue (Fig. 5). His86 is a highly conserved residue in RKIP’s ligand binding pocket.
FIGURE 5.
Locostatin modifies His86, a highly conserved residue in RKIP’s ligand-binding pocket. RKIP (20 µM) was incubated with locostatin (100 µM) or DMSO alone (0.1%) as control at 37°C for 24 h and then subjected to LC-MS/MS. Mass spectra are shown for the fragment 81-YREWHHFLVVNMK-93 from tryptic digestion of (A) the control RKIP sample and (B) the locostatin-treated RKIP sample. Tandem mass spectra are shown for the same tryptic fragment from (C) the control RKIP sample and (D) the locostatin-treated RKIP sample, with modified ions underlined.
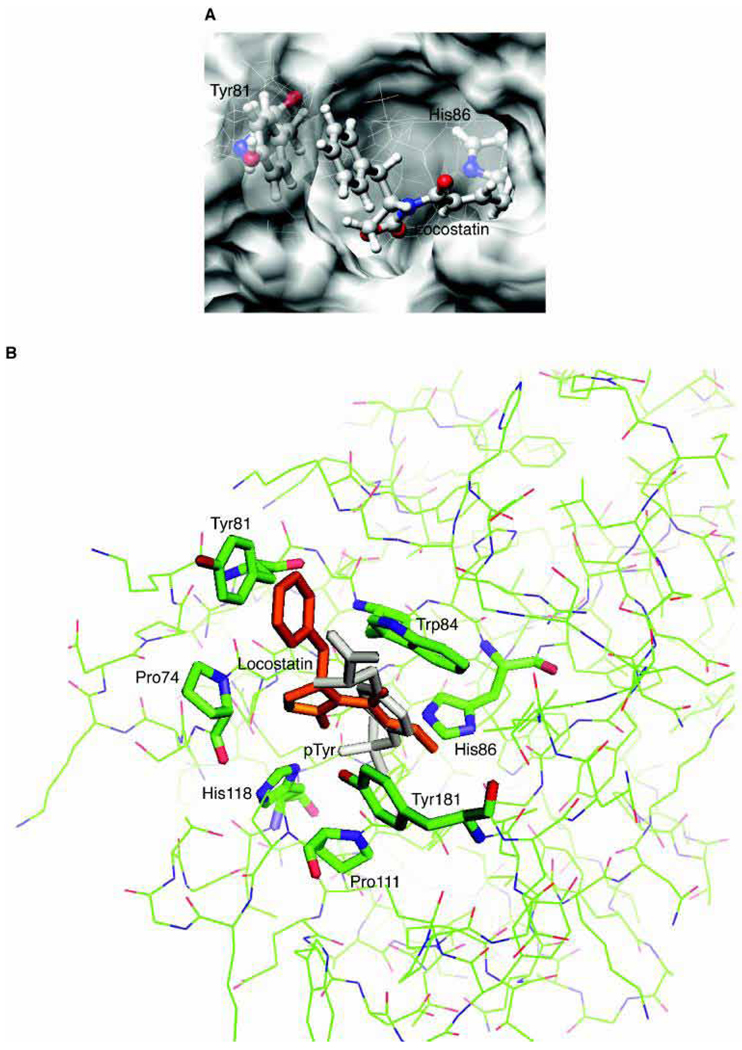
Computational docking and subsequent quantum mechanical refinement of locostatin to RKIP revealed that the lowest-energy interaction for the recognition step (not considering the subsequent alkylation reaction) results in locostatin bound in such a way that N3 and N1 of His86 are 5 Å and 5.6 Å, respectively, from the β-carbon of the α,β-unsaturated acyl group (Fig. 6). This Michael acceptor functionality on locostatin is thus well situated to alkylate His86.
FIGURE 6.
Computational modeling of the recognition interaction between locostatin and RKIP results in locostatin well positioned to modify His86 in RKIP’s ligand-binding pocket. The structure of human RKIP bound to pTyr (PDB code 2QYQ) was utilized for the docking calculations, after removing the pTyr coordinates to allow for docking of locostatin. (A) The docked structure for the calculated lowest-energy interaction of locostatin for the recognition step (not considering the subsequent alkylation reaction). (B) Superimposition of the docked locostatin molecule onto the RKIP-pTyr cocrystal structure, with locostatin in orange and pTyr in light gray. [Images for Figures 6A and 6B were, respectively, rendered with UCSF Chimera (http://www.cgl.ucsf.edu/chimera/) and PyMOL (http://www.pymol.org/).]
The lowest-energy docked structure has locostatin in a trans configuration, and the phenyl ring of its benzyl substituent closely abuts the phenyl ring of Tyr81, with a centroid-centroid distance of 4.16 Å and a distance of the Tyr81 centroid to the locostatin phenyl plane of 3.65 Å (Fig. 6). This raises the possibility of a pi-pi stacking interaction between the locostatin’s phenyl group and Tyr81. One face of the hydrophobic phenyl ring of locostatin is still positioned partly out of the ligand-binding pocket and would be exposed to the aqueous environment. This may account for the tendency of locostatin to cause aggregation of RKIP. Alternatively, and not mutually exclusively, aggregation may be driven by modification-induced conformational changes in the protein that expose hydrophobic residues.
Based on the RKIP-pTyr cocrystal structure (PDB code 2QYQ), binding of locostatin would be expected to sterically block pTyr binding (Fig. 6B). It is not yet clear whether the ligand-binding pocket of RKIP functions as a pTyr-binding site under physiological conditions. While there is evidence that phosphorylation of Ser388, Ser389, and Tyr341 on Raf-1 increases affinity of Raf-1 for RKIP (Ref. 18 and R. Leo Brady, personal communication), there are also results to suggest that interaction of Raf-1 with RKIP instead blocks the phosphorylation of these same residues on Raf-1.26 The ligand-binding pocket could also conceivably accommodate Tyr and Asp residues, though with lower expected affinity than pTyr.18 It is possible that phosphorylation of Tyr residues on Raf-1 such as its Tyr341 residue (Ref. 18 and R. Leo Brady, personal communication) and GRK2 could modulate the affinity of binding of these proteins to RKIP.
Alkylation of His86 on RKIP by locostatin would sterically hinder binding of proteins whose association is dependent on interaction in the ligand-binding pocket. There are, of course, other possibilities for how binding of locostatin to RKIP disrupts binding to Raf-1 and GRK2. Locostatin could cause a conformational change in RKIP that affects protein-protein interactions. In addition, precipitation of RKIP on alkylation by locostatin may itself be a part of the mechanism, although the fact that locostatin disrupts interactions with Raf-1 and GRK2 but not IKKα and TAK1 suggests a protein-selective mechanism, more in line with steric hindrance or a specific conformational effect. The fact that interactions of RKIP with certain proteins but not others are disrupted by locostatin also implies very different modes of binding between different proteins. Some proteins such as Raf-1 and GRK2 may bind within the ligand-binding pocket as a critical part of their overall association with RKIP, although elements outside of the ligand-binding pocket are also important, such as the phosphorylation state of Ser153.6 Other proteins such as IKKα and TAK1 may be less sensitive to competitive occupancy of the pocket, possibly binding another part of RKIP entirely. The present study greatly enhances an understanding of the molecular mechanism of action of locostatin and demonstrates how this small molecule can be useful for the dissection of specific RKIP interactions and functions.
ACKNOWLEDGMENTS
We thank Prof. Kam C. Yeung (University of Toledo) for DNA constructs for RKIP-binding proteins. This work was supported by National Institutes of Health Grant No. GM077622 (to GF) and National Science Foundation Career Award Grant No. CHE-0847340 (to JAG).
ABBREVIATIONS
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- ESI
electrospray ionization
- GRK2
G protein-coupled receptor kinase 2
- HA
hemagglutinin
- HPLC
high-performance liquid chromatography
- IKKα
inhibitor of κB kinase α
- LC
liquid chromatography
- MM
molecular mechanics
- MS
mass spectrometry
- PAGE
polyacrylamide gel electrophoresis
- PB
Poisson-Boltzmann
- PDB
Protein Data Bank
- PEBP
phosphatidylethanolamine-binding protein
- pTyr
O-phosphotyrosine
- PVDF
polyvinylidene fluoride
- RKIP
Raf kinase inhibitor protein
- QM
quantum mechanics
- SDS
sodium dodecyl sulfate
- TAK1
transforming growth factor β-activated kinase 1
REFERENCES
- 1.Mc Henry KT, Ankala SV, Ghosh AK, Fenteany G. A non-antibacterial oxazolidinone derivative that inhibits epithelial cell sheet migration. ChemBioChem. 2002;3:1105–1111. doi: 10.1002/1439-7633(20021104)3:11<1105::AID-CBIC1105>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 2.Mc Henry KT, Montesano R, Zhu S, Beshir AB, Tang HH, Yeung KC, Fenteany G. Raf kinase inhibitor protein positively regulates cell-substratum adhesion while negatively regulating cell-cell adhesion. J Cell Biochem. 2008;103:972–985. doi: 10.1002/jcb.21470. [DOI] [PubMed] [Google Scholar]
- 3.Zhu S, Mc Henry KT, Lane WS, Fenteany G. A chemical inhibitor reveals the role of Raf kinase inhibitor protein in cell migration. Chem Biol. 2005;12:981–991. doi: 10.1016/j.chembiol.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 5.Yeung K, Janosch P, McFerran B, Rose DW, Mischak H, Sedivy JM, Kolch W. Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the Raf kinase inhibitor protein. Mol Cell Biol. 2000;20:3079–3085. doi: 10.1128/mcb.20.9.3079-3085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorenz K, Lohse MJ, Quitterer U. Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature. 2003;426:574–579. doi: 10.1038/nature02158. [DOI] [PubMed] [Google Scholar]
- 7.Yeung KC, Rose DW, Dhillon AS, Yaros D, Gustafsson M, Chatterjee D, McFerran B, Wyche J, Kolch W, Sedivy JM. Raf kinase inhibitor protein interacts with NF-κB-inducing kinase and TAK1 and inhibits NF-κB activation. Mol Cell Biol. 2001;21:7207–7217. doi: 10.1128/MCB.21.21.7207-7217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang H, Park S, Sun SC, Trumbly R, Ren G, Tsung E, Yeung KC. RKIP inhibits NF-κB in cancer cells by regulating upstream signaling components of the IκB kinase complex. FEBS Lett. 2010;584:662–668. doi: 10.1016/j.febslet.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernier I, Tresca JP, Jolles P. Ligand-binding studies with a 23 kDa protein purified from bovine brain cytosol. Biochim Biophys Acta. 1986;871:19–23. doi: 10.1016/0167-4838(86)90128-7. [DOI] [PubMed] [Google Scholar]
- 10.Schoentgen F, Saccoccio F, Jolles J, Bernier I, Jolles P. Complete amino acid sequence of a basic 21-kDa protein from bovine brain cytosol. Eur J Biochem. 1987;166:333–338. doi: 10.1111/j.1432-1033.1987.tb13519.x. [DOI] [PubMed] [Google Scholar]
- 11.Grandy DK, Hanneman E, Bunzow J, Shih M, Machida CA, Bidlack JM, Civelli O. Purification, cloning, and tissue distribution of a 23-kDa rat protein isolated by morphine affinity chromatography. Mol Endocrinol. 1990;4:1370–1376. doi: 10.1210/mend-4-9-1370. [DOI] [PubMed] [Google Scholar]
- 12.Tohdoh N, Tojo S, Agui H, Ojika K. Sequence homology of rat and human HCNP precursor proteins, bovine phosphatidylethanolamine-binding protein and rat 23-kDa protein associated with the opioid-binding protein. Brain Res Mol Brain Res. 1995;30:381–384. doi: 10.1016/0169-328x(95)00029-r. [DOI] [PubMed] [Google Scholar]
- 13.Goumon Y, Muller A, Glattard E, Marban C, Gasnier C, Strub JM, Chasserot-Golaz S, Rohr O, Stefano GB, Welters ID, Van Dorsselaer A, Schoentgen F, Aunis D, Metz-Boutigue MH. Identification of morphine-6-glucuronide in chromaffin cell secretory granules. J Biol Chem. 2006;281:8082–8089. doi: 10.1074/jbc.M502298200. [DOI] [PubMed] [Google Scholar]
- 14.Atmanene C, Laux A, Glattard E, Muller A, Schoentgen F, Metz-Boutigue MH, Aunis D, Van Dorsselaer A, Stefano GB, Sanglier-Cianferani S, Goumon Y. Characterization of human and bovine phosphatidylethanolamine-binding protein (PEBP/RKIP) interactions with morphine and morphine-glucuronides determined by noncovalent mass spectrometry. Med Sci Monit. 2009;15:BR178–BR187. [PubMed] [Google Scholar]
- 15.Schoentgen F, Seddiqi N, Bucquoy S, Jolles P, Lemesle-Varloot L, Provost K, Mornon JP. Main structural and functional features of the basic cytosolic bovine 21 kDa protein delineated through hydrophobic cluster analysis and molecular modelling. Protein Eng. 1992;5:295–303. doi: 10.1093/protein/5.4.295. [DOI] [PubMed] [Google Scholar]
- 16.Dadvar P, Kovanich D, Folkers GE, Rumpel K, Raijmakers R, Heck AJ. Phosphatidylethanolamine-binding proteins, including RKIP, exhibit affinity for phosphodiesterase-5 inhibitors. ChemBioChem. 2009;10:2654–2662. doi: 10.1002/cbic.200900452. [DOI] [PubMed] [Google Scholar]
- 17.Pikielny CW, Hasan G, Rouyer F, Rosbash M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron. 1994;12:35–49. doi: 10.1016/0896-6273(94)90150-3. [DOI] [PubMed] [Google Scholar]
- 18.Park S, Rath O, Beach S, Xiang X, Kelly SM, Luo Z, Kolch W, Yeung KC. Regulation of RKIP binding to the N-region of the Raf-1 kinase. FEBS Lett. 2006;580:6405–6412. doi: 10.1016/j.febslet.2006.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banfield MJ, Barker JJ, Perry AC, Brady RL. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure. 1998;6:1245–1254. doi: 10.1016/s0969-2126(98)00125-7. [DOI] [PubMed] [Google Scholar]
- 20.Serre L, Vallee B, Bureaud N, Schoentgen F, Zelwer C. Crystal structure of the phosphatidylethanolamine-binding protein from bovine brain: a novel structural class of phospholipid-binding proteins. Structure. 1998;6:1255–1265. doi: 10.1016/s0969-2126(98)00126-9. [DOI] [PubMed] [Google Scholar]
- 21.Rath O, Park S, Tang HH, Banfield MJ, Brady RL, Lee YC, Dignam JD, Sedivy JM, Kolch W, Yeung KC. The RKIP (Raf-1 Kinase Inhibitor Protein) conserved pocket binds to the phosphorylated N-region of Raf-1 and inhibits the Raf-1-mediated activated phosphorylation of MEK. Cell Signal. 2008;20:935–941. doi: 10.1016/j.cellsig.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Granovsky AE, Clark MC, McElheny D, Heil G, Hong J, Liu X, Kim Y, Joachimiak G, Joachimiak A, Koide S, Rosner MR. Raf kinase inhibitory protein function is regulated via a flexible pocket and novel phosphorylation-dependent mechanism. Mol Cell Biol. 2009;29:1306–1320. doi: 10.1128/MCB.01271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frisch MJT, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JJA, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09, Revision A.2. Wallingford, CT: 2009. [Google Scholar]
- 24.Guo J, Wu HW, Hu G, Han X, De W, Sun YJ. Sustained activation of Src-family tyrosine kinases by ischemia: a potential mechanism mediating extracellular signal-regulated kinase cascades in hippocampal dentate gyrus. Neuroscience. 2006;143:827–836. doi: 10.1016/j.neuroscience.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 25.Shemon AN, Eves EM, Clark MC, Heil G, Granovsky A, Zeng L, Imamoto A, Koide S, Rosner MR. Raf Kinase Inhibitory Protein protects cells against locostatin-mediated inhibition of migration. PLoS One. 2009;4:e6028. doi: 10.1371/journal.pone.0006028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trakul N, Menard RE, Schade GR, Qian Z, Rosner MR. Raf kinase inhibitory protein regulates Raf-1 but not B-Raf kinase activation. J Biol Chem. 2005;280:24931–24940. doi: 10.1074/jbc.M413929200. [DOI] [PubMed] [Google Scholar]