Abstract
Background & objectives
In drug resistant, especially multi-drug resistant (MDR) tuberculosis, fluoroquinolones (FQs) are used as second line drugs. However, the incidence of FQ-resistant Mycobacterium tuberculosis is rapidly increasing which may be due to extensive use of FQs in the treatment of various other diseases. The most important known mechanism i.e., gyrA mutation in FQ resistance is not observed in a significant proportion of FQ resistant M. tuberculosis isolates suggesting that the resistance may be because of other mechanisms such as an active drug efflux pump. In this study we evaluated the role of the efflux pumps in quinolone resistance by using various inhibitors such as carbonyl cyanide m-chlorophenyl hydrazone (CCCP), 2,4-dinitrophenol (DNP) and verapamil, in clinical isolates of M. tuberculosis.
Methods
A total of 55 M. tuberculosis clinical isolates [45 ofloxacin (OFL) resistant and 10 ofloxacin sensitive] were tested by Resazurin microtitre assay (REMA) to observe the changes in ofloxacin minimum inhibitory concentration (MIC) levels in presence of efflux inhibitors as compared to control (without efflux inhibitor).
Results
The MIC levels of OFL showed 2-8 folds reduction in presence of CCCP (16/45; 35.5%), verapamil (24/45; 53.3%) and DNP (21/45; 46.6%) while in case of isolates identified as OFL sensitive these did not show any effect on ofloxacin MICs. In 11 of 45 (24.5%) isolates change in MIC levels was observed with all the three inhibitors. Overall 30 (66.6%) isolates had reduction in OFL MIC after treatment with these inhibitors. A total of eight isolates were sequenced for gyrA gene, of which, seven (87.5%) showed known mutations. Of the eight sequenced isolates, seven (87.5%) showed 2 to 8 fold change in MIC in presence of efflux inhibitors.
Interpretation & conclusions
Our findings suggest the involvement of active efflux pumps of both Major Facilitator Super Family (MFS) family (inhibited by CCCP and DNP) and ATP Binding Cassette (ABC) transporters (inhibited by verapamil) in the development of OFL resistance in M. tuberculosis isolates. Epidemiological significance of these findings needs to be determined in prospective studies with appropriate number of samples / isolates.
Keywords: Drug resistance, efflux pump inhibitors, MIC, Mycobacterium tuberculosis, ofloxacin, tuberculosis
Mycobacterium tuberculosis, the aetiological agent of tuberculosis (TB), has re-emerged as a killer pathogen in western countries after the increase of HIV-AIDS cases and development of resistance to various anti-tubercular drugs. This increase in the number of multi-drug resistant M. tuberculosis isolates has drawn the attention towards the identification of alternate drugs like fluoroquinolones (FQs) for the treatment of TB. It is known that M. tuberculosis commonly acquires drug resistant phenotype by accumulation of mutations in the structural genes encoding the drug target or the enzymes involved in drug activation. Other known cause of drug resistance in mycobacteria is efflux of drug molecules1. The principal cellular target of the FQs is the DNA gyrase encoded by gyrA and gyrB genes. Mutation in the quinolone resistance determining region (QRDR) of gyrA was the most common cause of FQ resistance in various organisms2,3. However, studies carried out in India have reported that only 11.74 and 45 per cent5 of ofloxacin resistant M. tuberculosis isolates harbour mutations in their gyrA gene and no mutation was found in gyrB gene. As mutations in DNA gyrase alone do not account for the mechanism(s) of resistance in a significant proportion of FQs resistant M. tuberculosis isolates, it suggests the need to investigate the role of alternate mechanisms, like efflux pumps. The upregulation of efflux systems can significantly decrease the intracellular concentration of many antibiotics, reducing their clinical efficacy. For this reason attention has been focused on identifying inhibitors of the efflux systems of Gram-negative and Gram-positive bacteria that could potentially be used in combination with antibiotics to improve efficacy and abolish resistance1. Banerjee and co-workers6 observed that carbonyl cyanide m chlorophenyl hydrazone (CCCP), verapamil and 2,4-dinitro phenol (DNP) increased the accumulation of drug possibly due to inhibition of active efflux. Several mycobacterial efflux pumps associated with FQs resistance have been described. These efflux pumps include the pumps of Major Facilitator Superfamily (MFS) family (lfrA, Rv1634 and Rv1258c) and ATP Binding Cassette (ABC) transporters (DrrAB, PstB and Rv2686c-2687c-2688c)1. For better understanding of drug resistance and to find out the newer drugs and/or identify suitable drug targets for better treatment of TB, there is a need to understand the exact mechanism(s) of resistance to FQs in M. tuberculosis. In the present study we have studied the effect of certain efflux inhibitors on in vitro susceptibility levels in ofloxacin (OFL)-resistant clinical M. tuberculosis isolates.
Material & Methods
M. tuberculosis isolates: The study was performed in the National JALMA Institute for Leprosy & Other Mycobacterial Diseases, Agra. A total of 55 clinical isolates of M. tuberculosis along with M. tuberculosis reference strain H37Rv were included in the present study. Isolates were obtained from Mycobacterial Repository Centre of the Institute, which were deposited in the repository from July 2004 through January 2008. These included isolates from Agra (n=45), Delhi (n=3), Kanpur (n=3), Varanasi (n=2), Allahabad (n=1) and Jaipur (n=1). Ofloxacin-resistant M. tuberculosis isolates (n=45) had ofloxacin MIC of ≥4 mg/l tested by Lowenstein-Jensen (L-J) method. Of the 45 OFL-resistant isolates, 31 belonged to the MDR group. Ten M. tuberculosis isolates were ofloxacin-sensitive with MIC < 2-4 mg/l. All the M. tuberculosis isolates were biochemically identified7.
Effect of efflux inhibitors on minimum inhibitory concentration (MIC) levels of OFL: To determine the extent of the efflux pump mediated ofloxacin resistance in M. tuberculosis isolates, MIC levels for ofloxacin were determined using Resazurin microtitre assay8 in the presence or absence of efflux pump inhibitors [CCCP and DNP and verapamil (Sigma, USA)]. CCCP and DNP are the proton motive force inhibitors whereas verapamil is a calcium channel blocker for ABC transporters6. Stock solution of CCCP and DNP was prepared in DMSO while verapamil was dissolved in distilled water. Final concentrations used in Resazurin microtiter assay CCCP (1 mg/l), verapamil (5 mg/l) and DNP (20 mg/l).
A total of 100 μl volume of Middlebrook 7H9 broth (Difco, USA) supplemented with 10 per cent oleic acid, albumin, dextrose and catalase (OADC) and 0.2 per cent glycerol was dispensed in the wells of a 96-well cell culture plate (Nunc, Denmark). Different concentrations of ofloxacin prepared in Middlebrook 7H9 medium were: 0.25, 0.5, 1, 2, 4, 8, 16, 32 and 64 mg/l. M. tuberculosis growth was taken from L-J slope, homogenized bacterial suspension of No.1 McFarland standard was prepared and diluted to 1:20 in 7H9 broth. This diluted suspension (100 μl) was used to inoculate each well of the plate. Plates were sealed and incubated at 37°C for one week. Resazurin dye (Sigma, USA) (0.02%, 25 μl) was added to each well; plates were reincubated for two more days. A change in colour from blue to pink indicated the growth of bacteria and the MIC was read as the minimum ofloxacin concentration that prevented the colour change in resazurin dye.
DNA extraction, PCR, and DNA sequencing: Genomic DNA from mycobacterial isolate (log phase on L-J slant) was extracted using phsiochemical procedure as described by van Soolingen et al9. A DNA fragment of 320 bp of quinolone resistant determining region (QRDR) of gyrA gene in M. tuberculosis isolates was amplified using the following primers: 5’CAG CTA CAT CGA CTA TGC GA 3’ and 5’GGG CTT CGG TGT TAC CTC AT 3’ as described earlier4. Amplification reactions consisted of a denaturation step of 3 min at 95°C, followed by 35 cycles of 1 min at 94°C, 1 min at 51°C, and 2 min at 72°C, and a final extension step of 10 min at 72°C. The amplification product was used as the template in direct nucleotide sequencing. Out of 45, ofloxacin- resistant isolates, eight were sequenced for QRDR of gyrA. Amplified PCR products were purified from 1 per cent agarose gel using QIAEX II Gel Extraction Kit (Qiagen, Germany). All purified PCR products were sequenced with ABI PRISM 310 automated DNA sequencer (Applied Biosystem, USA) as per manufacturer's instructions. Sequences generated were confirmed using BLAST tool (available online at www.ncbi.nih.gov/BLAST) and compared with M. tuberculosis H37Rv strain using ClustalW multiple sequence alignment (www.ebi.ac.uk/Tools/clustalw2).
Results
MICs of ofloxacin determined in absence of efflux inhibitors were compared with those determined in presence of efflux inhibitors. Two fold or more reduction in MIC levels was considered as an indication of presence of efflux activity in ofloxacin-resistant M. tuberculosis isolates10. It was observed that the MIC levels of ofloxacin decreased in 16 of 45 (35.5%) isolates in the presence of CCCP, in 24 (53.3%) isolates in the presence of verapamil and in 21 (46.6%) isolates in the presence of DNP. All three efflux inhibitors (CCCP, DNP and verapamil) showed the MIC reduction in 11 (24.5%) isolates (Table I). Efflux inhibitors did not have any effect on ofloxacin MICs in 10 (100%) ofloxacin-sensitive isolates. Of the 10 ofloxacin sensitive isolates, three were inhibited at 1 μg/ml and seven were inhibited at 2 μg/ml concentration.
Table I.
Summary of effect of efflux inhibitors on ofloxacin MICs in resistant M. tuberculosis isolates
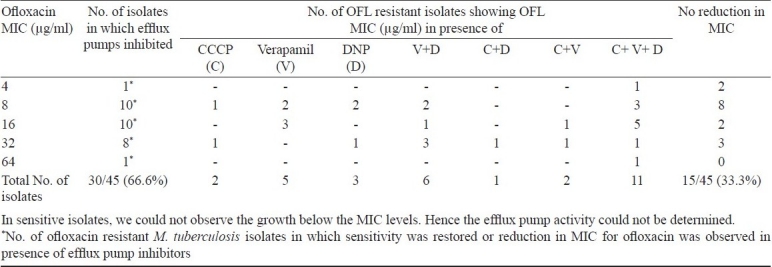
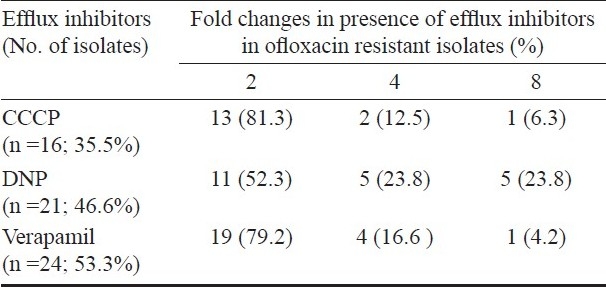
In the presence of DNP, the MIC values for ofloxacin were found to decrease 4-folds in 10 (22.2%) and 2-folds in 11 (24.4%) isolates, verapamil showed 4-folds inhibition in five (11.1%) isolates and 2-folds in 19 (42.2%) isolates. In presence of CCCP efflux inhibitor, MIC levels for ofloxacin were lowered 4-folds in three (6.6%) and 2-folds in 13 (28.8%) ofloxacin-resistant isolates (Table II).
Table II.
Fold changes in ofloxacin MIC of M. tuberculosis isolates (n=45) in presence of efflux inhibitors (CCCP, DNP and verapamil)
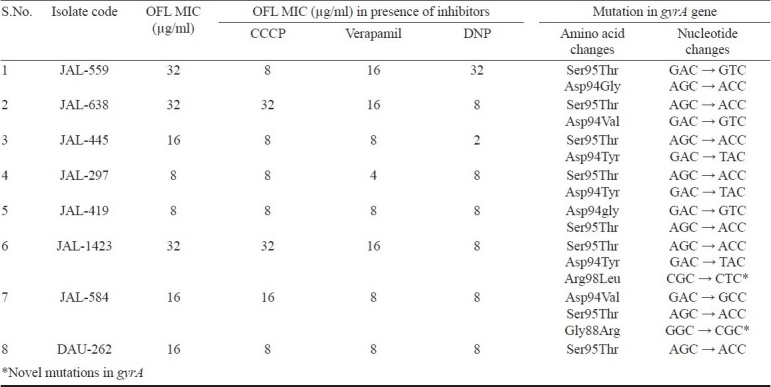
Of the 30 ofloxacin resistant isolates, eight were sequenced for QRDR of gyrA. It was observed that all eight isolates showed single mutation at codon Ser95Thr. Seven isolates (87.5%) showed the mutations at codon Asp94Tyr (JAL -445, JAL-297 and JAL -1423), Asp94Val (JAL -638 and JAL -584), Asp94Gly (JAL -559 and JAL -419), Gly88Arg (JAL -584) and Arg98Leu (JAL -1423) in ofloxacin resistant isolates. Mutations in gyrA gene and its relation to MIC levels of ofloxacin were described in Table III. Two novel mutations in different codons (Arg98Leu and Gly88Arg) of gyrA were also found along with known mutations. Except mutation at Ser95thr codon, no other mutation was observed in one isolate (DAU-262) in gyrA gene.
Table III.
Showing change in MIC levels for OFL in the presence of inhibitors and presence of mutations in gyrA gene
Discussion
Data from the present study revealed that the MICs of maximum number of isolates were affected in the presence of verapamil suggesting the importance of efflux pumps of ABC transporter family in ofloxacin resistance in M. tuberculosis. It was also observed that MIC (0.5 mg/l) of H37Rv was not affected in presence of efflux pump inhibitors. Piddock and Ricci11 had also reported that CCCP and reserpine do not change the MICs of FQs for the reference strain M. tuberculosis H37Rv.
Overall inhibitory effect of efflux inhibitors on ofloxacin MICs was observed in 66.6 per cent M. tuberculosis isolates showing the contribution of active efflux pumps in the development of ofloxacin resistance in these isolates. In these isolates, MIC levels decreased two to eight fold after treatment with efflux pump inhibitors but only in 20 per cent isolates microbiological classification changed from resistant to sensitive (out of 6, 1 isolate was blocked by all the three inhibitors, 2 were blocked by DNP, 2 by DNP and verapamil whereas 1 isolate was blocked by verapamil). Hence pumps appear to be contributing to increase in the level of resistance majority of isolates. It has been reported that in the presence of reserpine and MC 207.110 efflux pump inhibitors two-fold reduction in MIC was seen in 57-100 per cent M. tuberculosis isolates resistant to FQs (ciprofloxacin, moxifloxacin, levofloxacin, ofloxacin, gatifloxacin) and 57 per cent to linezolid10. Low activity of these inhibitors was also reported in ofloxacin-sensitive isolates10. Reduction in MIC level in presence of CCCP, DNP and verapamil inhibitors provide evidence for the presence of both type proton motive force and ATP dependent extrusion system involved in FQs resistance.
From the earlier studies, DNA sequencing of gyrA showed that all the strains possessed a natural mutation at codon Ser95Thr (AGC→ACC) polymorphism, which did not have a significant impact on fluoroquinolone susceptibility12,13. The present study also revealed the same. Mutation at Ser95Thr codon was previously reported as a marker for evolutionary genetics and does not correlate with drug resistance14 or it has no direct role in the development of drug resistance, as it also occurs in drug-sensitive strains4,14. Seven of the eight OFL resistant isolates, sequenced possessed mutations other than Ser95Thr in the gyrA gene. Of which, two mutations at codon Gly88Arg (GGC → CGC), Arg98Leu (CGC → CTC) were novel and were not reported previously. In FQ resistant isolates, the mutations at codon 88 i.e., Gly88Cys and Gly88Ala mutations were reported earlier15,16.
We also observed AGC → ACC point mutation at codon Ser95Thr in one OFL resistant isolates (DAU-262). In this isolate only efflux mediated drug resistance mechanism (PMF and ATP dependent) were involved in which OFL MIC was decreased 2-fold (from 16 to 8 μg/ml) in presence of CCCP, DNP and verapamil inhibitors. Only one OFL resistant isolate (JAL-419) showed a point mutation at codon Asp94gly (GAC → GTC) responsible for OFL resistance and OFL MIC level did not decrease in presence of tested efflux inhibitors. In the remaining six isolates both mechanisms i.e., point mutation and efflux pumps were involved in OFL resistance. The OFL MIC for these isolates was found to be between 8 and 32 μg/ml. Contribution to degree of quinolone resistance thus appears to be significant.
Gupta and co-workers17 have also reported the reversal of resistance to all major anti-tuberculous drugs (rifampicin, isoniazid, streptomycin, ofloxacin) in presence of efflux pump inhibitors (CCCP and verapamil) which vary in different mycobacterial species and isolates. Results of the present study provide further evidence that efflux mechanism plays an important role in the development of ofloxacin resistance in M. tuberculosis isolates. It may further be speculated that as the drugs of FQ group show cross-resistance, these efflux pumps might be a common mechanism contributing to this phenomenon. As the isolates were not selected by any acceptable sampling procedure, it will not be appropriate to discuss the epidemiological relevance and statistical significance of these findings.
This preliminary study shows the role of efflux pumps inhibitor (CCCP, DNP and verapamil) in conferring ofloxacin resistance phenotype in M. tuberculosis isolates. There is a need to further investigate the changes in mRNA levels of genes encoding efflux pumps as well as detection of mutations in gyrA prospectively by selecting the isolates using appropriate sampling procedure.
Acknowledgments
Authors acknowledge the Department of Biotechnology, and Central TB Division, New Delhi, for providing financial support and thank Shriyut Jitendra, Rajesh, Sanjay and Dhaniram for technical support. The first two authors (MS and GPSJ) acknowledge the Council of Scientific and Industrial Research, New Delhi for providing Senior Research Fellowship.
References
- 1.De Rossi E, Ainsa JA, Riccardi G. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol Rev. 2006;30:36–52. doi: 10.1111/j.1574-6976.2005.00002.x. [DOI] [PubMed] [Google Scholar]
- 2.Kocagoz T, Hackbarth CJ, Unsal I, Rosenberg EY, Nikaido H, Chambers HF. Gyrase mutations in laboratory- selected, fluoroquinolone-resistant mutants of Mycobacterium tuberculosis H37Ra. Antimicrob Agents Chemother. 40:1768–74. doi: 10.1128/aac.40.8.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takiff HE, Salazar L, Guerrero C, Philipp W, Huang WM, Kreiswirth B, et al. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–80. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqi N, Shamim M, Hussain S, Choudhary RK, Ahmed N, Prachee S, et al. Molecular characterization of multidrug-resistant isolates of Mycobacterium tuberculosis from patients in North India. Antimicrob Agents Chemother. 2002;46:443–50. doi: 10.1128/AAC.46.2.443-450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sulochana S, Narayanan S, Paramasivan CN, Suganthi C, Narayanan PR. Analysis of fluoroqunolone resistance in clinical isolates of Mycobacterium tuberculosis from India. J Chemother. 2007;19:89–93. doi: 10.1179/joc.2007.19.2.166. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee SK, Bhatt K, Rana S, Misra P, Chakraborti PK. Involvement of an efflux system in mediating high level of fluoroquinolone resistance in Mycobacterium smegmatis. Biochem Biophys Res Commun. 1996;226:362–8. doi: 10.1006/bbrc.1996.1362. [DOI] [PubMed] [Google Scholar]
- 7.Vestal AL. Procedure for the isolation and identification of mycobacteria. Atlanta, Georgia: Cente for Disease Control and Prevention; 1977. US Department of Health, Education and Welfare Pub no. (CDC)77 - 8230; pp. 65–98. [Google Scholar]
- 8.Martin A, Camacho M, Portaels F, Palomino JC. Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: Rapid, simple and inexpensive method. Antimicrob Agents Chemother. 2003;47:3616–9. doi: 10.1128/AAC.47.11.3616-3619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Soolingen D, de Hass PEW, Hermans PWM, Groenen MA, van Embden JOA. Comparision of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of M. tuberculosis. J Clin Microbiol. 1993;31:1987–95. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escribano I, Rodríguez JC, Llorca B, García-Pachon E, Ruiz M, Royo G. Importance of the efflux pump systems in the resistance of Mycobacterium tuberculosis to fluoroquinolones and linezolid. Chemotherapy. 2007;53:397–401. doi: 10.1159/000109769. [DOI] [PubMed] [Google Scholar]
- 11.Piddock LJ, Ricci V. Accumulation of five fluoroquinolones by Mycobacterium tuberculosis H37Rv. J Antimicrob Chemother. 2001;48:787–91. doi: 10.1093/jac/48.6.787. [DOI] [PubMed] [Google Scholar]
- 12.Ginsburg AS, Grosset JH, Bishai WR. Fluoroquinolones, tuberculosis, and resistance. Lancet Infect Dis. 2003;3:432–42. doi: 10.1016/s1473-3099(03)00671-6. [DOI] [PubMed] [Google Scholar]
- 13.Shi R, Zhang J, Li C, Kazumi Y, Sugawara I. Emergence of Ofloxacin resistance in Mycobacterium tuberculosis clinical isolates from China as determined by gyrA mutation analysis using denaturing high-pressure liquid chromatography and DNA sequencing. J Clin Microbiol. 2006;44:4566–8. doi: 10.1128/JCM.01916-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sreevatsan S, Pan X, Stockbauer KE, Connell N, Kreiswirth BN, Whittam TS, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionary recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–74. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitaksajjakul P, Wongwit W, Punprasit W, Eampokalap B, Peacock S, Ramasoota P. Mutations in gyrA and gyrB genes of fluoroquinolone-resistant Mycobacterium tuberculosis from TB pateints in Thailand. Southeast Asian J Trop Med Public Health. 2005;36:228–37. [PubMed] [Google Scholar]
- 16.Huang TS, Kunin CM, Shin-Jung Lee S, Chen YS, Tu HZ, Liu YC. Trends in fluoroquinolone resistance of Mycobacterium tuberculosis complex in a Taiwanese medical centre: 1995-2003. J Antimicrob Chemother. 2005;56:1058–62. doi: 10.1093/jac/dki353. [DOI] [PubMed] [Google Scholar]
- 17.Gupta AK, Chauhan DS, Srivastava K, Das R, Batra S, Mittal M, et al. Estimation of efflux mediated multi-drug resistance and its correlation with expression levels of two major efflux pumps in mycobacteria. J Commun Dis. 2006;38:246–54. [PubMed] [Google Scholar]


