Abstract
Background & objectives
Limited information is available on shiga toxin producing Escherichia coli (STEC) in animals and birds from India. An outbreak of acute diarrhoea in poultry birds at Aizawl, Mizoram was investigated for detection and characterization of STEC and enteropathogenic E. coli (EPEC).
Methods
E. coli was isolated and identified from rectal swabs, intestinal contents, heart blood and spleen of 19 poultry birds that died due to acute diarrhoea during the outbreak. Phenotypic characterization was done by standard bacteriological and biochemical techniques. All the isolates were serotyped based on their somatic antigens. Virulence genes (stx1, stx2, eaeA and hlyA) were detected by multiplex PCR assay.
Results
A total of 42 E. coli isolates were obtained, of which 24 belonged to 3 serogroups (O64, O89 and O91) and the remaining 18 were untypable (UT). Altogether, 14 (33.33%) isolates carried at least 1 virulence gene, of which 10 (23.81%) and 4 (9.52%) were recorded as STEC and EPEC, respectively. Of the 10 STEC isolates, one carried only stx2, one carried stx2 and hlyA, four carried stx1, stx2 and hlyA, two carried stx1, eaeA and hlyA genes and two carried stx1 and eaeA. Of the four EPEC isolates, two carried eaeA and hlyA, one carried only eaeA gene and 1 carried only hlyA gene.
Interpretation & conclusions
This is the first report on the involvement of STEC in poultry in India.
Keywords: EPEC, India, poultry, STEC, virulence genes
The morbidity and mortality associated with several large outbreaks of gastrointestinal diseases caused by Shiga toxin-producing Escherichia coli (STEC) indicating the threat of these organisms to public health1. These are commonly recovered from food animals and were found responsible for severe gastrointestinal and systemic diseases such as haemorrhagic colitis (HC) and haemolytic uremic syndrome (HUS) leading to diarrhoea, especially among the infants in the developing countries1,2. STEC strains produce one or both of two major types of Shiga toxins, designated Stx1 and Stx2. The Stx2 is associated with an increased risk of developing HUS3. Although, in India, reports are available on isolation, identification and characterization of STEC in human and animals4–10, there appears to be no information on association of STEC in poultry.
An outbreak of acute diarrhoea in broiler chickens aged 6-8 wk was reported in Aizawl, Mizoram in 2007. We investigated this outbreak for detection and characterization of pathogenic organism in the broiler chickens with diarrhoea.
Material & Methods
Collection of samples: An outbreak of diarrhoeal disease in a flock of 150 broiler chickens housed under intensive care system at Aizawl, Mizoram in 2007 was attended. A total of 49 birds were affected, of which 19 died. These 19 dead birds were brought to the pathology/microbiology laboratory, College of Veterinary Sciences & Animal Husbandry, Aizawl, Mizoram. During post-mortem examination, rectal swabs, intestinal contents, heart blood and pieces of spleen were collected aseptically for isolation and identification of causative agents.
Screening of the specimens for Coccidia and Group A rotavirus: In addition to screening for Coccidia, the faecal samples were also screened for the presence of group A rotavirus by RNA-PAGE analysis11 with certain modifications. In brief, RNA was extracted from faecal samples followed by electrophoresis in 7.5 per cent polyacrylamide gel using Laemmli's discontinuous buffer system. Silver staining of the gel was done as described by Svensson et al12.
Bacteriological screening of clinical specimens: The clinical samples (heart blood, liver, spleen, rectal swab, intestinal contents) were immediately inoculated on 10 per cent sheep blood agar and MacConkey's agar (HiMedia, Mumbai, India) plates and incubated at 37°C for 18-24 h. Pure and a single population of bacterial colonies were recorded from heart blood and spleen samples. Five randomly selected colonies from MacConkey's agar and 10 per cent sheep blood agar plates were picked up and subcultured on eosin methylene blue (EMB) agar (HiMedia, Mumbai, India) plates to observe the characteristic metallic sheen of E. coli. The well separated pure colonies were picked up on nutrient agar slants as pure culture and subjected for standard morphological and biochemical tests13.
Serotyping of E. coli: The 42 E. coli isolates were serotyped based on their somatic (O) antigens at National Salmonella and Escherichia Centre, Central Research Institute, Kasauli, India.
Preparation of E. coli DNA for PCR assay: For rapid detection of virulence genes, isolated bacterial cultures were inoculated into 2 ml Luria Bartani (L-B) broth and incubated at 37°C under constant shaking for 18 h. After incubation, 1 ml broth culture was taken in a 1.5 ml microcentrifuge tube and centrifuged at 5867 g for 10 min. The pellet was washed twice in sterile normal saline solution (NSS) (0.85% NaCl) and resuspended in 400 μl of nuclease-free sterile distilled water and boiled for 10 min followed by immediate chilling. Cell debris was removed by centrifugation at 2292 g for 5 min. The supernatant was used as template DNA for PCR.
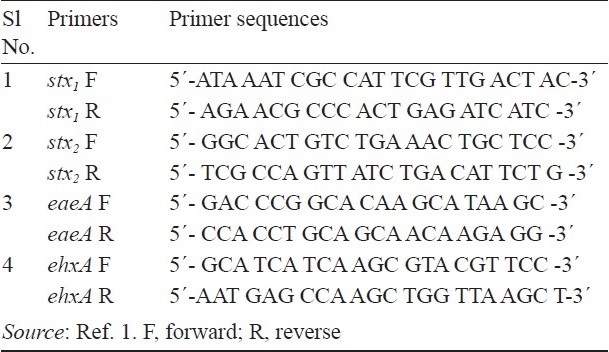
Detection of virulence genes by multiplex PCR: A multiplex PCR was carried out using 4 sets of oligonucleotide primers for stx1, stx2, eaeA and hlyA genes (Table I). The PCR protocol was followed as per the method described by Paton and Paton1 with some modifications. In brief, the multiplex PCR mixture of 25.0 μl contained 1X PCR buffer, 1.5 mM of MgCl2, each primer within the 4 primer sets at a concentration of 40 nM, 200 μM each of dNTPs, 1.0 U of Taq DNA polymerase and 2.0 μl of template DNA. The PCR reaction was performed in a thermal cycler (Thermo Electron, Germany) using the following standard cycling procedure: an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 94°C for 45 sec, primer annealing at 59°C for 45 sec and extension at 72°C for 1 min and a final extension at 72°C for 6 min.
Table I.
Oligonucleotide primers used in multiplex PCR reaction
Amplified products were separated by agarose gel (2% agarose in 1X Tris-borate-EDTA buffer) electrophoresis at 5v/cm for 2 h and stained with ethidium bromide (0.5 μg/ml)14. Standard molecular size marker (100 bp DNA ladder) was included in each gel. DNA fragments were observed by ultraviolet transilluminator and photographed in a gel documentation system (Alpha Imager, Germany).
The PCR was performed three times to ensure the repeatability of the technique and to make sure that isolates were correctly assigned to respective patterns.
Results
Epidemiological details and post mortem observations: Out of 150 birds in the flock, 49 were affected and 19 died within one week time with an overall mortality and case fatality rate of 12.67 per cent (19/150) and 38.78 per cent (19/49), respectively. Prior to death, the affected birds were anorexic and emaciated, dull and depressed with ruffled feathers and showed progressive somnolence with closed eyes. Majority of the birds were shivering and huddled near the source of heat. Clinically ill birds showed profuse watery diarrhoea and severe dehydration. On post- mortem, besides the generalized septicaemic lesions, severe lesions of enteritis accompanied with focal necrotic lesions in the mucosa of the small intestine were prominent in majority of the cases. Spleens and livers were swollen and congested with haemorrhagic or necrotic foci. Major parasites including round worm, tape worm and Coccidia sp. were not recorded during post-mortem examination/screening of faecal samples.
RNA-PAGE: All the faecal and rectal swab samples screened by RNA-PAGE for detection of group A rotavirus were also found negative.
Bacterial isolation and characterization: The bacteriological examination of heart blood, liver, spleen and intestinal contents revealed the presence of Gram-negative bacilli. In biochemical tests, the isolates were identified as E. coli. The haemolytic pattern in 10 per cent sheep blood agar showed narrow zone of haemolysis, typical for the STEC. A total of 42 isolates of E. coli were recorded from 19 clinical samples, which belonged to 4 serogroups, viz., O64 (16), O89 (3), O91 (5) and UT (18).
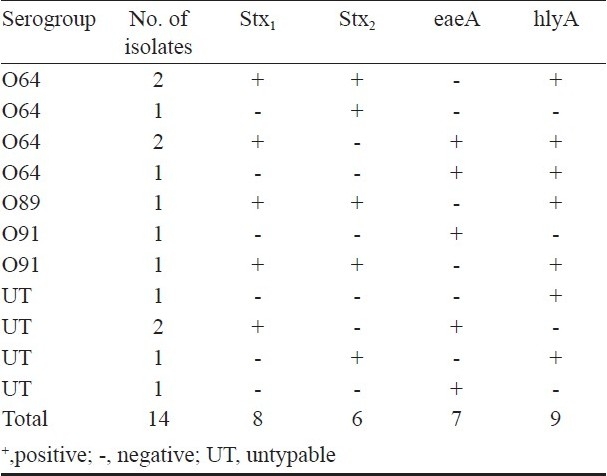
Multiplex PCR for virulence genes: Multiplex PCR assay yielded amplified products of ~180 bp, ~255bp, ~384bp and ~534 bp specific for stx1, stx2, eaeA and hlyA genes, respectively. Out of 42 isolates, 14 (33.33%) carried at least one virulence gene, of which 10 (23.81%) and 4 (9.52%) were detected as STEC and EPEC, respectively. Of the 10 STEC isolates, one carried only stx2, one carried stx2 and hlyA, four carried stx1, stx2 and hlyA, two carried stx1, eaeA and hlyA genes and two carried stx1 and eaeA . Similarly, out of 4 EPEC isolates, one carried eaeA and hlyA, two carried only eaeA gene and one carried only hlyA gene (Table II).
Table II.
Virulence genes profile of E. coli strains isolated from poultry birds with diarrhoea
Discussion
During the investigation of the present outbreak of acute diarrhoea in a poultry flock, the clinical symptoms and post-mortem study indicated the involvement of systemic infection by some enteric pathogens. Because of absence of group A rotavirus, Salmonella spp. or any other diarrhoea causing parasites and isolation of pure haemorrhagic E. coli from heart blood as well as intestinal contents warranted for further investigation of virulence genes of E. coli isolates.
In India, there is limited information regarding the STEC in animals including cattle5, sheep4,9, fish7, beef8 and human faeces6,8 available. Wani et al4 and Farooq et al10 reported the association of EPEC in chickens in Kashmir, India.
In the present study, a total of four different serogroups of E. coli were isolated, of which O64 was the predominant. Contrary to our result, other workers4,15from Jammu and Kashmir reported the predominance of O9, O8, O60 and O25. Mishra et al16 found O2, O19, O20 and O78 as predominant serogroups amongst the E. coli isolates from 250 clinical specimens from poultry in India. These findings indicate the variable distribution of different serogroups of E. coli in different geographical regions in Inida.
The present study showed a higher percentage of E. coli isolates carrying at least one virulence gene. Farooq et al10 reported 19.81 per cent E. coli isolates from different avian species containing at least one virulence gene. In another study, Wani et al4 reported that only 12 out of 426 (2.82%) E. coli isolates from poultry and pigeon carried at least one virulence gene. In both the cases, the apparently healthy birds or birds with no history of diarrhoea were tested, which may be the reason for recording of low percentage of E. coli with virulence genes. However, in the present study, all the E. coli isolates were recovered from clinically infected birds. It also justifies the association of STEC and/or EPEC isolates as the main cause of present outbreak.
Detection of STEC in poultry in the present study is probably the first report in India. Absence of STEC is also reported by Kobayashi et al17 in faecal samples from 199 broiler chickens, 32 pigeons and 86 gulls in Finland. Schroeder et al18 also failed to isolate any STEC in retail chicken and turkey obtained from Washington, DC, USA. In all these cases, E. coli was recovered from apparently healthy birds, either from live birds or after slaughter. Parreira and Gyles19 reported the association of STEC with 53 per cent of avian pathogenic E. coli (APEC) isolates: 35 per cent from lesions of avian cellulitis, 32 per cent from avian septicemia, 13 per cent from swollen head syndrome (SHS) in chickens and 20 per cent from diseased turkeys. None of the 5 isolates from healthy chickens were positive for stx genes19. These reports indicate that STEC may be closely associated with diarrhoeal disease in poultry in the present study.
Of the 14 E. coli isolates positive for at least one virulence gene, 7 (50) possessed eaeA, which is in agreement with the findings of other workers17, who reported a higher percentage (57 and 40% of chicken and gulls, respectively) of E. coli isolates carrying eaeA gene. However, in contrast to our study, Wani et al4 reported only 2.49 per cent of E. coli isolates from chicken carrying eaeA gene.
Prevalence of EPEC was 9.52 per cent among E. coli isolates from chickens in the present study, while other studies reported 2.74 per cent4 and 15.56 per cent10. Of the four EPEC isolates in this study, one carried both eaeA and hlyA genes under the serogroup O64. Wani et al4 reported 1.49 per cent isolates of E. coli from chicken carrying both eaeA and hlyA genes. Kobayashi et al17 reported that eae E. coli strains were highly prevalent among gulls (40.0%), pigeons (7.0%) and broiler chickens (57.0%), which lacked hlyA gene. All these reports indicate the variable distribution of eaeA and hlyA gene in E. coli isolates from birds.
Our result is in agreement with that of by Parreira and Gyles19 reporting 53 per cent APEC with stx gene sequences: one isolate carried stx2 sequence, two carried both stx1 and stx2 sequences, and the remaining 49 isolates carried only stx1 sequences. In conclusion, our findings provide the information about the involvement of STEC in diarrhoea in poultry in India.
Acknowledgments
The authors thank the Director, National Salmonella & Escherichia Center, CRI, Kasauli, HP, India, for serotyping the isolates; the Dean, CVSc&AH, CAU, Selesih, Aizawl, Mizoram for providing necessary funds and facilities to carry out the investigation.
References
- 1.Paton AW, Paton JC. Detection and characterization of shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohaemorrhagic E. coli hlyA, rfbO111 and rfbO157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narato JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnson RP, Gyles CL. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol. 1999;37:497–503. doi: 10.1128/jcm.37.3.497-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wani SA, Samanta I, Bhat MA, Nishikawa Y. Investigation of Shiga toxin- producing Escherichia coli in avian species in India. Lett Appl Microbiol. 2004;39:389–94. doi: 10.1111/j.1472-765X.2004.01586.x. [DOI] [PubMed] [Google Scholar]
- 5.Pal A, Ghosh S, Ramamurthy T, Yamasaki S, Tsukamoto T, Bhattacharya SK, et al. Shiga-toxin producing Escherichia coli from healthy cattle in a semi urban community in Calcutta, India. Indian J Med Res. 1999;110:83–5. [PubMed] [Google Scholar]
- 6.Chattopadhayay UK, Dutta S, Deb A, Pal D. Verotoxin producing Escherichia coli – an environment induced emerging zoonosis in and around Calcutta. Int J Environ Health Res. 2001;1:107–12. doi: 10.1080/09603120020019692. [DOI] [PubMed] [Google Scholar]
- 7.Sanath Kumar H, Otta SK, Karunasagar I, Karunasagar I. Detection of Shiga-toxigenic Escherichia coli (STEC) in fresh seafood and meat marketed in Mangalore, India by PCR. Lett Appl Microbiol. 2001;33:334–8. doi: 10.1046/j.1472-765x.2001.01007.x. [DOI] [PubMed] [Google Scholar]
- 8.Khan A, Yamasaki S, Sato T, Ramamurthy T, Pal A, Datta S, et al. Prevalence and genetic profiling of virulence determinants of non-O157 Shiga-toxin producing Escherichia coli isolated from cattle, beef and humans, Calcutta, India. Emerg Infec Dis. 2002;8:54–62. [PubMed] [Google Scholar]
- 9.Bhat MA, Nishikawa Y, Wani SA. Prevalence and virulence gene profiles of Shiga toxin - producing Escherichia coli and enteropathogenic Escherichia coli from diarrhoeic and healthy lambs in India. Small Ruminant Res. 2008;75:65–70. [Google Scholar]
- 10.Farooq S, Hussain I, Mir MA, Bhat MA, Wani SA. Isolation of atypical Enteropathogenic Escherichia coli and shiga toxin1 and 2f-producing Escherichia coli from avian species in India. Lett Appl Microbiol. 2009;48:692–7. doi: 10.1111/j.1472-765X.2009.02594.x. [DOI] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single step RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Svensson L, Uhnoo I, Grandein M, Wadell G. Molecular epidemiology of roptavirus infections on Uppsala, Sweden (1981): disappearance of a predominant electropherotype. J Med Virol. 1986;18:101–11. doi: 10.1002/jmv.1890180202. [DOI] [PubMed] [Google Scholar]
- 13.Edwards PR, Ewing WH. Identification of enterobacteriaceae. 3rd ed. Minneapolis, MN: Burgess Publishing Company; 1972. [Google Scholar]
- 14.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning-A laboratory manual. 2nd ed. New York: Cold Spring Harbour laboratory Press, Cold Spring Harbour; 1989. [Google Scholar]
- 15.Malik AR, Rashid A, Qureshi AM, Wani SA. Prevalence of enterobacteria in migratory and non-migratory water birds in Srinagar. J Res- SKUAST-J. 2002;1:75–8. [Google Scholar]
- 16.Mishra A, Sharda R, Chhabra D, Moghe MN. Escherichia coli isolates from domestic poultry. Indian J Anim Sci. 2002;72:727–9. [Google Scholar]
- 17.Kobayashi H, Pohjanvirta T, Pelkonen S. Prevalence and characteristics of intimin- and shiga toxin-producing Escherichia coli from gulls, pigeons and broilers in Finland. J Vet Med Sci. 2002;64:1071–3. doi: 10.1292/jvms.64.1071. [DOI] [PubMed] [Google Scholar]
- 18.Schroeder CM, White DG, Ge B, Zhang Y, Mcdermort PF, Ayers S, et al. Isolation of antimicrobial-resistant Escherichia coli from retail meats purchased in greater Washington, DC, USA. Int J Food Microbiol. 2003;85:197–202. doi: 10.1016/s0168-1605(02)00508-1. [DOI] [PubMed] [Google Scholar]
- 19.Parreira VR, Gyles CL. Shiga toxin genes in avian Escherichia coli. Vet Microbiol. 2002;87:341–2. doi: 10.1016/s0378-1135(02)00084-6. [DOI] [PubMed] [Google Scholar]


