Figure 4.
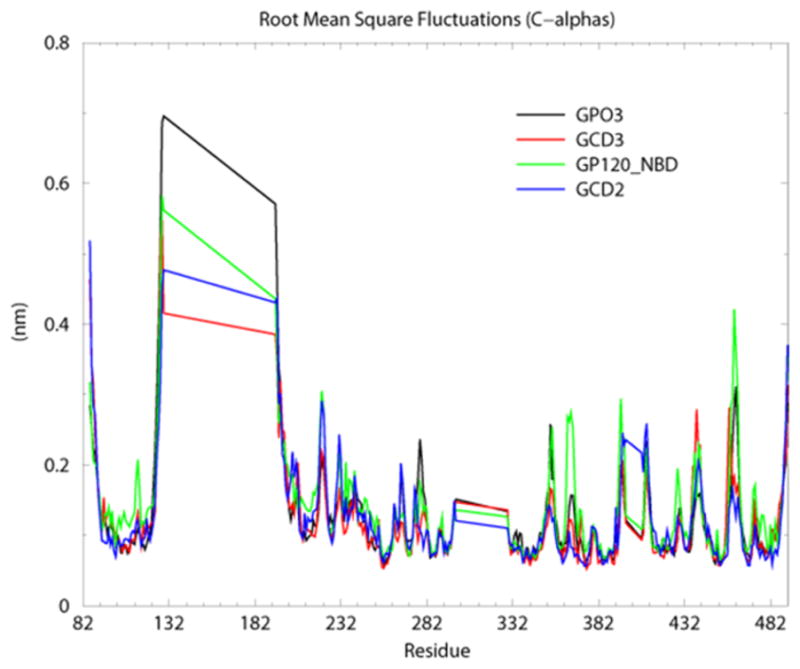
Calculated root mean square fluctuations (RMSFs) from the average, for gp120 residue backbone C-α atoms for the four constructs, GPO3 (black), GCD2 (red), GCD3 (blue) and GPO3_NBD (green). Residue numbering for the core gp120 crystal structures maintains the numbering from the full-length gp120 sequence (residues 82–492). A gap in the numbering exists from residues (127 to 194 and 296 to 330) corresponding to the substitution of the V1/V2 loops with the tripeptide and the deletion the V3 loops. The peaks at residue 127 and 296 correspond to these gaps.
