Summary
Background
During endocytosis in yeast, a choreographed series of discrete local events at the plasma membrane lead to a rapid burst of actin polymerization and then internalization of an endocytic vesicle. What initiates Arp2/3-dependent actin polymerization in this process is not well understood.
Results
The Schizosaccharomyces pombe WISH/DIP/SPIN90 orthologue dip1p is an actin patch protein that regulates the temporal sequence of endocytic events. dip1Δ mutants exhibit a novel phenotype in which early events such as WASp localization occur normally, but arrival of Arp2/3, actin polymerization and subsequent steps are delayed and occur with apparently random timing. In studying this mutant, we demonstrate that positive feedback loops of WASp, rapid actin assembly, and Arp2/3 contribute to switch-like behavior that initiates actin polymerization. In the absence of dip1p, a subset of patches is activated concurrently with the “touch” of a neighboring endocytic vesicle.
Conclusions
These studies reveal a switch-like mechanism responsible for the initiation of actin assembly during endocytosis. This switch may be activated in at least two ways, through a dip1p-dependent mechanism and through contact with another endocytic vesicle.
Introduction
Temporal and spatial regulation of actin dynamics drives processes such as cell migration, cell morphogenesis, membrane trafficking and cell division. Endocytosis in yeast has been established as a powerful system to study the activation and function of Arp2/3 dependent actin assembly in vivo. During this process, over 50 proteins gather in a sequential manner at discrete sites on the plasma membrane to coordinate membrane invagination, and scission to form an endocytic vesicle. Time-lapse imaging of these “actin patches” has defined a stereotyped series of events that occur in about 30 sec: arrival of clathrin and adapter proteins at the plasma membrane, followed by recruitment of the actin polymerization machinery including WASp, myosin type I and Arp2/3 complex, as well as a large set of other F-actin binding proteins, phospholipids, and F-bar proteins [1–3]. An attractive property of this system is that it is possible to analyze in a quantitative manner many discrete local events occurring at different sites on the plasma membrane, all in the presence of a common cytoplasm [4–6].
Actin filaments, which only arrive part way through this process, are an essential component of endocytosis in yeast. Actin polymerization has been proposed to provide force to overcome the high internal turgor pressure for membrane invagination [7], and contribute to the scission process by generating tension along the endocytic pit with amphiphysins or with myosin I [8, 9]. Actin is also needed for the recruitment of many endocytic proteins to the site [10, 11].
The explosive nucleation and formation of branched actin filaments at the patch are mediated by the Arp2/3 complex [12, 13]. How the initiation of actin assembly is triggered is still not clear. Although several conserved Arp2/3 activators have been identified, a key factor that has been proposed is the Arp2/3 activator, wsp1p (the WASp orthologue) [12, 14]. However, it is not known whether wsp1p activity itself is regulated, as unlike mammalian WASp/ WAVE proteins, yeast WASp is not regulated by autoinhibition and does not contain a CRIB domain for cdc42 GTPase interaction [15].
The WISH/DIP family of proteins (which also includes SPIN90 in humans and Ldb17 in budding yeast [16, 17]) has poorly understood functions in regulating actin dynamics. In mammalian cells, this protein is implicated both as a positive regulator of Arp2/3 and a negative regulator of the formin mDia2, and has been shown to interact directly with the Arp2/3 complex, WASp, G-actin and F-actin, and the FH2 domain of mDia [16, 18–22]. In vivo, reducing WISH/DIP expression leads to defective cell migration, formation of prominent filopodia, and reduced levels of receptor-mediated endocytosis [19, 23]. Its potentially complex functions, especially in vivo, remain to be defined rigorously.
Here, we study the mechanism of initiation of actin assembly at the endocytic patch. We find that the S. pombe WISH/DIP orthologue dip1p has a novel role in the temporal regulation of Arp2/3-based actin assembly at the patch; it does not appear to be a formin regulator. dip1Δ mutants exhibit an unusual phenotype in that actin assembly in patches is delayed for highly variable amounts of time. Through the study of these mutant cells, we find that the mechanism for initiation of actin polymerization has switch-like properties. In this switch, positive feedback loops between WASp, Arp2/3 and rapid actin polymerization may work together to produce a local burst of actin assembly for endocytosis.
Results
Dip1p regulates actin patch formation and endocytosis
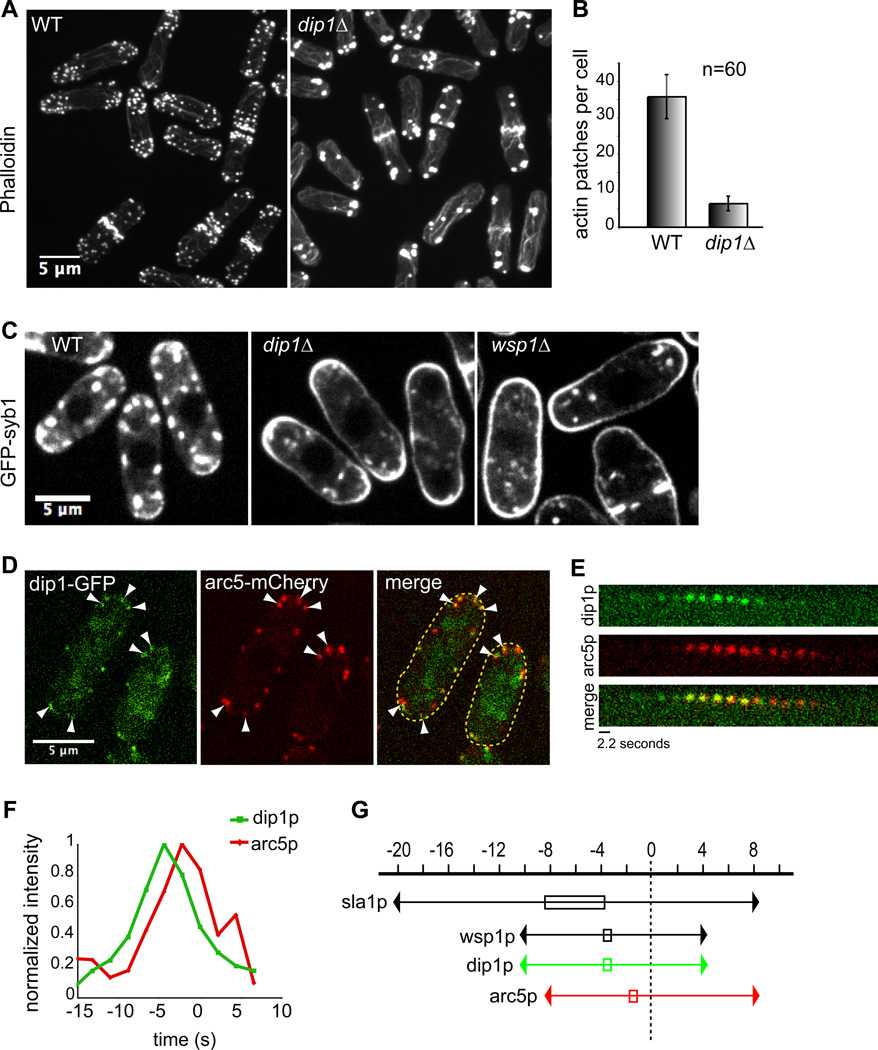
The sequence of S. pombe dip1p (SPBC24c6.10c) predicts that it is a 43.6 kDa protein that contains the conserved leucine-rich region (Pfam domain of unknown function 2013), but is missing the SH3 and proline-rich domains found in other family members [21]. S. pombe dip1Δ cells were viable and grew at wildtype rates at a range of conditions (Figure S1). They were morphologically indistinguishable from wildtype cells and exhibited normal cell polarization programs such as “new end take off”. AlexaFluor 488-phalloidin staining, however, revealed profound defects in F-actin organization. dip1Δ cells had fewer patches that contained F-actin: wildtype cells contained an average of 36 ± 6 actin patches per cell, whereas dip1Δ mutants had 6.5 ± 2.0 per cell (n = 60 cells for each strain)(Figure 1A,B). These patches localized normally to cell tips and septa, but appeared larger and stained more intensely with Phalloidin than in wildtype. Actin cables and rings were more numerous and robust in dip1Δ (Figure S3A), and actin bundles in cables and rings were often abnormally long, looping around cell tips and emanating out from the contractile ring respectively.
Figure 1. dip1 mutants have defects in actin organization and endocytosis.
(A) AlexaFluor 488-phalloidin stained wildtype (left) and dip1Δ (right) cells. Maximum intensity projections of confocal images are shown. (B) Quantification of the average number of actin patches per cell from images as in A. (n = 60 cells for each strain). Patches within clusters were counted as individuals only if they could be visually distinguished. (C) Images of cells of indicated genotype expressing the integral membrane protein GFP-syb1. GFP-syb1 accumulates at the plasma membrane in endocytic mutants. (D) Dip1p co-localizes with the Arp2/3 marker arc5p in actin patches. Wildtype cells expressing arc5-mCherry and dip1-GFP from their endogenous promoters were imaged in a medial focal plane. White arrowheads indicate sites where dip1p and arc5p co-localize. (E) Time-lapse images of dip1-GFP and arc5-mCherry in a single patch. Arrow indicates patch internalization. GFP and mCherry images were acquired sequentially with exposure times of 2s and 0.2s respectively. (F) Normalized fluorescence intensities of dip1-GFP (green) and arc5-mCherry (red) within a single patch in WT (left). Time = 0s indicates the time of patch internalization. (G) Schematic of dip1p dynamics relative to actin patch proteins sla1p, wsp1p and arc5p. Time = 0s indicates the time of patch internalization. Boxed area indicates time at which protein concentration peaks at the patch. Scale bars = 5µm.
To test whether dip1p functions in endocytosis, we used an endocytosis assay based upon the localization pattern of GFP-syb1 (synaptobrevin), an integral membrane protein that normally localizes on the plasma membrane and cytoplasmic vesicles, which are likely to be endocytic (Figure 1C). In endocytic mutants such as wsp1Δ (WASp), myo1Δ (Myosin I) and end4Δ (Sla2 orthologue) (Figure 1C and unpublished data), GFP-syb1 accumulated on the plasma membrane, reflecting a decrease in the rate of internalization. In dip1Δ, GFP-syb1 also accumulated all around the plasma membrane (Figure 1C), indicating a defect in endocytosis. As an additional assay for endocytosis, we also found that dip1 mutants exhibited a delay in the internalization of the dye FM4-64 (Figure S2A).
Dip1p localizes to sites of endocytosis
We determined the localization of dip1p by imaging a functional dip1-GFP expressed at endogenous levels from the dip1 locus on the chromosome. Dip1-GFP appeared as dim cortical dots that co-localized with the actin patch marker arc5-mCherry (Arp2/3 subunit), showing that dip1p is a component of the actin patch (Figure 1D). Dip1p arrived at the patch at −8 to −10 seconds (relative to time of patch internalization = 0), peaked in intensity at − 2 to − 4 seconds, internalized with other patch components and disassociated from the patch 2– 4 seconds after internalization. The peak of dip1p accumulation precedes the peak in Arc5p by 2–4 seconds (Figure 1E,F) and is very similar to that of wsp1p (WASp) [4] (Figure 1G, Figure 3B). Quantitation of peak dip1-GFP fluorescence in patches showed that it was present in about 20 molecules/patch similar to clc1p (clathrin light chain; estimated at 30–40 molecules/ patch) and is much less bright than wsp1p or arc5p (about 150 molecules/ patch) [6]; although the absolute numbers in this analysis can be subject to caveats, these ratios indicate that dip1p is sub-stoichiometric to wsp1p and Arp2/3 complex.
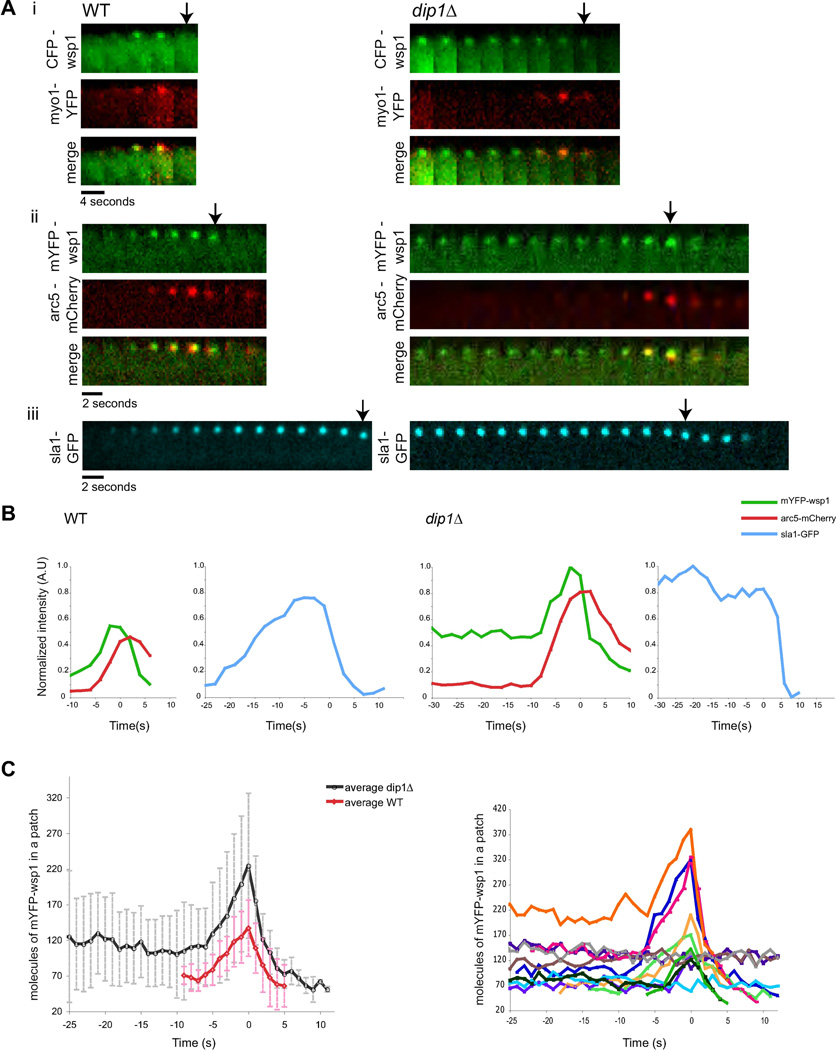
Figure 3. Dynamics of patch components in wildtype and dip1Δ mutants.
(A) Time-lapse images of individual patches in WT and dip1Δ cells labeled with the indicated markers. Widefield (i) and confocal images (ii, iii) were acquired through the medial focal section of the cell at 2s intervals. Arrows denote time of patch internalization. (B) Normalized fluorescence intensities of patch proteins at a single patch over time in WT (left) and dip1Δ (right). Time = 0s indicates the time of patch internalization. Arc5-mCherry and mYFP-wsp1 intensities are normalized to mYFP-wsp1 in dip1Δ and sla1-GFP intensities are normalized to sla1-GFP in dip1Δ. (C) Left: Average number of mYFP-wsp1 molecules per patch over time in WT (n=11) and dip1Δ (n=12). Numbers were estimated based on fluorescence intensity ratios (see Supplementary Methods). Right: Numbers of mYFP-wsp1 molecules in individual patches in dip1Δ cells are each plotted in different colors.
To test if dip1p localization is actin-dependent, we treated cells with 200 µM Latrunculin A (an actin monomer sequestering drug). Dip1p accumulated in dots at the cortex, indicating that the cortical localization of dip1p is primarily F-actin independent (Figure S3).
Dip1p may not affect formin activity in vivo
As the mammalian orthologue WISH/DIP has been found to act as an inhibitor of the mDia formin in vitro [21], we tested if dip1p regulates for3p, a formin that assembles actin cables at cell tips [24]. Dip1p localization and dynamics were independent of for3p and actin cables, and did not exhibit appreciable co-localization with for3p (Figure S3). For3p localization was not affected in dip1 mutants. Further, the rate of for3p dot movement, which corresponds to the rate of formin-based actin polymerization in the cable [25], was normal. Thus, although there is an increase in actin cable structures and an apparent increase in actin filaments in the cytokinetic ring in dip1Δ mutants, similar to an increase in filopodia seen in WISH/DIP knockdown cells, this effect could be a secondary effect of the decrease in the amount of actin polymerized by Arp2/3. Similar increases in cables are also seen in wsp1 and arp2/3 fission yeast mutants, for instance (our unpublished observations) and in cells overexpressing the patch protein gmf1p (an ADF-related factor) [26]. Our results suggest that dip1p has a primary function in regulating Arp2/3-dependent actin polymerization and not formins.
Dip1 regulates the timing of an actin assembly switch
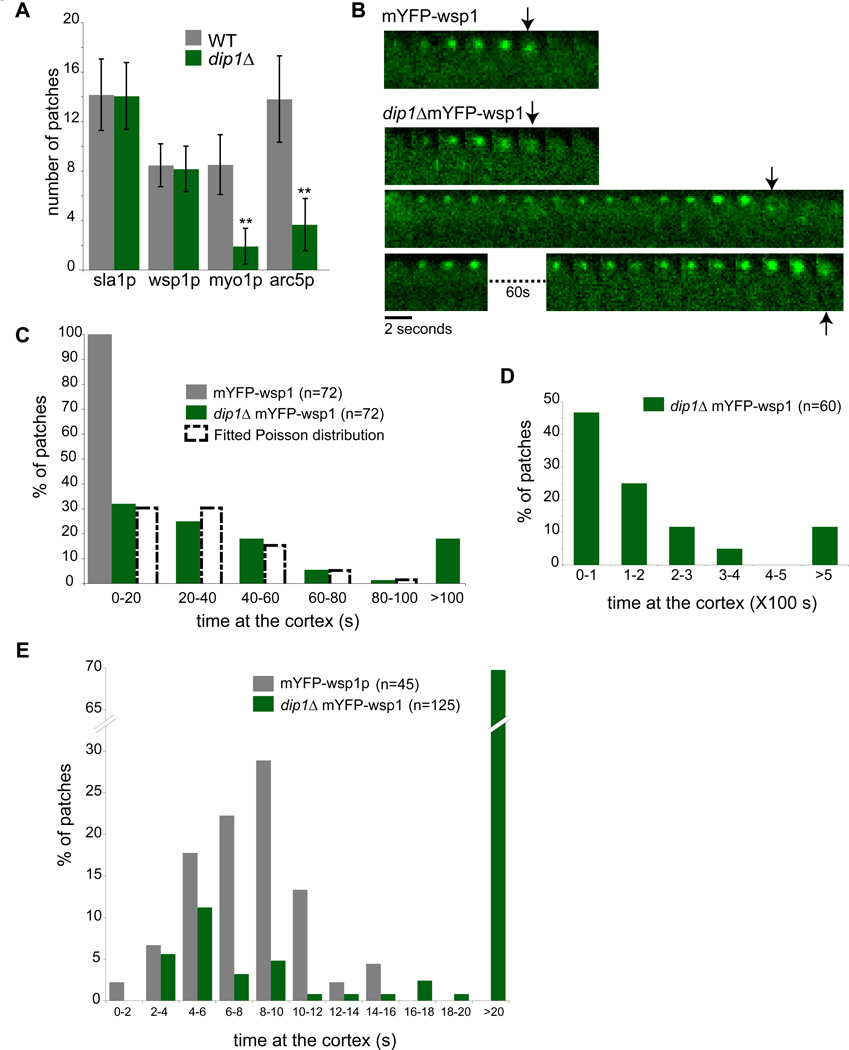
In order to determine which step in the actin patch assembly pathway is compromised in dip1Δ cells, we examined the localization of several patch components. Patch assembly factors that appear early: the endocytic adapter protein sla1p, and actin nucleation promoting factor, wsp1p (WASp), localized to a similar number of cortical sites in wildtype and dip1Δ cells (Figure 2A,S4). In contrast, the number of patches containing cortical myo1p (myosin I) and arc5p (arp2/3 subunit) were reduced by over 70% (n= 20 cells). Thus, together with the phalloidin staining data, these results suggest that dip1p is needed for the efficient recruitment of F-actin, Arp2/3, and other patch components downstream of wsp1p.
Figure 2. Dip1p regulates the timing of patch activation.
(A) Quantification of number of patches of the indicated markers in WT and dip1Δ cells. n=20 cells each. Error bars represent standard deviations. ** Significant difference (p< 0.0001) between dip1Δ and WT strains. (B) Time-lapse images of individual patches marked by mYFP-wsp1 in WT and dip1Δ cells. The montages track the maturation of a single representative patch over time. Each interval is 2 sec. Arrows mark the time of internalization of the endocytic vesicle. In the dip1Δ cells, the lifetimes of the mYFP-wsp1 containing patch is highly variable, as depicted by the three patches shown. (C) Quantitation of the lifetimes of mYFP-wsp1 patches. Images were acquired every 2 sec for 100 sec. Histograms of patches in WT (grey bars) and dip1Δ (green bars) are shown. n = 72 patches in 5 cells in WT; N= 72 patches in 15 cells in dip1Δ. The distribution of mYFP-wsp1 in dip1Δ was fitted to a Poisson distribution (dashed histogram). Mean and variance for Poisson distribution (λ) = 1, covariance (r2) = 0.948. (D) Images were acquired every 10 sec for 500 sec to track patch behavior over a longer time scale. n = 60 patches in 6 cells. (E) Images were acquired every 1 sec for 100 sec to track patch behavior over a short time scale. n = 45 patches in 5 cells in WT, and 125 patches in 25 cells in dip1Δ. Scale bar = 5µm.
To assay the dynamic behavior of these patches, we imaged mYFP-wsp1 at individual patches using time-lapse microscopy. In wildtype cells, wsp1p appeared at the plasma membrane and resided for an average of 8–10 seconds before moving into the cell interior and disappearing; all wsp1p-labeled patches were internalized in < 20 sec (n = 72 patches)(Figure 2B). In contrast, in dip1Δ, wsp1p remained on the cortex for highly variable periods of time (Figure 2B). We measured the lifetimes of these patches using different acquisition periods to characterize a range of time periods (Figure 2C,D,E), as photo-bleaching limited the number of images that we could acquire. In some patches, wsp1p displayed rapid dynamics and internalized with similar kinetics as the wildtype patches (in < 20 sec). Other patches took on the order of hundreds of seconds (over 10 fold longer than wildtype). By 400 seconds, however, 90% of the patches internalized (n = 72 patches). The distribution of patch lifetimes at the cortex could be fitted to a Poisson distribution, suggesting that a stochastic process governs the lifetime of the wsp1p-containing patch in dip1Δ cells.
Time-lapse microscopy showed that the recruitment of factors downstream of wsp1p was delayed. In wildtype cells, sla1p arrived at −20 sec, wsp1p peaked in intensity at −8–10 sec, and then arc5p peaked at about −2 sec (n= 20 patches)(Figure 1G and Figure 3A,B). In dip1Δ cells, wsp1p and sla1p resided at the patch for variable periods of time at a steady concentration without any detectable myo1p or arc5p (Figure 3A,B). Then, arc5p and myo1 arrived at the patch and 4–6 seconds later, the patch internalized (n= 20 patches). The concentration of wsp1p also often increased, 5 sec before internalization. In contrast, the adapter protein sla1p did not exhibit a peak before internalization, showing that only a subset of patch components increased at this stage. Subsequent steps of endocytosis were not delayed and proceeded with slightly faster dynamics than in wildtype cells (Figure S5), possibly due to the increased cytoplasmic concentration of certain patch components. Thus, dip1p is required for timing the initiation of events in actin polymerization, but is not needed for a subsequent robust response once the process has been initiated.
To further quantitate patch behavior, we counted the number of wsp1p proteins at each patch using fluorescence intensity (Figure 3C). In wildtype cells, wsp1p peaked to an average of 137 ± 39 molecules (n = 11 patches) just prior to internalization. The average number of wsp1p proteins in the stable inactive patches in dip1Δ cells was 120 ± 53 molecules and then peaked to 224 ± 102 proteins before internalization (n = 12 patches), though with substantial variability (Figure 3C, right). As the number of wsp1p proteins in the inactive patches in the dip1Δ cell was often equal or higher than the peak of wsp1p in wildtype cells, this analysis suggests that the number of wsp1p molecules in the patch is not the rate-limiting step for activation.
These findings suggest that dip1p regulates the timing of a switch, in which a patch containing low levels of wsp1p suddenly recruits more wsp1p, Arp2/3 complex, myo1p and F-actin, which then ultimately lead to internalization. In wildtype cells this transition occurs immediately after wsp1p arrival. In dip1 cells, this transition is delayed, and occurs at stochastic time intervals, but then subsequent events proceed with near normal kinetics. These data are not consistent with a model in which the slow indolent rise of an initiator reaches a threshold. Rather, the stochastic timing and the sharp transitions are indicative of a switch-like behavior that is triggered by a single (or small number) of events.
Turning on the switch is dependent on rapid actin polymerization
A switch behavior suggests the process is regulated by positive feedback loop(s). Properties of WASp, Arp2/3 and actin are likely to contribute to such a feedback loop. For instance, WASp and actin filaments can help activate Arp2/3, which nucleates actin filaments off the side of an actin filament, which then recruit more Arp2/3 and WASp [13, 27]. Thus one key element in this feedback may be actin polymerization.
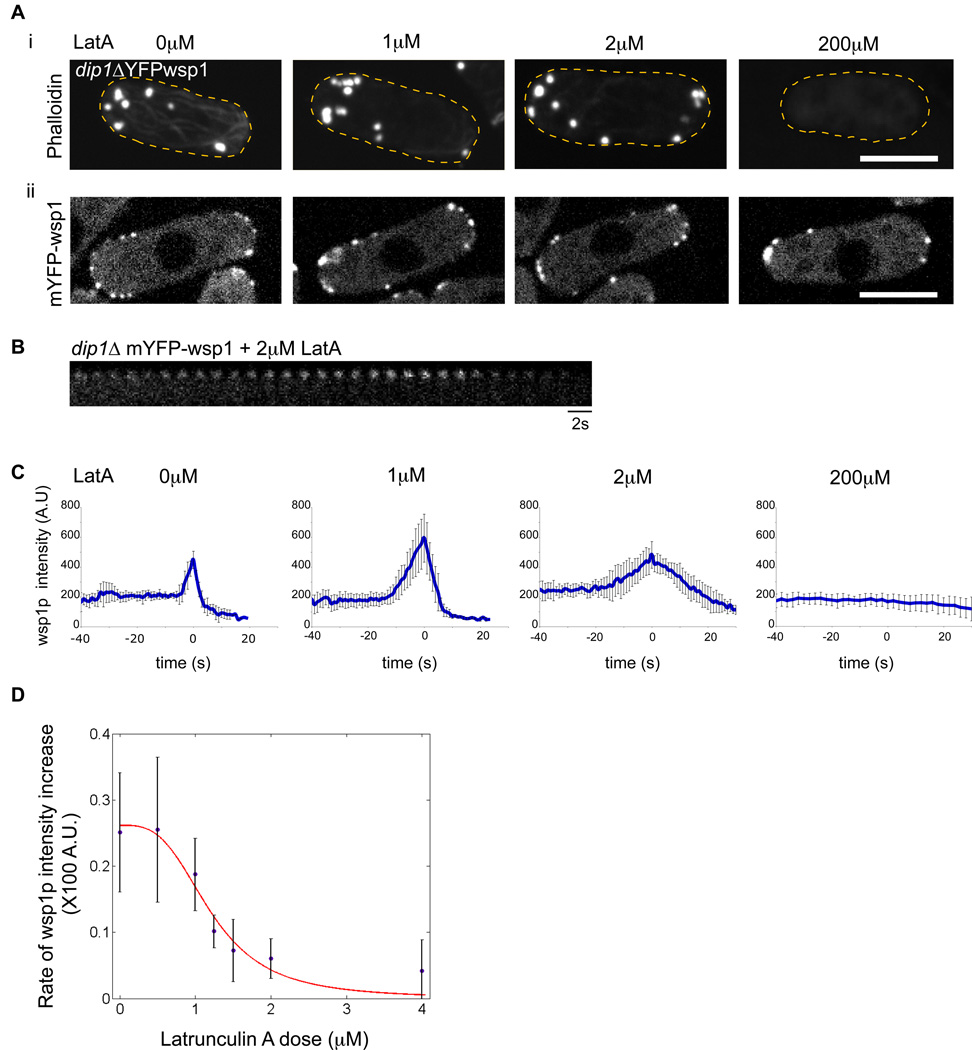
To probe the switch behavior in vivo, we measured the effects of attenuating actin polymerization rates using different concentrations of Latrunculin A, an actin-monomer sequestering drug [28]. As an assay for this switch, we measured mYFP-wsp1 intensities in individual patches over time. A high saturating dose (200 µM) of LatA causes complete depolymerization of all F-actin structures [29]. Wsp1p still localized to patches, but unlike in untreated cells (Figure 3), its intensity profiles over time did not show any peak, and the patches did not internalize (Figure 4A,C). Interestingly, at 2µM LatA, patches exhibited similar F-actin staining as that in wildtype cells. The rise in mYFP-wsp1 intensity leading up to its peak, however, was slower than in untreated cells; patches did not internalize and gradually disassembled over 30 sec (Figure 4B). We propose that this low dose of LatA decreases the rate of actin polymerization in the cell without causing total actin depolymerization. Intermediate doses of LatA (0.5–2.0µM) showed intermediate effects. To estimate the rate of “switch activation,” we measured how rapidly mYFP-wsp1 intensity increased to its peak (the slope of the upward part of the curves in the graphs in Figure 4C). A plot of these rates as a function of LatA concentration showed a profile that could fit to a sigmoidal curve with a Hill coefficient of 3.1 (Figure 4D). These data suggest that a rapid rate of actin polymerization, rather than merely the presence of actin filaments, is a critical component in this switch; at attenuated rates of actin polymerization, the switch occurs only slowly, or not at all. Further, the sigmoidal shape of the graph (Figure 4D) provides a possible indication of an all or none switch, suggestive of underlying cooperative mechanisms involving positive feedback with actin polymerization.
Figure 4. A switch for patch activation requires rapid actin polymerization.
(A) i) dip1Δ cells were treated with indicated concentrations of Latrunculin A (LatA) for 2 min, and then fixed and stained for F-actin with AlexaFluor 488-phalloidin. Maximum intensity projection images are shown. Note that at 1µM and 2µM doses of LatA, actin cables are disrupted, but actin patches remain. (ii) Images of mYFP-wsp1 in live dip1Δ cells treated with indicated concentrations of LatA for 2 min. Single focal plane confocal images are shown. (B) Time-lapse images of a mYFP-wsp1 patch in dip1Δ cell treated with 2µM LatA. Note that the patch does not internalize and gradually disappears. (C) Effect of LatA on the dynamic behavior of mYFP-wsp1 patches over time in dip1Δ cells. Cells were treated with indicated concentrations of LatA for 2 min and then imaged. Graphs show mean fluorescence intensities plotted over time, where t=0 at the peak of intensity. n=8 patches. (D) Effect of LatA on the rate of increase of mYFP-wsp1. Cells were treated with doses of LatA for 2 minutes and then imaged. Rates of increase of mYFP-wsp1 intensity leading up to peak at t=0 (e.g. the slopes of the upward part of the curves in C) were measured for each patch (n=8 patches at each dose). The average rates were used to fit a Hill curve using the formula f(x)= c*((Cλ/(Cλ + xλ))) with the following parameters C (critical LatA concentration) = 1.201, c (fitting parameter) = 0.2616 and λ (Hill co-efficient) = 3.18. Error bars = SD. Scale bars = 5µm.
Effect of WASp on patch dynamics
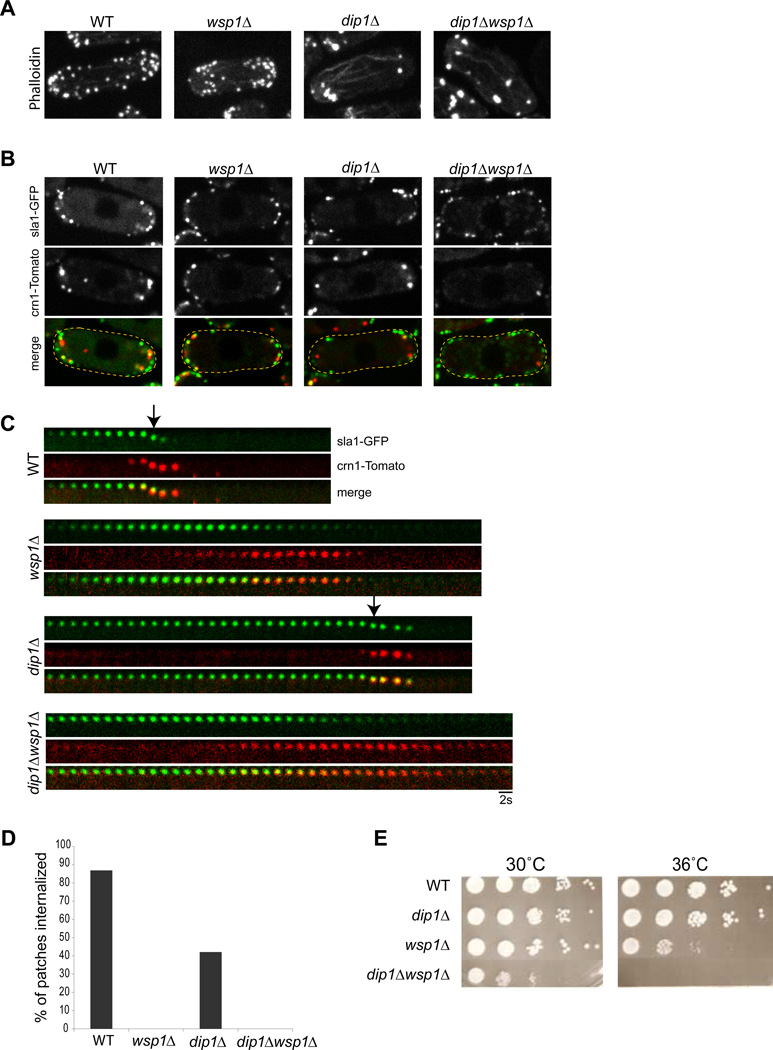
We examined the effect of wsp1p on the switch-like behavior in patch dynamics. wsp1Δ mutants are viable but have defects in endocytosis [30] (Figure 1). In these cells, actin filaments still assemble at patches: there were a normal number of patches that contain sla1p, actin and coronin (crn1-Tomato, an actin-dependent patch protein)(Figure 5A, B). However, these patches never internalized and instead gradually disassembled (Figure 5 C,D), similar to patches treated with 2µM LatA (Figure 4B). Thus, wsp1 mutants may have an inadequate actin polymerization rate to activate the switch. These data suggest that wsp1p is normally a critical component of the switch.
Figure 5. The effect of wsp1p on patch dynamics and its genetic interaction with dip1p.
(A) Alexa Fluor 488 phalloidin staining of cells of indicated genotype. Maximum intensity projection confocal images are shown. (B) Single focal planes images of these mutants expressing patch markers crn1p and sla1p. (C) Time lapse images of single patches. Arrows mark patch internalization, which fails in wsp1Δ and dip1Δwsp1Δ mutants. (D) Percentage of sla1-GFP marked patches that internalized over 50s in the indicated genotypes. n=76 patches in 10 cells for each strain. (E) Spot assay for growth. Cells of indicated geneotypes were spotted at different dilutions onto agar plates and incubated for 5 days at the indicated temperature. dip1Δ and wsp1Δ show a synthetic sick genetic interaction.
Functional relationship between Dip1 and WASp
As the mammalian orthologue WISH/DIP has been shown to bind directly to N-WASp [22], we tested whether dip1p functions by regulating wsp1p. We probed the genetic relationship between wsp1+ and dip1+ by characterizing a double wsp1Δ dip1Δ mutant. wsp1Δ dip1Δ mutants were synthetic sick for growth and nonviable at 36°C (Figure 5E). These double mutant cells exhibited a normal number of sla1-GFP positive patches, but had a worse defect in the patch localization of the actin-dependent marker coronin than either single mutant (Figure 5B). The patches that eventually recruited crn1p failed to internalize and then disassembled (Figure 5C). Thus the wsp1Δ dip1Δ mutant has properties of both single mutants. These results indicate that dip1p has functions apart from regulating WASp and may thus participate in a WASp-independent pathway.
Activation of a patch may be triggered by another endocytic vesicle
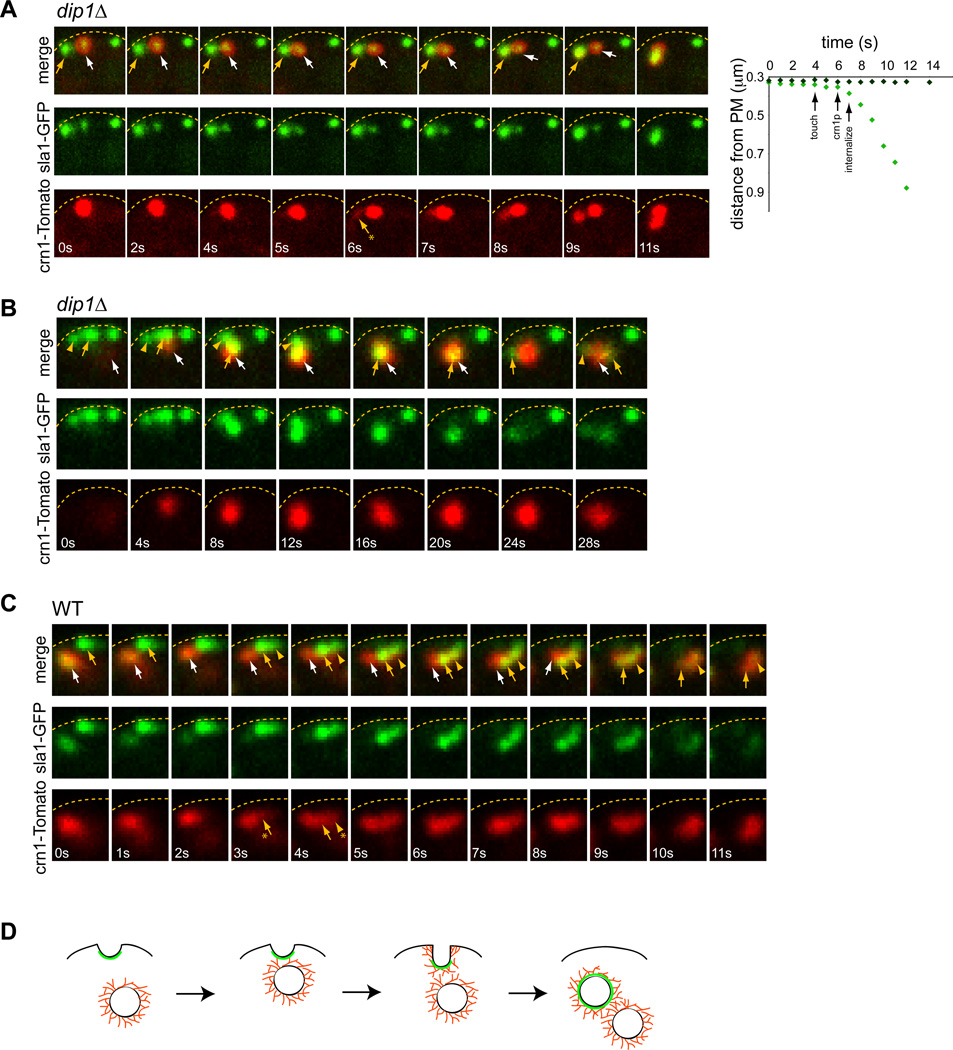
One current model for initiation of Arp2/3 actin polymerization is an actin “seed” model, in which pre-existing actin filaments prime the burst of actin polymerization [31]. Although individual actin filaments cannot be easily visualized in live cells, we did find evidence that activation of an actin patch is triggered by an actin-rich structure: another endocytic vesicle. Crn1-Tomato is an actin-dependent marker that labels the endocytic vesicle for significant periods of time after internalization [29]. Dual imaging of sla1-GFP and crn1-Tomato revealed that the activation and internalization of a patch was often preceded by a transient “touch” of an internalized endocytic vesicle (marked by crn1-Tomato). In dip1Δ cells, about 30% (n= 97 patches in 16 cells) of activation events were preceded by the arrival of another endocytic vesicle in its vicinity (Figure 6A,B; Figure S6A). In a typical case, upon the touch, the patch began to accumulate crn1-Tomato in 1–3 sec, signifying actin assembly, and then exhibited rapid internalization, often moving with the priming vesicle (Figure 6A graph). 70% of all touch events resulted in activation and internalization (n=45 patches in 15 cells; Figure S6B), although because of limitations in the resolution of light microscopy, it was not possible to determine in the remaining cases whether the vesicle came close enough. In some cases, a patch triggered the activation and internalization of multiple patches at once (Figure 6B). Because the activation of the patches is so infrequent in dip1Δ cells, the tight temporal correlation of patch touching and activation was highly significant. The stochastic nature of these visits provides one explanation for variable timing of patch activation in dip1 mutant cells. Similar events were also observed in a subset of wildtype cells at similar frequencies (80% of touch events; n= 18 patches in 4 cells; Figure 6C; Figure S6A, B), but the significance of these touches was less measurable as all patches were activated in any event.
Figure 6. Activation of a patch may be triggered by another endocytic vesicle.
(A) Left: Time-lapse images of dip1Δ cells expressing crn1-Tomato (red) and sla1-GFP (green). The sequence shows the movement of an internalized endocytic vesicle (white arrow) moving towards and touching an immature patch marked only with sla1-GFP (yellow arrow). This touch correlates with the recruitment of crn1p to the second patch (yellow arrow with asterisk) and its subsequent internalization. Right: Graph showing position of the sla1p-marked endocytic pit or vesicle relative to the plasma membrane (marked by the dashed lines) in this sequence, and the relative timings of the touch, recruitment of crn1p, and internalization. (B) Image sequence showing the movement of an endocytic vesicle (white arrow) to a cluster of sla1-GFP marked patches at the cortex (yellow arrow and arrowhead), and then the internalization of this entire cluster in a dip1Δ cell. (C) In a wildtype cell, a similar movement of an endocytic vesicle (white arrow) to a cluster of sla1-GFP positive patches (yellow arrow and arrowhead) leading to activation of the whole cluster. (D) Schematic drawing showing the activation of a patch after being “touched” by an endocytic vesicle. Sla1p is represented in green, and actin filaments are in red. Note that the organization of actin filaments is not known, and elements are not drawn to scale.
As one test of this model, we predicted that patch activation would be reduced if patches cannot move towards each other. Consistent with this model, patches do not move in LatA-treated cells and wsp1 mutants have severe defects in actin initiation in a dip1Δ background (Figure 5; [29]). Although internalized endocytic vesicles associate with actin cables [29], analyses of for3Δ dip1Δ mutant show that actin cables do not contribute to patch priming or to targeting of patches towards each other. These findings thus identify this “activation by touching” mechanism as an additional way to activate a patch.
Discussion
A switch for triggering actin assembly
The yeast endocytic patch serves as a model system for studying the regulation of actin assembly by the Arp2/3 complex in vivo. Here, we provide evidence that a switch-like mechanism is responsible for the initiation of actin assembly at each patch. The rapid kinetics, the stochastic timing of the switch in the absence of a trigger, and the sigmoidal curve for the dose-dependent effects of Lat A (Figure 4D) all support the existence of a switch-like mechanism for initiating actin polymerization downstream of wsp1p. Key elements of this switch include positive feedback relationships between actin assembly, accumulation of wsp1p and recruitment and activation of Arp2/3. In this perspective, dip1p is a necessary element of a timer that triggers the switch, without being a necessary component of the switch or the actin polymerization machinery itself. In the absence of dip1p, the activation of the switch no longer occurs within seconds, but appears with random timing; once activated, endocytosis then proceeds with normal (or slightly elevated) kinetics.
These studies provide a working model for the initiation of Arp2/3-dependent actin assembly in vivo. As clearly demonstrated in the dip1 mutant, WASp may localize initially on the cortex prior to actin assembly and arrival of Arp2/3. Upon a triggering event, actin, Arp2/3, and myosin type I are rapidly recruited for a highly local burst of actin polymerization. This observation is consistent with studies in wildtype fission and budding yeasts [4, 8, 10]. A key question is what triggers this burst of actin polymerization? Our quantitative data are not consistent with a current model [5], which proposes that the patch is triggered by a threshold concentration of wsp1p. Instead, one initiation event could be the addition of an F-actin seed to the system, which acts to recruit and activate Arp2/3 [31–33].
A critical element contributing to this switch may be the rapid assembly of actin filaments. At low doses of LatA (2 µM) or in a wsp1 mutant, the patch contains F-actin, but the actin lacks the necessary dynamics for more explosive actin assembly needed to drive endocytosis. One potential explanation for this effect is a preference of Arp2/3 to bind recently polymerized actin rich in ATP-actin [34, 35].
Our observations also suggest that there exists a program for patch disassembly that is independent of internalization. When the rate of actin polymerization is insufficient for internalization, for instance in wsp1 mutants or cells treated with 2µM LatA, the patch disassembles in less than a minute. This disassembly does not occur at the “arrest point” in dip1Δ mutants, as these patches persist at that point for many minutes, but occurs in dip1wsp1 double mutants soon after a patch has attempted actin assembly. Thus this disassembly occurs after the patch lifecycle has reach a certain stage, suggesting that disassembly is also a regulated process.
Dip1, a key factor in initiation of actin assembly
We identify dip1p as an important element in triggering this actin assembly switch at the actin patch. In contrast to components such as wsp1p, dip1p is not needed for the burst of actin polymerization per se, but appears to have a primary function in its proper timing. One speculative model consistent with several lines of data is that dip1p, along with the WASp, facilitates the presentation of an F-actin seed for initiation. Although the biochemical function of S. pombe dip1p remains to be defined, numerous interactions have been documented in its orthologues. Mammalian WISH/DIP/SPIN90 has been shown to bind directly to N-WASp via its SH3 domain to activate Arp2/3p mediated actin polymerization [22], and to interact with F- and G-actin, and other components of the endocytic machinery such as syndapin, dynamin, and the Arp2/3 complex [18, 19, 23]. In S. cerevisiae, Ldb17 localizes faintly to the patch with similar kinetics as dip1p [17]. ldb17 mutants also exhibit a small number of abnormally large actin patches and have endocytic defects. A C-terminal proline-rich domain (PRD) of Ldb17 binds to the SH3 domains of Sla1 and Bzz1. S. pombe dip1p, however, lacks the SH3 and PRD domains required for many of these interactions, but retains a conserved Leucine Rich Region (LRR) that has been implicated in the binding of the Arp2/3 complex, F- and G-actin, and formins in the mammalian orthologue [19, 21]. Thus it will be informative to compare the functions and molecular interactions among these family members.
One curious aspect of the dip1 mutants is how healthy and polarized these cells are, despite strongly reduced levels of endocytosis. wsp1Δ mutants, which have no detectable endocytic internalization as measured by patch imaging, are viable but have growth defects [30]. These observations suggest that clathrin-mediated endocytosis may not be essential for viability in S. pombe, and that even strongly reduced rates are sufficient for cell growth and polarization in the laboratory.
Priming a patch with another patch
Our direct observations suggest that an actin polymerization event can be primed by the touch of another endocytic vesicle. A correlation between the activation of a patch and the touch of another visiting patch appeared to be highly significant in dip1 cells, and similar events were observed in wildtype cells as well. We propose that these visiting vesicles, which are actively polymerizing actin [29], provide ATP-rich actin seeds for triggering actin polymerization at the patch site or some other critical component. It is not clear whether these events are analogous to those observed in budding yeast, where the early endosome, a larger component that is not actin-rich, appears to move and “eat up” patches [36].
Analogies to the cell cycle
These studies highlight similarities of endocytosis to the cell division cycle: both are characterized by the orchestrated progression of events that are temporally regulated by switch-like mechanisms at key transition points [37–39]. In this light, dip1p is analogous to a cell cycle regulatory factor that controls the transition from one cell cycle phase to the next. Although cell cycle mutants in which cell cycle periods are either slower or faster are well known [40], curiously, to our knowledge, no cell cycle mutant has been identified quite like dip1Δ, in which the length of a cell cycle phase (like G1 phase) is rendered highly variable with a random distribution. Additional contrasts and comparisons between endocytosis and the cell cycle are likely to yield general conceptual insights into the regulation of complex biological programs.
Experimental Procedures
Yeast Methods
The S. pombe strains used in this study are listed in Supplemental Table 1. Yeast cells were generally grown to log phase in YE5S media at 25°C and imaged on glass coverslips. See Supplementary Methods for details of endocytosis assays and other yeast methods.
Microscopy and image analysis
Microscopy was performed using either a widefield fluorescence microscope with a Hamamatsu ORCA100 CCD digital camera or a spinning disc confocal upright microscope with a Hamamatsu EM-CCD camera [29] using a 100X N.A. 1.4 oil objective. Images were acquired, processed and analyzed with the OpenLab 5.0.2 software (Improvision, Coventry, United Kingdom) or with ImageJ software (National Institute of Health, Bethesda, MD). See Supplementary Methods for details of image analyses.
Highlights.
-
-
The initiation of actin polymerization at endocytic sites is regulated by a switch-like mechanism that involves WASp, Arp2/3 and rapid actin assembly.
-
-
Dip1p, a WISH/DIP/SPIN90 orthologue, regulates the timing of this switch.
-
-
A second mode of initiation of actin polymerization may be through the touch of another endocytic vesicle.
Supplementary Material
Acknowledgements
We thank Bruce Goode and Melissa Chesarone for sharing preliminary data and discussion, Dennis Vitkup for discussion, members of the Chang Laboratory for helpful comments and discussions, and in particular Mark A. Ellis for providing strains and intellectual input. This work was supported by grants from the National Institute of Health GM056836 and GM069670 to F.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 2.Galletta BJ, Cooper JA. Actin and endocytosis: mechanisms and phylogeny. Curr Opin Cell Biol. 2009;21:20–27. doi: 10.1016/j.ceb.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merrifield CJ. Seeing is believing: imaging actin dynamics at single sites of endocytosis. Trends Cell Biol. 2004;14:352–358. doi: 10.1016/j.tcb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Sirotkin V, Beltzner CC, Marchand JB, Pollard TD. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J Cell Biol. 2005;170:637–648. doi: 10.1083/jcb.200502053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berro J, Sirotkin V, Pollard TD. Mathematical modeling of endocytic actin patch kinetics in fission yeast: disassembly requires release of actin filament fragments. Mol Biol Cell. 2010;21:2905–2915. doi: 10.1091/mbc.E10-06-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD. Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol Biol Cell. 2010;21:2894–2904. doi: 10.1091/mbc.E10-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol. 2009;11:1039–1042. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonsdottir GAand, Li R. Dynamics of yeast Myosin I: evidence for a possible role in scission of endocytic vesicles. Curr Biol. 2004;14:1604–1609. doi: 10.1016/j.cub.2004.08.055. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Sun Y, Drubin DG, Oster GF. The mechanochemistry of endocytosis. PLoS Biol. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson SM, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, Destaing O, Ko G, Takasaki J, Cremona O, et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgs HN, Pollard TD. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem. 2001;70:649–676. doi: 10.1146/annurev.biochem.70.1.649. [DOI] [PubMed] [Google Scholar]
- 13.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 15.Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- 16.Lim CS, Park ES, Kim DJ, Song YH, Eom SH, Chun JS, Kim JH, Kim JK, Park D, Song WK. SPIN90 (SH3 protein interacting with Nck, 90 kDa), an adaptor protein that is developmentally regulated during cardiac myocyte differentiation. J Biol Chem. 2001;276:12871–12878. doi: 10.1074/jbc.M009411200. [DOI] [PubMed] [Google Scholar]
- 17.Burston HE, Maldonado-Baez L, Davey M, Montpetit B, Schluter C, Wendland B, Conibear E. Regulators of yeast endocytosis identified by systematic quantitative analysis. J Cell Biol. 2009;185:1097–1110. doi: 10.1083/jcb.200811116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y, Kim S, Lee S, Kim SH, Park ZY, Song WK, Chang S. Interaction of SPIN90 with dynamin I and its participation in synaptic vesicle endocytosis. J Neurosci. 2005;25:9515–9523. doi: 10.1523/JNEUROSCI.1643-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim DJ, Kim SH, Lim CS, Choi KY, Park CS, Sung BH, Yeo MG, Chang S, Kim JK, Song WK. Interaction of SPIN90 with the Arp2/3 complex mediates lamellipodia and actin comet tail formation. J Biol Chem. 2006;281:617–625. doi: 10.1074/jbc.M504450200. [DOI] [PubMed] [Google Scholar]
- 20.Kim DJ, Kim SH, Kim SM, Bae JI, Ahnn J, Song WK. Factin binding region of SPIN90 C-terminus is essential for actin polymerization and lamellipodia formation. Cell Commun Adhes. 2007;14:33–43. doi: 10.1080/15419060701225010. [DOI] [PubMed] [Google Scholar]
- 21.Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol. 2007;17:579–591. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Fukuoka M, Suetsugu S, Miki H, Fukami K, Endo T, Takenawa T. A novel neural Wiskott-Aldrich syndrome protein (N-WASP) binding protein, WISH, induces Arp2/3 complex activation independent of Cdc42. J Cell Biol. 2001;152:471–482. doi: 10.1083/jcb.152.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SH, Choi HJ, Lee KW, Hong NH, Sung BH, Choi KY, Kim SM, Chang S, Eom SH, Song WK. Interaction of SPIN90 with syndapin is implicated in clathrin-mediated endocytic pathway in fibroblasts. Genes Cells. 2006;11:1197–1211. doi: 10.1111/j.1365-2443.2006.01008.x. [DOI] [PubMed] [Google Scholar]
- 24.Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- 25.Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 26.Nakano K, Kuwayama H, Kawasaki M, Numata O, Takaine M. GMF is an evolutionarily developed Adf/cofilin-super family protein involved in the Arp2/3 complex-mediated organization of the actin cytoskeleton. Cytoskeleton (Hoboken) 2010;67:373–382. doi: 10.1002/cm.20451. [DOI] [PubMed] [Google Scholar]
- 27.Padrick SB, Cheng HC, Ismail AM, Panchal SC, Doolittle LK, Kim S, Skehan BM, Umetani J, Brautigam CA, Leong JM, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelham RJ, Jr, Chang F. Role of actin polymerization and actin cables in actin-patch movement in Schizosaccharomyces pombe. Nat Cell Biol. 2001;3:235–244. doi: 10.1038/35060020. [DOI] [PubMed] [Google Scholar]
- 30.Lee WL, Bezanilla M, Pollard TD. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J Cell Biol. 2000;151:789–800. doi: 10.1083/jcb.151.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Achard V, Martiel JL, Michelot A, Guerin C, Reymann AC, Blanchoin L, Boujemaa-Paterski R. A "primer"-based mechanism underlies branched actin filament network formation and motility. Curr Biol. 20:423–428. doi: 10.1016/j.cub.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 32.Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amann KJ, Pollard TD. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat Cell Biol. 2001;3:306–310. doi: 10.1038/35060104. [DOI] [PubMed] [Google Scholar]
- 34.Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys J. 2006;91:3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toshima JY, Toshima J, Kaksonen M, Martin AC, King DS, Drubin DG. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc Natl Acad Sci U S A. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novak B, Vinod PK, Freire P, Kapuy O. Systems-level feedback in cell-cycle control. Biochem Soc Trans. 2010;38:1242–1246. doi: 10.1042/BST0381242. [DOI] [PubMed] [Google Scholar]
- 38.Ferrell JE, Jr, Pomerening JR, Kim SY, Trunnell NB, Xiong W, Huang CY, Machleder EM. Simple, realistic models of complex biological processes: positive feedback and bistability in a cell fate switch and a cell cycle oscillator. FEBS Lett. 2009;583:3999–4005. doi: 10.1016/j.febslet.2009.10.068. [DOI] [PubMed] [Google Scholar]
- 39.Charvin G, Oikonomou C, Siggia ED, Cross FR. Origin of irreversibility of cell cycle start in budding yeast. PLoS Biol. 2010;8:e1000284. doi: 10.1371/journal.pbio.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nurse P. Regulation of the eukaryotic cell cycle. Eur J Cancer. 1997;33:1002–1004. doi: 10.1016/s0959-8049(97)00091-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.