SUMMARY
Mycobacterium tuberculosis uses the ESX-1 secretion system to deliver virulence proteins during infection of host cells. Here we report a novel post-transcriptional control mechanism of ESX-1 mediated by MycP1, a serine protease. We show that MycP1 is required for ESX-1 secretion and has unusual substrate specificity. Unexpectedly, inhibition of protease activity increases secretion of ESX-1 substrates. We demonstrate that EspB, an ESX-1 substrate required for secretion, is a target of MycP1 in vitro and in vivo. During macrophage infection, an inactive MycP1 protease mutant causes hyper-activation of ESX-1 stimulated innate signaling pathways. MycP1 is required for growth in mice during acute infection, while protease inhibition leads to attenuated virulence during chronic infection. As the key ESX-1 substrates ESAT-6 and CFP-10 are highly immunogenic, fine-tuning of their secretion by MycP1 may balance virulence and immune detection and be essential for successful maintenance of long-term M. tuberculosis infection.
INTRODUCTION
Mycobacterium tuberculosis, the causative agent of human tuberculosis, infects two billion people, causing two million deaths every year (Hingley-Wilson et al., 2003). One reason for its success as a pathogen is its ability to manipulate its environment by exporting molecules that allow it to evade or control host immune responses. A key M. tuberculosis virulence determinant is the “Type VII” ESX-1 secretion system, which transports protein virulence factors into host cells (Stanley et al., 2003; Lewis et al., 2003; Guinn et al., 2004; Abdallah et al., 2007). ESX-1 secretion is required for early replication and full virulence in macrophages and mice, and has multiple effects on host cells.
The identity of all ESX-1 substrates and the mechanism by which they affect host cells is not well understood. Various activities have been ascribed to the ESX-1 substrates ESAT-6 and CFP-10, encoded by the genes esxA and esxB, including inhibition of phagosome maturation and cytokine signaling by infected macrophages (Stanley et al., 2003; Hsu et al., 2003; Pathak et al., 2007), interaction with the macrophage immune receptor TLR2 and inhibition of TLR signaling (Pathak et al., 2007), and formation of pores in mycobacterial phagosomes, perhaps allowing bacterial spread (Hsu et al., 2003; Smith et al., 2008). ESX-1 is clearly implicated in early stages of infection, including activation of the cytosolic signaling response (Weiden et al., 2000; Giacomini et al., 2001; Lewinsohn et al., 2006; O’Riordan et al., 2002). Gene targets of this signaling pathway include Type I IFNs, which function in antiviral defense, regulation of the immune response, control of cell growth and modulation of apoptosis (Taki, 2002). Importantly, Type I IFN induction is dependent on the ESX-1 secretion system (Stanley et al., 2007). M. tuberculosis elicits Type I IFN production via an as yet unidentified receptor, leading to phosphorylation of the IFN regulatory factor (IRF-3) transcription factor, which then promotes transcription of genes such as IFN-β and interferon-induced protein with tetratricopeptide repeats 1 (IFIT1).
Studies in M. tuberculosis and related mycobacteria have identified components and substrates of the ESX-1 system. The proteins EccCa1 (Rv3870), EccCb1 (Rv3871) and EccD1 (Rv3877) are essential for ESX-1 secretion in M. tuberculosis, while in M. smegmatis homologs of the proteins EspG (Rv3866), EccB (Rv3869), EccE1 (Rv3882c) and MycP1 are also required (Figure 1A; Stanley et al., 2003; Guinn et al., 2004; Hsu et al., 2003; Converse and Cox, 2005; Bitter et al., 2009). The machine components are predicted to be either cytosolic or membrane-bound and to interact with each other, but are not themselves secreted. However, specific roles for individual components of the ESX-1 system machinery have not been characterized. In addition to ESAT-6 and CFP-10, four other substrates of the ESX-1 system are known: EspA, EspB, EspC and EspR (Fortune et al., 2005; MacGurn et al., 2005; McLaughlin et al., 2007; Gao et al., 2004; Xu et al., 2007; Raghavan et al., 2008; J. A. MacGurn and J. S. Cox, unpublished data). An unusual feature that distinguishes ESX-1 from other systems is that secretion of all substrates is mutually dependent. For example, secretion of EspA is blocked in an ESAT-6 mutant, and vice versa (Fortune et al., 2005).
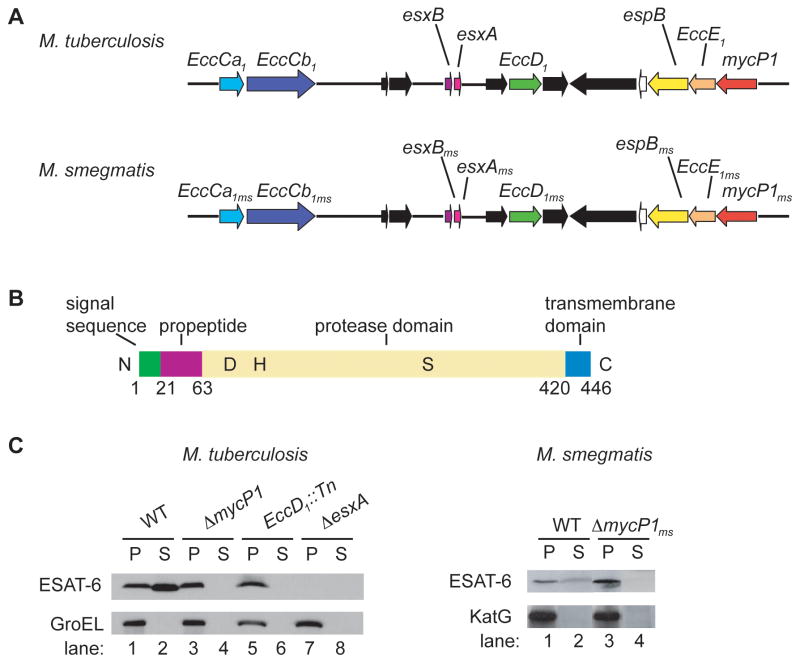
Figure 1. M. tuberculosis and M. smegmatis ΔmycP1 mutants fail to secrete ESAT-6.
(A) Schematic representation of ESX-1 locus in M. tuberculosis and M. smegmatis. (B) Diagram of M. tuberculosis MycP1 showing domains and active site residues D90, H131 and S332. (C) Pellets (P) and cell culture supernatants (S) were generated from indicated strains and ESAT-6, GroEL and KatG were detected by Western blot. GroEL and KatG served as lysis controls. See Figure S1 for a description of the M. tuberculosis ΔmycP1 strain construction.
Despite being essential for virulence, ESAT-6 is also a highly immunogenic T cell antigen (Brandt et al., 2000; Colangeli et al., 2000; Coler et al., 2001; Dietrich et al., 2006). Restoration of ESAT-6 secretion to the M. bovis BCG vaccine strain, which lacks the ESX-1 system, conferred enhanced protection against M. tuberculosis challenge but also increased virulence (Pym et al., 2003). Thus, while ESAT-6 is an important virulence factor, it also works against M. tuberculosis infection by stimulating the immune response. Thus it may be essential for M. tuberculosis to tightly regulate the amount of ESAT-6 being exported in order to maintain an optimal balance between virulence and immunogenicity. ESX-1 is under the transcriptional control of EspR, a DNA-binding protein that promotes transcription of the genes encoding EspA and EspC (Raghavan et al., 2008). This system is under negative feedback control as EspR is also secreted via the ESX-1 system, leading to downregulation of espA and espC transcription. In addition, the regulatory protein PhoP promotes transcription of espA, espC and Rv3614-3612c, thus affecting ESX-1 secretion (Frigui et al., 2008; Gonzalo-Asensio et al., 2008).
There are clues that indicate another potential control mechanism involving the ESX-1 substrate EspB. Upon secretion, EspB is cleaved near its C-terminus, but the protease responsible for the cleavage is unknown (McLaughlin et al., 2007; Xu et al., 2007). Western blotting experiments detected EspB as a 61 kDa band in cell lysates, but a 50 kDa band in secreted fractions, while the C-terminal 11 kDa band appeared to be unstable and was not detected (Xu et al., 2007). The EspB C-terminus is dispensable for its own secretion, as expression of a truncated form of EspB in an espB transposon mutant led to normal secretion of EspB. However, the C-terminus is essential for interaction of EspB with ESAT-6, maintenance of intracellular levels of ESAT-6, and secretion of ESAT-6 and CFP-10, suggesting that cleavage of EspB could have a regulatory function in ESX-1 secretion (Xu et al., 2007).
Given that EspB is processed, it is notable that one component of the ESX-1 secretion machine, MycP1, is a putative subtilisin-like serine protease (Brown et al., 2000; Dave et al., 2002). Bacterial subtilases are typically secreted and degrade proteins non-specifically to provide cells with readily importable peptides (Gupta et al., 2002). In contrast, eukaryotic subtilases typically cleave substrates after specific basic residues: for instance, yeast Kex2p is required for certain proteolytic processing steps during the biogenesis of the α mating pheromone (Bergeron et al., 2000; Julius et al., 1984). MycP1 has not been extensively studied but is likely important for ESX-1 secretion as, like other system components, it is encoded by a gene located within the ESX-1 locus (Figure 1A; Gey van Pittius et al., 2001). MycP1 localizes to the cell wall/membrane fraction and is expressed constitutively during growth in culture, but not in the vaccine strain M. bovis BCG (Brown et al., 2000; Dave et al., 2002). It has been reported that MycP1 expression is increased during growth in macrophages, and its M. leprae homolog is transcribed during human infection (Brown et al., 2000; Ribeiro-Guimarães et al., 2007).
In this work, we show that MycP1 is essential for ESX-1 function in M. tuberculosis and is required for early replication in macrophages and full virulence in mice. Surprisingly, MycP1 plays a dual role in regulating secretion activity of the ESX-1 system. Whereas the MycP1 protein is required for secretion, abolition of MycP1 protease activity by mutagenesis of the active site leads to increased secretion. The increase in abundance of ESX-1 substrates is sensed by infected macrophages, which induce a heightened cytosolic surveillance response. We find that, unusually for bacterial subtilases, MycP1 has a defined substrate specificity and cleaves substrates following proline residues. In addition, we identify EspB as a substrate of MycP1. We conclude that the MycP1 protein is required for ESX-1 secretion but that its protease activity negatively regulates secretion via EspB, a protein substrate that is required for the function of the ESX-1 system. Identification of a second level of regulation of the ESX-1 secretion system supports the notion that M. tuberculosis has evolved remarkably tight controls on ESX-1 secretion capacity through distinct and novel molecular mechanisms.
RESULTS
MycP1 is required for ESX-1 secretion in M. tuberculosis
To determine the role of MycP1 in ESX-1 secretion, we created a mycP1 deletion mutant in M. tuberculosis via homologous recombination and verified that the deletion had occurred using Southern blot analysis (Supplementary Figure S1). To test if MycP1 is required for ESAT-6 secretion, we probed filtered culture supernatants from exponentially growing wild-type, ΔmycP1, EccD1::Tn and ΔesxA M. tuberculosis strains using ESAT-6-specific antibodies. We observed that ESAT-6 was present in supernatants of wild-type M. tuberculosis but not in the ΔmycP1 mutant, a phenotype identical to that of the known ESX-1 mutant, EccD1::Tn (Figure 1C). ESAT-6 was present in extracts from cell pellets of all three strains and, as expected, no ESAT-6 was detectable in the ΔesxA mutant cells. We also confirmed that wild-type M. smegmatis secretes ESAT-6 but the ΔmycP1ms mutant does not (Figure 1C). Thus, MycP1 is required for ESX-1 secretion.
Purification, protease activity and substrate specificity profiling of MycP1
Analysis of the amino acid sequence of MycP1 suggests that it is a member of the subtilase class of serine proteases (Brown et al., 2000; Dave et al., 2002). MycP1 contains an N-terminal Sec signal sequence and a C-terminal transmembrane domain, and protein topology prediction programs (TMHMM, PSORTb) suggest that it is likely anchored in the cell membrane with its active site in the extracytoplasmic space (Sonnhammer et al., 1998; Gardy et al., 2005). In addition, MycP1 has a putative propeptide and a catalytic triad of Asp, His and Ser residues, all hallmarks of the subtilase family (Figure 1B).
To determine whether MycP1 has protease activity, we expressed the portion of M. smegmatis MycP1ms containing its propeptide and protease domain as an MBP fusion protein in E. coli and purified it using amylose affinity chromatography (Figure 2A, lane 1). The fusion protein also contained a cleavage site for the protease Factor Xa between MBP and the MycP1ms propeptide, allowing removal of MBP. Subtilases are known to require their propeptides for correct folding, with subsequent removal of the propeptide by the protease domain, resulting in an active protease (Fu et al., 2000). Though the Factor Xa-cleaved MycP1ms propeptide-protease domain fusion was initially inactive, prolonged incubation with the protease led to cleavage at a site between the MycP1ms propeptide and protease domain, removing the propeptide and activating MycP1ms. We activated subsequent preparations of MBP- MycP1ms by incubating with active MycP1ms, and purified active MycP1ms using hydroxyapatite affinity chromatography (Figure 2A, lane 2).
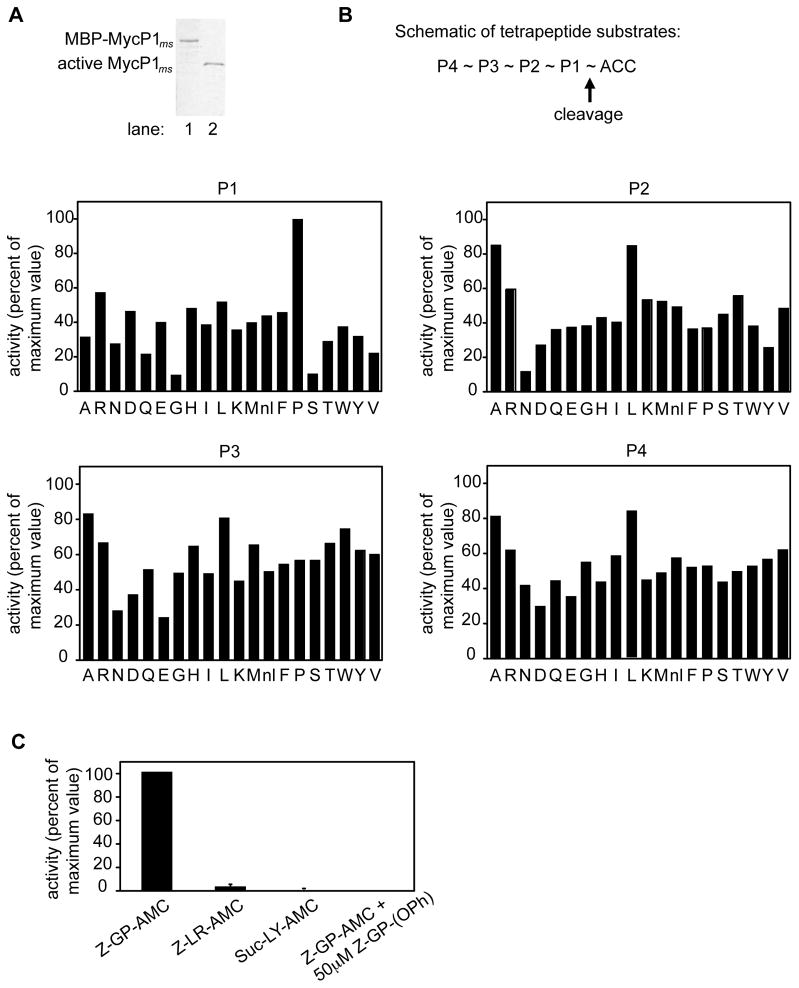
Figure 2. Purification and substrate specificity profiling of M. smegmatis MycP1.
(A) MBP-MycP1 (lane 1) and active MycP1 (lane 2) were purified using amylose and hydroxyapatite affinity chromatography respectively, and resolved by SDS-PAGE. (B) MycP1 protease activity was measured against fluorogenic tetrapeptide substrate pools, in each of which the amino acid at one position was held constant while the other positions were varied. The x axis indicates which amino acid was held constant (nl = norleucine), while the y axis shows activity plotted relative to the highest activity of the library. (C) Activity of MycP1 against individual fluorogenic dipeptide substrates, in the absence or presence of specific inhibitor.
Since the substrate specificity of MycP1ms was unknown, we tested its activity against a positional scanning synthetic combinatorial library, which comprised tetrapeptides of every possible amino acid composition (Choe et al., 2006). We found that MycP1ms had activity against a wide variety of substrates with different amino acids in the P1 position but has an unusual preference for proline at this site (Figure 2B). MycP1ms shows greater promiscuity in the P2-P4 positions, though there is a preference for small hydrophobic amino acids such as alanine and leucine at these sites. The preference of MycP1ms for proline at P1 was confirmed using individual substrates (Figure 2C). Although the degeneracy of the MycP1sm recognition sequence made it difficult to find potential substrates with this information alone, these results allowed us to design a specific substrate, Z-Gly-Pro-AMC, and a specific phosphonate inhibitor, Z-Gly-Pro-(OPh)2, which completely abrogated the activity of MycP1ms in vitro (Figure 2C).
Protease activity of MycP1 negatively regulates ESX-1 secretion
Since the ΔmycP1 strain failed to secrete ESAT-6, we reasoned that protease activity would also be required for MycP1 function and ESX-1 secretion. To begin to determine the role of MycP1 protease activity, we expressed and purified a protease mutant (MycP1ms S334A) in which the active-site serine codon in M. smegmatis mycP1ms was replaced with an alanine codon using site-directed mutagenesis. Recombinant MycP1ms S334A had no significant activity against Z-Gly-Pro-AMC (Figure 3A). We then examined ESAT-6 secretion in M. smegmatis ΔmycP1sm bacteria expressing either wild-type MycP1ms or MycP1ms S334A. Expression of wild-type MycP1ms in ΔmycP1ms cells restored ESAT-6 secretion, although not to wild-type levels. Surprisingly, despite the fact that MycP1ms is essential for secretion, expression of MycP1ms S334A significantly increased ESAT-6 secretion above wild-type levels (Figure 3B). We observed identical results when we expressed a different active site mutant (D92A, data not shown) of MycP1sm in ΔmycP1ms cells, indicating that the phenotype is the direct result of the loss of proteolytic activity.
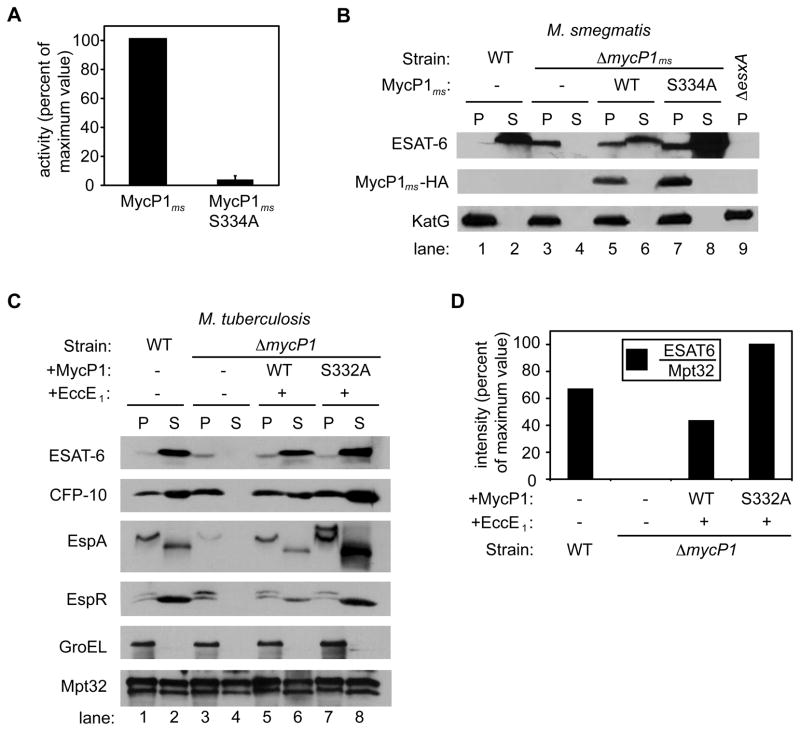
Figure 3. Protease activity of MycP1 negatively regulates ESX-1 secretion.
(A) MycP1 protease activity was measured against the Z-GP-AMC substrate. Activity of protease-inactive MycP1 (MycP1 S334A), expressed relative to wild-type MycP1. (B) Western blot detection of ESAT-6, MycP1-HA and KatG in M. smegmatis pellets (P) and supernatants (S) from wild-type and mutant strains. (C) Western blot detection of ESAT-6, CFP-10, EspA, EspR, GroEL and Mpt32 in M. tuberculosis pellets (P) and supernatants (S) from wild-type and mutant cells. See Figure S2 for data showing that the ΔmycP1 mutation did not exert a polar effect on transcription of the downstream gene, eccE1. (D) Quantitation of ESAT6 in M. tuberculosis supernatants from wild-type and mutant strains, expressed relative to Sec-dependent substrate Mpt32.
We observed a similar phenotype in M. tuberculosis, in which expression of inactive MycP1 (MycP1 S332A) led to increased secretion of ESAT-6 (Figure 3C). In M. tuberculosis we complemented the ΔmycP1 strain with wild-type or inactive MycP1 in an operon with the downstream gene, EccE1, to achieve complete complementation (see Supplementary Methods and Supplementary Figure S2). In addition to ESAT-6, secretion of other known ESX-1 substrates – CFP-10, EspA and EspR – was also increased relative to wild-type. However, Mpt32, a substrate of the Sec pathway that is exported independently of ESX-1, was secreted to equal levels in all strains, demonstrating that the MycP1 mutant specifically modulates ESX-1 secretion (Figure 3C). Expression of wild-type MycP1 restored ESAT-6 secretion nearly to levels observed in wild-type M. tuberculosis. Since the difference in ESAT-6 secretion observed between ΔmycP1 expressing wild-type MycP1 versus MycP1 S332A was not as robust in M. tuberculosis as in M. smegmatis, we further examined ESAT-6 in culture supernatants by quantitative western blotting. Using this technique we determined that the amount of ESAT-6 relative to Mpt32 in culture supernatants was approximately twofold greater in the MycP1 S332A strain than the wild-type strain (Figure 3D). Therefore, the MycP1 protein is required for ESX-1 secretion but its protease activity inhibits the system.
MycP1 cleaves EspB, an ESX-1 substrate
We were curious to understand the mechanism by which MycP1 proteolysis negatively regulates protein secretion. Since MycP1 is itself required for secretion in M. tuberculosis, a simple model is that MycP1 self-proteolysis leads to its degradation, thus controlling ESX-1 secretion. However, quantitative western blotting showed that the levels of wild-type and S332A mutant MycP1 were equivalent and that the half-lives of the proteins were also identical (not shown).
An alternative hypothesis is that MycP1 cleaves an activator of ESX-1 secretion. A likely candidate is EspB (Rv3881c), which is secreted in an ESX-1-dependent manner and is required for the function of the ESX-1 system (McLaughlin et al., 2007; Gao et al., 2004; Xu et al., 2007). EspB accumulates in culture supernatants in an apparent proteolytically processed form, though the protease responsible has not been identified (McLaughlin et al., 2007). Furthermore, previous work has suggested that cleavage of EspB occurs after Proline 332 (Xu et al., 2007), a result consistent with our susbtrate specificity profiling results. We therefore used an antibody specific for EspB to test whether cleavage requires MycP1 in vivo. This antibody recognizes a 100 residue fragment of EspB comprising amino acids 234 to 333, and thus only detects full-length protein and the N-terminal cleavage products (Figure 4C, McLaughlin et al., 2007). Full-length EspB (460 amino acids) was present in whole cell lysates of all strains (Figure 4A, lanes 1–4). We observed that EspB was secreted in its cleaved form by wild-type M. tuberculosis (Figure 4A, lane 5) and by M. tuberculosis ΔmycP1 cells expressing wild-type MycP1 (Figure 4A, lane 7), but was not secreted by the ΔmycP1 mutant (Figure 4A, lane 6). However, full-length EspB was secreted from the MycP1 S332A strain (Figure 4A, lane 8), consistent with the prediction that MycP1 directly cleaves EspB. We also observed that a fraction of the secreted EspB was still processed into a low molecular weight form. Using a higher resolution gel, we observed that the original EspB band in secreted fractions of wild-type cells and the complemented strain resolved into three different EspB cleavage products (Figure 4A, lower panel, lanes 5 and 7), implying that EspB is cleaved at a minimum of three sites during secretion. Importantly, two of the three cleavage products were absent in the supernatant of the MycP1 S332A strain, while full-length EspB was detected (Figure 4A, lane 8). This suggests that MycP1 is responsible for cleaving EspB at two sites.
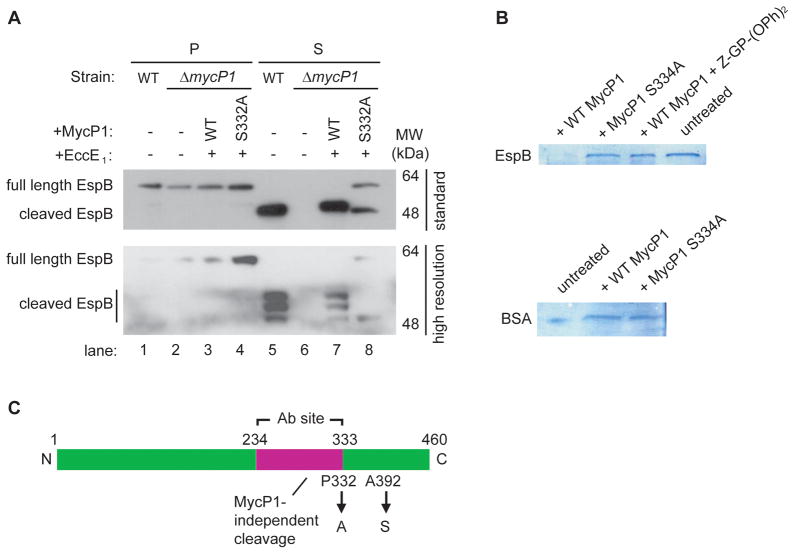
Figure 4. MycP1 directly cleaves EspB, an ESX-1 substrate.
(A) Western blot detection of EspB in M. tuberculosis pellets (P) and supernatants (S) from wild-type and mutant strains. The same samples were run on a standard 13 × 8 cm gel (W x L, top panel) and a higher resolution 16 × 16 cm gel (bottom panel) to reveal the different cleaved forms of EspB. (B) In vitro cleavage assay of MBP-EspB (top panel) and BSA (bottom panel) by WT MycP1, MycP1 S332A, and WT MycP1 in presence of specific inhibitor. (C) Schematic representation of EspB showing cleavage sites and the region used to generate the EspB antibody. See Figure S3 for MS results used to identify the EspB cleavage products.
To determine whether MycP1 directly cleaves EspB, recombinant EspB was subjected to in vitro cleavage by MycP1sm. Wild-type MycP1sm efficiently cleaved EspB but not the specificity control protein BSA (Figure 4B). However, neither protease-dead MycP1sm or wild-type MycP1sm treated with Z-Gly-Pro-(OPh)2 inhibitor could cleave EspB, indicating that EspB is a proteolytic substrate of MycP1.
To identify the sites at which EspB is cleaved in vivo, we used mass spectroscopy to determine the exact molecular weight of cleaved EspB purified from M. tuberculosis secreted protein fractions (Supplementary Figure S3). Although we were unable to identify the Pro332 cleaved form, we identified Ala392 as a second cleavage site. Our substrate specificity profiling studies indicated that Ala is tolerated at this site, though not most preferred. Based on the sizes of the fragments on the SDS-PAGE gel (Figure 4A, lane 5), it is likely that the top band represents the Ala392 product and the middle band the Pro332 fragment, both of which are dependent on MycP1.
To study the role of EspB cleavage by MycP1, we created Ala392Ser and Pro332Ala mutant alleles of espB by site-directed mutagenesis (Figure 4C). Unfortunately, repeated attempts to transform either wild-type M. tuberculosis or an M. marinum espB::Tn mutant strain with integrating and episomal vectors expressing these mutants yielded extremely poor transformation efficiencies, indicating that these mutations were toxic to the cells.
Effects of increased ESX-1 secretion during macrophage infection
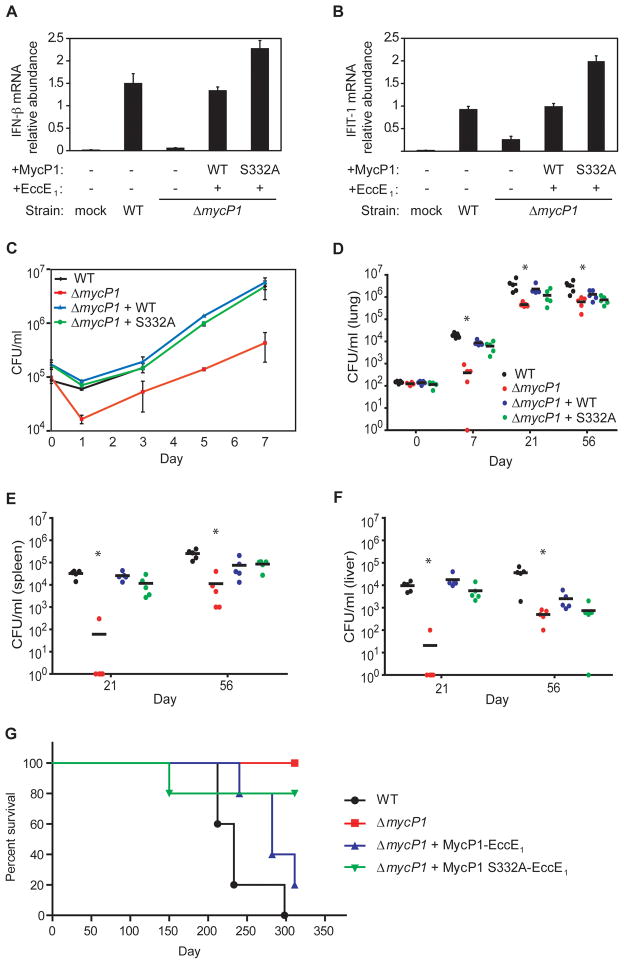
Since fine control of virulence factors is important for bacterial pathogens, we sought to determine if increased secretion of ESX-1 substrates in the MycP1 S332A mutant impacts M. tuberculosis virulence. Since ESX-1 is required for stimulating innate immune responses, we tested whether ESX-1 hyper-activation in the MycP1 S332A strain modified these responses. In macrophages and dendritic cells, M. tuberculosis induces the cytosolic surveillance response, characterized by the transcription of IRF-3-responsive genes such as IFN-β and IFIT-1, in an ESX-1-dependent manner (Stanley et al., 2007; Lewinsohn et al., 2006). We infected murine bone marrow-derived macrophages with wild-type, ΔmycP1, complemented and MycP1 S332A strains, extracted macrophage RNA and quantified IFN-β and IFIT-1 mRNA expression using quantitative real-time PCR. As expected, wild-type bacteria elicited a strong cytosolic response, but ΔmycP1 mutant cells induced the pathway poorly. Importantly, the MycP1 S332A strain induced the cytosolic response more robustly than wild-type (Figure 5A and B). These data indicate that the MycP1 S332A strain has hyper-activated ESX-1 secretion during macrophage infection, resulting in increased innate immune signaling.
Figure 5. Oversecretion of ESAT-6 by ΔmycP1 + MycP1 S332A mutant is sensed by macrophages, and MycP1 is required for virulence in macrophages and mice.
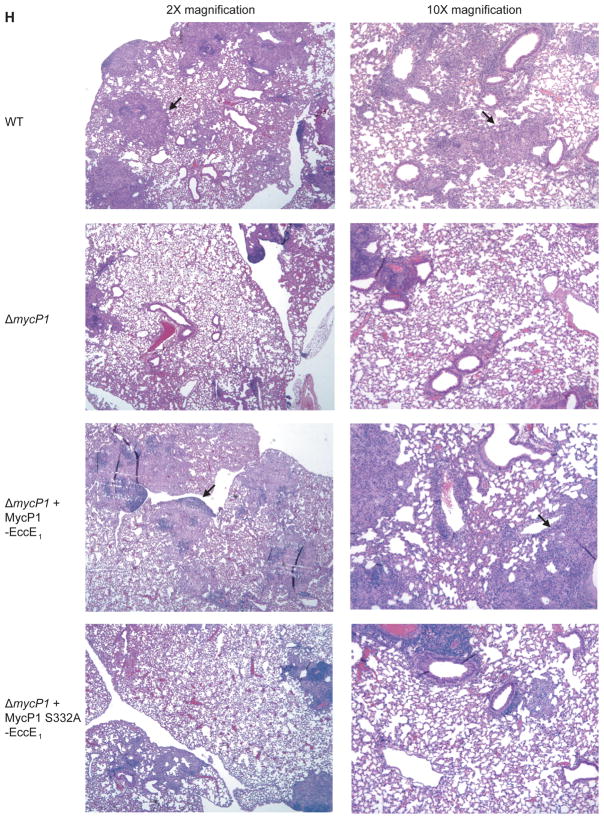
Bone-marrow-derived macrophages were infected with wild-type and mutant strains of M. tuberculosis. At indicated time points total RNA was harvested from macrophage monolayers, and IFN-β (A) and IFIT-1 (B) mRNA levels were measured by quantitative PCR. Values were normalized to actin mRNA levels and each sample was assayed in triplicate. (C) Macrophages were infected, lysed at the indicated time points, and bacterial CFU were enumerated by plating. (D) BALB/c mice were infected with 102 CFU of each strain by aerosol, and bacteria were isolated from lungs immediately following infection, as well as at 7 and 21 days post infection. Five mice were used per time point and asterisks indicate significant differences by Kruskal-Wallis test. CFU were also obtained from spleens (E) and livers (F) of infected mice at 21 days post infection. (G) Survival of infected BALB/c mice (n = 5 per group). Statistical analysis indicated significant differences between all four strains (P<0.05) and more subtle differences between ΔmycP1 + WT MycP1 and ΔmycP1 + MycP1 S332A strains (P<0.13). (H) Hematoxylin/eosin-stained lung sections of mice infected with indicated strains, harvested 56 days post infection. Lungs from mice infected with wild-type and ΔmycP1 + WT MycP1 bacteria showed numerous, consolidated granulomas (arrows) while lungs from mice infected with ΔmycP1 and ΔmycP1 + MycP1 S332A strains showed fewer granulomas and more alveolar space. See Figures S4A and S4B for more images and results of our quantitative histopathology analysis.
As ESX-1 is required for growth in macrophages and mice, we postulated that the enhanced secretion of ESX-1 substrates in the MycP1 S332A strain might alter the course or outcome of a M. tuberculosis macrophage infection (Stanley et al., 2003). We found that ΔmycP1 bacilli are attenuated for intracellular growth as compared to wild-type bacteria, consistent with other ESX-1 mutants such as EccD1::Tn, which also shows an initial drop in bacterial CFU followed by some restoration of bacterial loads, although never to the level of wild-type bacteria (Stanley et al., 2003). However, expression of wild-type or inactive MycP1 in the ΔmycP1 mutant strain restored growth to wild-type levels (Figure 5C). Thus, although innate immune signaling is hyper-activated by the MycP1 S332A strain, its effects do not alter M. tuberculosis replication in macrophages ex vivo.
Virulence of mycP1 mutants in mice
To test the role of MycP1 in pathogenesis in vivo, we infected mice via the aerosol route with wild-type, ΔmycP1, complemented and MycP1 S332A bacteria. Wild-type, complemented and MycP1 S332A bacteria grew normally during the acute stages of infection, leading to a 100-fold increase in colony-forming units (CFU) in mouse lungs within seven days and a 10,000-fold increase within 21 days post infection (Figure 5D). In contrast, ΔmycP1 mutant cells replicated poorly, leading to CFU levels 10-fold lower than the other strains at days seven and 21. Similar phenotypes have been observed with other ESX-1 mutants such as EccD1::Tn and ΔesxA, which are also attenuated for growth at these time points (Stanley et al., 2003). The attenuation of the ΔmycP1 strain was even more pronounced in spleens (Figure 5E) and livers (Figure 5F), in which ΔmycP1 bacterial CFU were 100-fold lower than those from wild-type, complemented and MycP1 S332A bacteria 21 days post infection. Survival curves indicated that all of the mice infected with wild-type M. tuberculosis succumbed to infection by 300 days post infection, while none of the mice infected with ΔmycP1 mutant bacteria succumbed to infection during that time (Figure 5G). Mice infected with the complemented strain also succumbed to infection, though with slightly slower kinetics than those infected with wild-type bacteria, consistent with the partial restoration of ESX-1 secretion observed in secretion assays. Interestingly, mice infected with MycP1 S332A bacteria showed similar survival rates as those infected with ΔmycP1 cells, with all surviving over the course of the experiment with the exception of one that succumbed very early, most likely to non-tuberculous disease. When sections of infected mouse lungs harvested 56 days post-infection were examined, it was apparent that the MycP1 S332A strain elicited a more muted inflammatory response than the wild-type and complemented strains, giving rise to histopathological lesions that resembled those induced by ΔmycP1 cells (Figure 5H and Supplemental Figure S4). Therefore, the MycP1 protein, like other ESX-1 components, is critical during the acute stages of infection and required for growth. While the regulation of ESX-1 secretion by MycP1 protease activity is dispensable for the early growth of the bacterium, it is important for immunopathology and virulence during the chronic stage of infection.
DISCUSSION
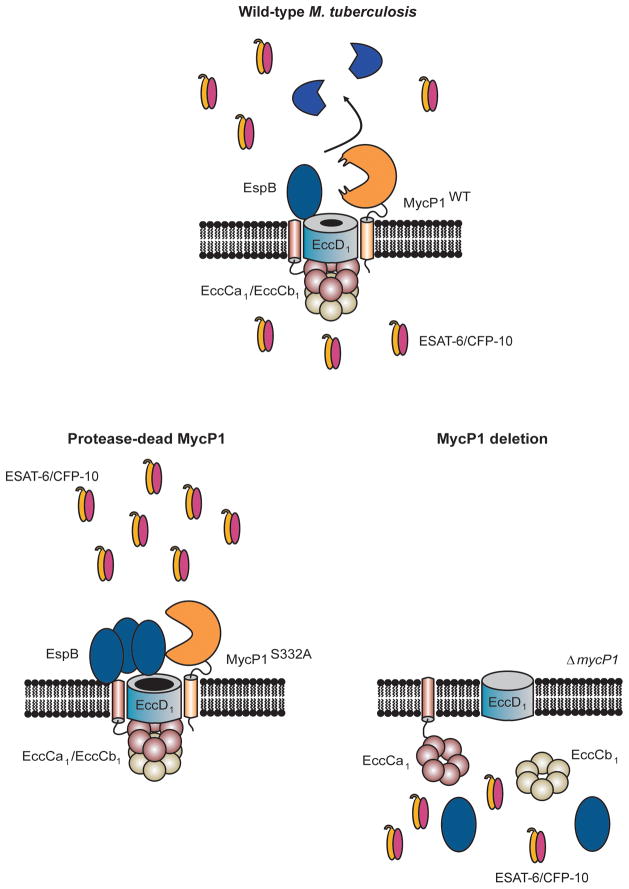
We have discovered that MycP1 is not only an obligatory component of the ESX-1 secretion system, but that this protease plays an unexpected dual role in controlling protein export. In particular, ESX-1 secretion is abolished in the absence of MycP1 but inhibition of MycP1 protease activity leads to enhanced secretion of ESX-1 substrates. How would protease activity modulate ESX-1 secretion? The identification of EspB, which itself if required for ESX-1 secretion, as a direct MycP1 substrate suggests a model in which full-length EspB functions in the periplasmic space to promote secretion but its proteolysis serves to limit secretion of other ESX-1 substrates (Figure 6). Since EspB cleavage products are absent from M. tuberculosis cell lysates, EspB is likely secreted as a full-length protein into the periplasm, where it is proteolyzed by MycP1, thus turning off secretion. This is consistent with our data that in the MycP1 S332A mutant, full-length EspB accumulates and ESX-1 secretion increases. Although its mechanism of action is unknown, full-length EspB could serve as a limiting component of the secretion pore, through which other ESX-1 substrates subsequently pass. Why does removal of MycP1 abolish secretion? We speculate that MycP1 interacts with multiple ESX-1 machine components and thus is required for complex formation, though other possibilities certainly exist. Examples of such positive and negative regulation of a pathway by a single protein are unusual in biology.
Figure 6. Model of MycP1-mediated regulation of the ESX-1 secretion system in M. tuberculosis.
In wild-type M. tuberculosis, MycP1 is part of a complex and proteolyzes EspB in the periplasm. Full-length EspB is an activator of ESX-1 secretion and its cleavage functions to inhibit secretion. In ΔmycP1 + MycP1 S332A cells, full-length EspB accumulates and therefore ESX-1 secretion is hyper-activated. In ΔmycP1 bacteria, secretion is abolished because the secretion complex fails to assemble in the absence of MycP1.
We found that EspB is proteolyzed at at least three sites, two of which (Pro332, Ala392) require MycP1. Both of these sites are consistent with our in vitro profiling studies. The protease responsible for cleaving EspB at the third site has not been identified. However, the M. tuberculosis genome contains four homologues of MycP1, each associated with a duplication of the ESX-1 secretion system, whereas no homologues of EspB exist (Gey van Pittius et al., 2001). One of these MycP1 homologues may proteolyze EspB at the third site. The combination of genetics and profiling has been important to understand the molecular details of MycP1 function.
Although the ESX-1 system is a key virulence determinant of M. tuberculosis, its exact role in virulence has not yet been elucidated. One consequence of ESX-1 secretion is that it stimulates a unique cytosolic surveillance pathway, perhaps utilizing the pore-forming activity of ESAT-6 to allow substrates to escape from the macrophage phagosome into the cytosol (Stanley et al., 2007; Weiden et al., 2000; Giacomini et al., 2001; Lewinsohn et al., 2006; O’Riordan et al., 2002). Although the role of this signal transduction pathway in antibacterial defense is unclear, the gene targets include a number of immuno-modulatory molecules, including Type I IFNs (Weiden et al., 2000; Giacomini et al., 2001). It is therefore suggestive that increased activity of ESX-1 may over-stimulate the cytosolic surveillance response, tipping the balance between bacterium and host control. Indeed, infection of macrophages with the MycP1 S332A mutant led to increased IFN-β production and mice infected with these bacteria displayed an altered inflammatory response.
Given that ESX-1 is under fine transcriptional control by EspR (Raghavan et al., 2008), it is curious that M. tuberculosis has evolved an additional layer of regulation mediated by MycP1. Whereas EspR activity is required for inducing the system, irreversible proteolysis by MycP1 may allow the bacillus to terminate secretion rapidly and completely. Further, the periplasmic location of MycP1 would allow the bacterium to sense and respond to changes in the environment. Although we do not understand the details of the overlap between transcriptional and post-transcriptional control of ESX-1 secretion, it is clear that the regulation mediated by MycP1 is important during infection.
A compelling rationale for why tight control of ESX-1 secretion is important for M. tuberculosis infection is that its key substrates, ESAT-6 and CFP-10, are both essential for virulence as well as highly immunogenic (Brandt et al., 2000; Colangeli et al., 2000; Coler et al., 2001; Dietrich et al., 2006). In particular, ESAT-6 is a highly protective antigen in experimentally infected animals (Pym et al., 2003). Thus it is possible that M. tuberculosis regulates the quantity and/or timing of ESAT-6 export in order to achieve an optimal balance between promoting its own virulence and stimulating the immune system. The inability of ΔmycP1 mutant bacteria to secrete ESAT-6 led to severe attenuation in macrophage and mouse models, consistent with previous reports demonstrating a key role of ESX-1 during the early phase of infection (Stanley et al., 2003). Although M. tuberculosis MycP1 S332A mutant cells grew to similar bacterial loads as wild-type, survival assays and histopathological analysis showed that it is attenuated compared to wild-type M. tuberculosis. Similar phenotypes have been observed in ΔsigC, ΔsigE, ΔsigH and ΔwhiB mutants of M. tuberculosis (Sun et al., 2004; Ando et al., 2003; Kaushal et al., 2002; Steyn et al., 2002). Wild-type bacteria may therefore shut off secretion later during infection to avoid detection by the immune system. The observation that the number of ESAT-6 reactive T cells decreases during chronic M. tuberculosis infection suggests that the bacterium does indeed shield these antigens from the immune system (Lazarevic et al., 2005). The constitutive ESAT-6 secretion caused by inactivation of MycP1 may promote ESAT-6 antigen presentation by antigen-presenting cells during chronic infection, priming T cells to generate a stronger immune response against M. tuberculosis. MycP1 could perhaps be an interesting drug target for latent M. tuberculosis infection, as it is predicted to be outside the cell membrane and hence accessible to small molecules. Perhaps more obviously, if inhibition of MycP1 activity uncovers hidden, potent antigens during latent infection, the incorporation of protease-dead MycP1 alleles into current live attenuated vaccine strains may increase immunogenicity.
EXPERIMENTAL PROCEDURES
A detailed description of all procedures and protocols is available as Supplemental Data.
Bacterial strains, plasmids and culture conditions
M. smegmatis (mc2155) and M. tuberculosis (Erdman) were grown as described (Cox et al., 1999). The M. tuberculosis mycP1 deletion mutant (ΔmycP1) was constructed by replacing the entire mycP1 open reading frame with the hygromycin resistance gene by homologous recombination. The deletion plasmid was delivered into M. tuberculosis using specialized phage, as described previously (Glickman et al., 2000). Deletion of mycP1 was confirmed by Southern blotting.
Protein Preparation and Analysis
For secretion assays M. smegmatis and M. tuberculosis were grown in Sauton’s medium as described (Converse et al., 2005; Stanley et al., 2003; Gao et al., 2003). Proteins were visualized by immunoblotting using antibodies against ESAT-6 (Assay Designs), HA (Covance), EspA, EspB, EspR, KatG, GroEL and Mpt32 (all from Colorado State University; EspA was also the kind gift of S. Fortune, Harvard University). For quantitative Western blots, all incubations of the nitrocellulose membranes were performed in Odyssey blocking reagent (Li-cor Biosciences) and infrared (IR)-dye conjugated secondary antibodies were used. Proteins thus labeled were visualized using the Odyssey infrared imaging system and band density quantitated with the accompanying Odyssey 3.0 software.
Expression and purification of wild-type and protease-dead MycP1
The transmembrane regions and topology of MycP1 were analyzed in silico using the protein prediction programs TMHMM (Sonnhammer et al., 1998) and PSORTb (Gardy et al., 2005). N-terminally maltose binding protein (MBP)-tagged M. smegmatis MycP1ms propeptide and active domain was expressed in E. coli BL21 codon plus cells. MBP-MycP1ms was purified from lysed cells using amylose resin (New England Biolabs), yielding approximately 2 mg of purified protein per liter of culture. MBP and the propeptide were removed by incubating with active MycP1ms and subsequent binding to 40μM Macro-Prep ceramic hydroxyapatite beads (Bio-Rad). Protease-dead M. tuberculosis and M. smegmatis MycP1 were generated by replacing the active-site Ser residue (Ser332 and Ser334 respectively) with Ala by site-directed mutagenesis, using pYO17 (M. tuberculosis) and pSEC82 and pYO1 (M. smegmatis) as templates.
Protease activity assays, substrate specificity profiling and inhibitor assays
MycP1ms activity assays were performed in 20 mM Tris pH 8, 100 mM NaCl, 2 mM CaCl2 and 0.01% Brij-35. Substrate specificity was determined using a library of fluorogenic tetrapeptide substrates, as described earlier (Choe et al., 2006). Individual activity assays were performed similarly on single dipeptide substrates diphenyl Nα-benzoxycarbonyl-Gly-Pro-amino methyl coumarin (Z-GP-AMC), Z-Leu-Arg-AMC (Z-LR-AMC) and Suc-Leu-Tyr-AMC (Suc-LY-AMC) (Bachem). The inhibitor Z-Gly-Pro-phosphonate (Z-GP-(OPh)2) was chemically synthesized as described previously (Gilmore et al., 2006).
Expression, purification and cleavage assay of recombinant EspB
N-terminally MBP-6x Histidine (His) tagged M. tuberculosis EspB was expressed in E. coli BL21 codon plus cells. After cell lysis, MBP-His-EspB was purified using amylose resin (New England Biolabs) followed by Ni-NTA agarose resin (Qiagen), yielding approximately 2 mg of purified protein per liter of culture. The MBP-His tag was removed by incubating with His-tagged AcTEV protease (Invitrogen) and subsequent binding to Ni-NTA agarose resin. EspB was treated with equal quantities of wild-type MycP1ms, protease-dead MycP1ms or wild-type MycP1ms plus Z- Gly-Pro-(OPh)2 in MycP1 activity buffer at room temperature for 10 minutes. Reactions were stopped by addition of the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) and SDS-PAGE sample buffer. Proteins were resolved on 4–20% polyacrylamide gels and visualized by Coomassie staining.
Purification of native EspB, cleavage site identification and characterization of EspB mutant
An M. tuberculosis strain expressing C-terminally His-FLAG-tagged EspB under the control of a Tet-inducible promoter was constructed and cultured in Sauton’s medium as described above. Secreted EspB-His-FLAG was purified from culture supernatants using anti-FLAG M2 affinity gel (Sigma). The exact molecular weight of EspB-His-FLAG was determined by liquid chromatography mass spectrometry of the intact protein at the Vincent Coates Foundation Mass Spectrometry Laboratory (Stanford University) and used to identify the EspB cleavage site.
Macrophage infections
All mice were housed under pathogen-free conditions, and all experiments were conducted using a University of California, San Francisco, Institutional Animal Care and Use Committee-approved protocol. Bone marrow-derived macrophages from C57BL/6 mice were harvested, cultured and infected as described (Stanley et al, 2003). Macrophages were infected with M. tuberculosis strains at a multiplicity of infection (MOI) of 1 for colony-forming unit (CFU) enumeration, and an MOI of 10 for RNA extraction.
Quantitative real-time PCR
Four hours post infection with M. tuberculosis, macrophage RNA was extracted as described (Stanley et al., 2007). One microgram of RNA was reverse-transcribed using the SuperScript III First Strand kit (Invitrogen) with oligo-dT primers. A 1:20 dilution of the cDNA was used for quantitative real-time PCR with SYBR green as label and primers for actin (ACTIN-F and ACTIN-R), interferon-beta (IFN-β) (IFNb-F and IFNb-R) and interferon induced protein with tetratricopeptide repeats 1 (IFIT-1) (IFIT-1F and IFIT-1R).
Mouse infection
BALB/c mice (Jackson Laboratories) were infected by aerosol with 1.5 ×102 colony-forming units as described (Kumar et al., 2007). Lungs were harvested, homogenized and plated for CFU at 0, 7, 21 and 56 days post infection, as described (Cox et al., 1999; Stanley et al., 2003). Spleens and livers were similarly harvested and plated at 21 and 56 days post infection, but not at 0 and 7 days post infection as bacterial loads would be below detection limits. In addition, lungs, spleens and livers collected 56 days after infection were sectioned and stained with hematoxylin/eosin at the Histology Core Laboratory of the Gladstone Institute. Sections were visualized on an upright Nikon Eclipse E600 epifluorescence microscope. For survival experiments, infected mice were sacrificed when they had lost 15% of their maximal body weight. Statistical analysis was performed using the Kruskal-Wallis test for CFU data and the Kaplan-Meier test for survival data, using GraphPad InStat Software.
Supplementary Material
Acknowledgments
We thank M. Tempesta for cloning of pYO36, P. Manzanillo for help with macrophage experiments, and H. Madhani for critical reading of the manuscript. K.C. was supported by an A.P. Giannini Family Foundation fellowship, M.U.S. by an NIAID KAI076632A grant, D.H.G. by National Research Service Award GM069243. This work was supported by National Institutes of Health grants AI63302 and AI51667 (J.S.C.) and GM56531 (C.S.C.). J.S.C. acknowledges the support of the Sandler Family Supporting Foundation and the W.M. Keck Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdallah AM, Gey van Pittius NC, Champion PAD, Cox JS, Luirink J, Vandenbroucke-Grauls CMJE, Appelmelk BJ, Bitter W. Type VII secretion – mycobacteria show the way. Nature Reviews. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- Ando M, Yoshimatsu T, Ko C, Converse PJ, Bishai WR. Deletion of Mycobacterium tuberculosis sigma factor E results in delayed time to death with bacterial persistence in the lungs of aerosol-infected mice. Infect Immun. 2003;71:7170–7172. doi: 10.1128/IAI.71.12.7170-7172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron F, Leduc R, Day R. Subtilase-like pro-protein convertases: from molecular specificity to therapeutic applications. J Mol Endocrinol. 2000;24:1–22. doi: 10.1677/jme.0.0240001. [DOI] [PubMed] [Google Scholar]
- Bitter W, Houben EN, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao LY, Liu J, Gey van Pittius NC, Pym AS, Rubin EJ, Sherman DR, Cole ST, Brosch R. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 2009;5:e1000507. doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt L, Elhay M, Rosenkrands I, Lindblad EB, Andersen P. ESAT-6 subunit vaccination against Mycobacterium tuberculosis. Infect Immun. 2000;68:791–795. doi: 10.1128/iai.68.2.791-795.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Dave JA, Gey van Pittius NC, Stevens L, Ehlers MR, Beyers AD. The mycosins of Mycobacterium tuberculosis H37Rv: a family of subtilisin-like serine proteases. Gene. 2000;254:147–155. doi: 10.1016/s0378-1119(00)00277-8. [DOI] [PubMed] [Google Scholar]
- Choe Y, Leonetti F, Greenbaum DC, Lecaille F, Bogyo M, Bromme D, Ellman JA, Craik CS. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J Biol Chem. 2006;281:12824–12832. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- Colangeli R, Spencer JS, Bifani P, Williams A, Lyashchenko K, Keen MA, Hill PJ, Belisle J, Gennaro ML. MTSA-10, the product of the Rv3874 gene of Mycobacterium tuberculosis, elicits tuberculosis-specific, delayed-type hypersensitivity in guinea pigs. Infect Immun. 2000;68:990–993. doi: 10.1128/iai.68.2.990-993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coler RN, Campos-Neto A, Ovendale P, Day FH, Fling SP, Zhu L, Serbina N, Flynn JL, Reed SG, Alderson MR. Vaccination with the T cell antigen Mtb 8.4 protects against challenge with Mycobacterium tuberculosis. J Immunol. 2001;166:6227–6235. doi: 10.4049/jimmunol.166.10.6227. [DOI] [PubMed] [Google Scholar]
- Converse SE, Cox JS. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J Bacteriol. 2005;187:1238–1245. doi: 10.1128/JB.187.4.1238-1245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave JA, Gey van Pittius NC, Beyers AD, Ehlers MR, Brown GD. Mycosin-1, a subtilisin-like serine protease of Mycobacterium tuberculosis, is cell wall-associated and expressed during infection of macrophages. BMC Microbiol. 2002;2:30–37. doi: 10.1186/1471-2180-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich J, Andersen C, Rappuoli R, Doherty TM, Jensen CG, Andersen P. Mucosal administration of Ag85B-ESAT-6 protects against infection with Mycobacterium tuberculosis and boosts prior bacillus Calmette-Guerin immunity. J Immunol. 2006;177:6353–6360. doi: 10.4049/jimmunol.177.9.6353. [DOI] [PubMed] [Google Scholar]
- Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, Sherman DR, Bloom BR, Rubin EJ. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci USA. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigui W, Bottai D, Majlessi L, Monot M, Josselin E, Brodin P, Garnier T, Gicquel B, Martin C, Leclerc C, Cole ST, Brosch R. Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog. 2008;4:e33. doi: 10.1371/journal.ppat.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Inouye M, Shinde U. Folding pathway mediated by an intramolecular chaperone. The inhibitory and chaperone functions of the subtilisin propeptide are not obligatorily linked. J Biol Chem. 2000;275:16871–16878. doi: 10.1074/jbc.275.22.16871. [DOI] [PubMed] [Google Scholar]
- Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol. 2004;53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- Gey Van Pittius NC, Gamieldien J, Hide W, Brown GD, Siezen RJ, Beyers AD. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C gram-positive bacteria. Genome Biol. 2001;2:0044.1–0044.18. doi: 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini E, Iona E, Ferroni L, Miettinen M, Fattorini L, Orefici G, Julkunen I, Coccia EM. Infection of human macrophages and dendritic cells with Mycobacterium tuberculosis induces a differential cytokine gene expression that modulates T cell response. J Immunol. 2001;166:7033–7041. doi: 10.4049/jimmunol.166.12.7033. [DOI] [PubMed] [Google Scholar]
- Gilmore BF, Carson L, McShane LL, Quinn D, Coulter WA, Walker B. Synthesis, kinetic evaluation, and utilization of a biotinylated dipeptide proline diphenyl phosphonate for the disclosure of dipeptidyl peptidase IV-like serine proteases. Biochem Biophys Res Commun. 2006;347:373–379. doi: 10.1016/j.bbrc.2006.06.113. [DOI] [PubMed] [Google Scholar]
- Glickman M, Cox JS, Jacobs WR., Jr A Novel Mycolic Acid Cyclopropane Synthetase Is Required for Cording, Persistence, and Virulence of Mycobacterium tuberculosis. Mol Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- Gonzalo-Asensio J, Mostowy S, Harders-Westerveen J, Huygen K, Hernández-Pando R, Thole J, Behr M, Gicquel B, Martín C. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One. 2008;3:e349. doi: 10.1371/journal.pone.0003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol. 2004;51:359–70. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Beg QK, Lorenz P. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol. 2002;59:15–32. doi: 10.1007/s00253-002-0975-y. [DOI] [PubMed] [Google Scholar]
- Hingley-Wilson SM, Sambandamurthy VK, Jacobs WR., Jr Survival perspectives from the world's most successful pathogen, Mycobacterium tuberculosis. Nature Immunol. 2003;4:949–955. doi: 10.1038/ni981. [DOI] [PubMed] [Google Scholar]
- Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, Eisenberg D, Russell RG, Derrick SC, Collins FM, Morris SL, King CH, Jacobs WR., Jr The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci USA. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- Kaushal D, Schroeder BG, Tyagi S, Yoshimatsu T, Scott C, Ko C, Carpenter L, Mehrotra J, Manabe YC, Fleischmann RD, Bishai WR. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc Natl Acad Sci USA. 2002;99:8330–8335. doi: 10.1073/pnas.102055799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Schelle MW, Jain M, Lin FL, Petzold CJ, Leavell MD, Leary JA, Cox JS, Bertozzi CR. PapA1 and PapA2 are acyltransferases essential for the biosynthesis of the Mycobacterium tuberculosis virulence factor sulfolipid-1. Proc Natl Acad Sci USA. 2007;104:11221–11226. doi: 10.1073/pnas.0611649104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Nolt D, Flynn JL. Long-term control of Mycobacterium tuberculosis infection is mediated by dynamic immune responses. J Immunol. 2005;175:1107–1117. doi: 10.4049/jimmunol.175.2.1107. [DOI] [PubMed] [Google Scholar]
- Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guérin attenuation. J Infect Dis. 2003;187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn DM, Grotzke JE, Heinzel AS, Zhu L, Ovendale PJ, Johnson M, Alderson MR. Secreted proteins from Mycobacterium tuberculosis gain access to the cytosolic MHC class-I antigen-processing pathway. J Immunol. 2006;177:437–442. doi: 10.4049/jimmunol.177.1.437. [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Raghavan S, Stanley SA, Cox JS. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol Microbiol. 2005;57:1653–1663. doi: 10.1111/j.1365-2958.2005.04800.x. [DOI] [PubMed] [Google Scholar]
- McLaughlin B, Chon JS, MacGurn JA, Carlsson F, Cheng TL, Cox JS, Brown EJ. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog. 2007;3:e105. doi: 10.1371/journal.ppat.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y, Ogawa K, Takeuchi Y, Tomoda S. One Step Synthesis and Structural Confirmation with 1-Pyrroline Trimer. Chem Lett. 1977;1977:693–696. [Google Scholar]
- O’Riordan M, Yi CH, Gonzales R, Lee KD, Portnoy DA. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc Natl Acad Sci USA. 2002;99:13861–13866. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak SK, Basu S, Basu KK, Banerjee A, Pathak S, Bhattacharyya A, Kaisho T, Kundu M, Basu J. Direct extracellular interaction between the early secreted antigen ESAT-6 of Mycobacterium tuberculosis and TLR2 inhibits TLR signaling in macrophages. Nat Immunol. 2007;8:610–618. doi: 10.1038/ni1468. [DOI] [PubMed] [Google Scholar]
- Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, Williams A, Griffiths KE, Marchal G, Leclerc C, Cole ST. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat Med. 2003;9:533–539. doi: 10.1038/nm859. [DOI] [PubMed] [Google Scholar]
- Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature. 2008;454:717–721. doi: 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Guimarães ML, Tempone AJ, Amaral JJ, Nery JA, Gomes Antunes SL, Pessolani MC. Expression analysis of proteases of Mycobacterium leprae in human skin lesions. Microb Pathog. 2007;43:249–254. doi: 10.1016/j.micpath.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Roback P, Beard J, Baumann D, Gille C, Henry K, Krohn S, Wiste H, Voskuil MI, Rainville C, Rutherford R. A predicted operon map for Mycobacterium tuberculosis. Nucleic Acids Res. 2007;35:5085–5095. doi: 10.1093/nar/gkm518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Manoranjan J, Pan M, Bohsali A, Xu J, Liu J, McDonald KL, Szyk A, Laronde-Leblanc N, Gao LY. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from vacuole. Infect Immun. 2008;76:5478–5487. doi: 10.1128/IAI.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer ELL, von Heijne G, Krogh A. Proc of Sixth Int Conf on Intelligent Systems for Molecular Biology. AAAI Press; 1998. A hidden Markov model for predicting transmembrane helices in protein sequences; pp. 175–182. [PubMed] [Google Scholar]
- Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- Steyn AJ, Collins DM, Hondalus MK, Jacobs WR, Jr, Kawakami RP, Bloom BR. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci USA. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Converse PJ, Ko C, Tyagi S, Morrison NE, Bishai WR. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol Microbiol. 2004;52:25–38. doi: 10.1111/j.1365-2958.2003.03958.x. [DOI] [PubMed] [Google Scholar]
- Taki S. Type I interferons and autoimmunity: lessons from the clinic and from IRF-2-deficient mice. Cytokine Growth Factor Rev. 2002;13:379–391. doi: 10.1016/s1359-6101(02)00023-0. [DOI] [PubMed] [Google Scholar]
- Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2004;2:1946–1956. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiden M, Tanaka N, Qiao Y, Zhao BY, Honda Y, Nakata K, Canova A, Levy DE, Rom WN, Pine R. Differentiation of monocytes to macrophages switches the Mycobacterium tuberculosis effect on HIV-1 replication from stimulation to inhibition: modulation of interferon response and CCAAT/enhancer binding protein beta expression. J Immunol. 2000;165:2028–2039. doi: 10.4049/jimmunol.165.4.2028. [DOI] [PubMed] [Google Scholar]
- Xu J, Laine O, Masciocchi M, Manoranjan J, Smith J, Du SJ, Edwards N, Zhu X, Fenselau C, Gao LY. A unique Mycobacterium ESX-1 protein co-secretes with CFP-10/ESAT-6 and is necessary for inhibiting phagosome maturation. Mol Microbiol. 2007;66:787–800. doi: 10.1111/j.1365-2958.2007.05959.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.