Abstract
Chromokinesins are microtubule-motor molecules that possess chromatin binding activity and are important for mitotic and meiotic regulation. The chromokinesin-member Kif4A is unique in that it localizes to nucleus during interphase of the cell cycle. Kif4 deletion by gene targeting in mouse embryonic cells was known to associate with DNA damage response. However, its precise role in DNA damage or repair pathway is not clear. Here we report that Kif4A associates with BRCA2 in a biochemical identification and that the interaction is mediated by the Kif4A C-terminal cargo-binding domain and BRCA2 C-terminal conserved region. Upon nucleus-specific laser micro-irradiation, Kif4A was rapidly recruited to sites of DNA damage. Significantly, the depletion of Kif4A from cells by shRNA impaired the ionizing-radiation induced foci (IRIF) formation of Rad51, both quantitatively and qualitatively. In contrast, the IRIF of γ-H2AX or NBS1 was largely intact. Moreover, Kif4A knockdown rendered cells hypersensitive to ionizing radiation in a colonogenic survival assay. We further demonstrated that Kif4A deficiency led to significantly decreased homologous recombination in an I-SceI endonuclease induced in vivo recombination assay. Together, our results suggest a novel role for a chromokinesin family member in the DNA damage response by modulating the BRCA2/Rad51 pathway.
Keywords: chromokinesin, Kif4A, BRCA2, DNA damage response
Introduction
Kinesin and dynein are two types of ATP-dependent mechano-chemical motors that are involved in the transportation of a variety of cytoplasmic cargos along microtubule fibers, the regulation of microtubule stability and the maintenance of centrosome integrity. During mitosis, kinesins are engaged in spindle assembly, microtubule attachment at the kinetochores, chromosome alignment and condensation, and cytokinesis.1–3 Among the kinesin family, chromoskinesins, including kinesin-4 (hKif4A/xKlp1) and kinesin-10 members (hKid/dmNod), stand out as a unique subclass in that they additionally possess a chromatin/DNA binding motif, which confers an additional dimension of cellular activity. Kif4A is a chromokinesin previously shown to have roles in regulating middle-spindle formation by virtue of interacting with the cytokinesis regulator PRC1, and chromosome condensation through the condensin complex.4–6 Kif4A also promotes neuronal survival through suppressing the enzymatic activity of PARP-1, known for the role in detecting and signaling DNA damage to allow strand breaks repair.7 Interestingly, genetic targeting of Kif4 in mouse embryonic stem cells was shown to promote transformation phenotype and tumor formation in a nude mice model.8 Thus, Kif4A is a candidate molecule involved in DNA damage response and/or DNA repair pathways.
Through a biochemical procedure, we identified that Kif4A associates with the breast cancer susceptibility gene product BRCA2. BRCA2 is critical for homologous recombination in higher eukaryotes in part by interacting with the recombinase Rad51. We showed that Kif4A is an early DNA damage response molecule by using laser micro-irradiation system. We provide further evidence that Kif4A plays a novel role in modulating BRCA2/Rad51 DNA damage response, in addition to its known function as a microtubule-based mitotic regulator.
Results
Kif4A associates with BRCA2
BRCA2 is essential for regulating Rad51 recombinase activity. To explore how it coordinates with additional cofactors to modulate Rad51 in various cellular contexts, we have employed a biochemical approach to identify novel BRCA2-associated proteins. We prepared nuclear extract (NEXT) from HeLa cells, which was then subjected to ion exchange chromatography and affinity-pulldown by using BRCA2 antibodies. The resulting BRCA2-containing protein complexes were separated by gradient SDS-PAGE analysis and the candidate BRCA2-associated polypeptides were identified by mass spectrometry. Through this approach, we identified a few proteins that are linked to DNA repair, chromatin remodeling and ubiquitin-pathway (data not shown). We chose to investigate one of the prominent candidates, namely Kif4A in this study because of its potential connection with DNA damages as described in the Introduction.
Kif4A associates with BRCA2 through the cargo domain
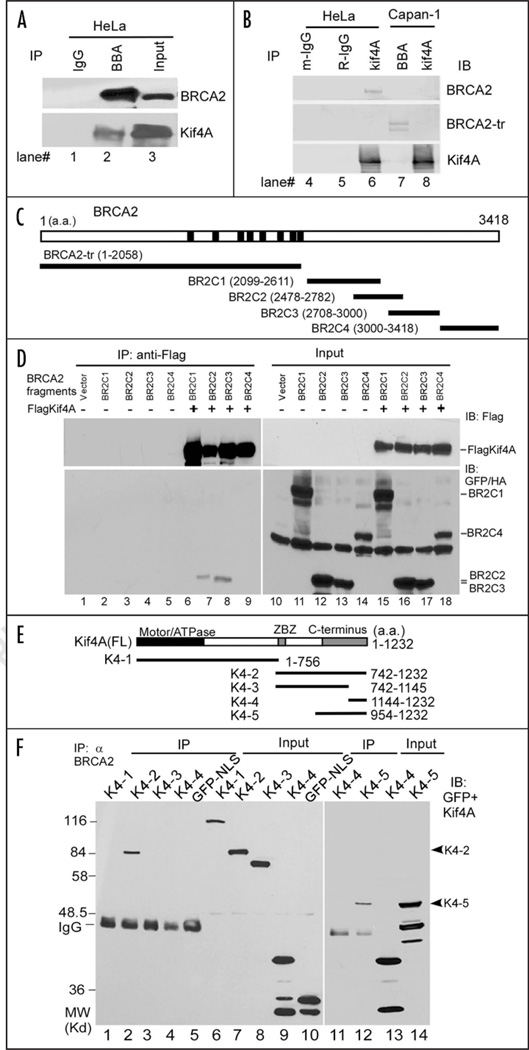
To validate the interaction between Kif4A and BRCA2, we performed a reciprocal co-immunoprecipitation (co-IP) using HeLa cell lysate. As shown in Figure 1A, Kif4A associated with BRCA2 in HeLa cells (BRCA2+/+), but not in Capan-1 cells (BRCA2 deficient) that only express a C-terminally truncated BRCA2 (Fig. 1B). This result suggested that the C-terminal region of BRCA2 might be required for binding to Kif4A. Hence, we generated several GFP-tagged BRCA2 fragments covering the C-terminal conserved portion of BRCA2 to further map the region in BRCA2 involved in binding to Kif4A (Fig. 1C). By co-expressing these BRCA2 mutants (namely BR2C1 through 4) together with Flag-tagged Kif4A in HeLa cells followed by a co-IP assay, we found that BR2C2 and BR2C3 are capable of Kif4A association, suggesting that the overlapping region (a.a. 2708–2782 of BRCA2) might be required for Kif4A/BRCA2 interaction (Fig. 1D). The association was intact when ethidium bromide was added in the cell extract to dissociate chromatin,9 thus excluding the possibility of DNA-mediated nonspecific binding (Fig. 1D). Kif4A and its homologues consist of several distinct modules, including a globular ATPase domain at the N-terminus, a long coiled-coil stalk in the middle, and a globular cargo-docking domain at the C-terminus10,11 (Fig. 1E). To explore the interrelationship between BRCA2 and Kif4A, we therefore determined the domain of Kif4A required for BRCA2 association. We generated a series of GFP-tagged Kif4A mutants for expression in HeLa cells (Fig. 1E) and tested their affinity towards BRCA2 by a similar co-IP assay. The result showed that the C-terminal cargo-binding domain of Kif4A is essential and sufficient for BRCA2 interaction (Fig. 1F). Taken together, these results demonstrate that a portion of the C-terminal conserved region of BRCA2 and the cargo-docking domain of Kif4A are responsible for BRCA2/Kif4A association in vivo. Notably, the overlapping sequence between BR2C2 and BR2C3 corresponds to part of the BRCA2 OB (oligo-binding) fold #1.12 Thus, it is possible that Kif4A could regulate the single-strand DNA binding activity of BRCA2 during the reaction of homologous recombination that intimately involves Rad51.
Figure 1.
Kif4A associates with BRCA2. (A and B) Co-immunoprecipitation and immunoblot assays for endogenous BRCA2 and Kif4A. Cells expressing wild-type (HeLa, lanes 1–6) or mutated BRCA2 (Capan-1, lanes 7 & 8) were immunoprecipitated with control IgG (lanes 1, 4 & 5), BBA antibody (lanes 2 & 7) or α Kif4A (lane 6 & 8), and immunoblotted with either BBA to detect the BRCA2 or α Kif4A to detect Kif4A. BRCA2-tr, truncated BRCA2 (~220 KD); m-IgG, mouse IgG; R-IgG, rabbit IgG. IB, immunoblot. (C) Structural illustration of BRCA2 fragments fused with GFP and a T-antigen derived NLS (nuclear localization signal) for mammalian expression. BRCA2-tr was the truncation mutant expressed in Capan-1 cells. (D) Co-immunoprecipitation followed by immunoblot assays for Flag-Kif4A and BRCA2 fragments ectopically co-expressed in HeLa cells. (E) Structural illustration of full length Kif4A and the GFP-tagged Kif4A mutants. ZBZ (amino acids 746–842), leucine zipper/basic/leucine zipper domain. K4-1 is additionally tagged with a NLS sequence since it cannot localize to nucleus by itself. (F) Co-immunoprecipitation assay for BRCA2 and the Kif4A mutants. Kif4A mutants and GFP-NLS were expressed in HeLa cells respectively and the cell extract was prepared for IP. Results were by immunoblot (IB).
Kif4A localizes to DNA damage sites upon laser miro-irradiation
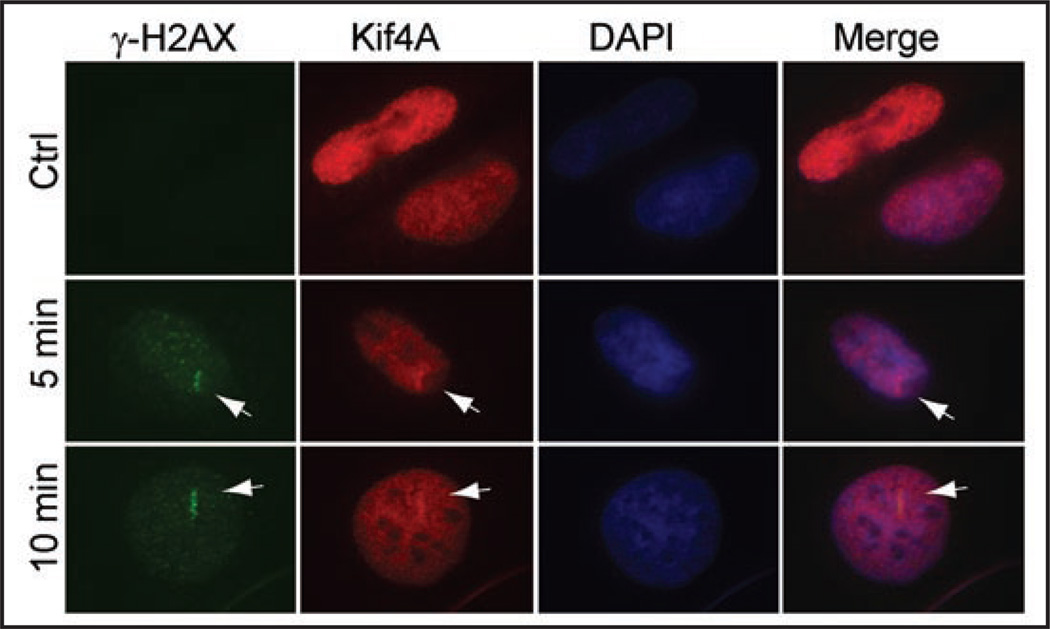
BRCA2 is proposed to play critical roles in DNA damage response and double strand breaks (DSBs) via the homologous recombination (HR) pathway.13,14 To test the potential role of Kif4A during DNA damage response and/or repair, we next sought to visualize the potential Kif4A nuclear focus formation induced by DNA damage treatment. Nonetheless, we found that conventional ionizing radiation or genotoxic chemical (adriamycin or mitomycin) challenges failed to elicit Kif4A nuclear foci formation (data not shown). That a DSB repair factor (DNA-PK, e.g.,) does not focus at DNA damage sites, indeed, has been well documented previously.15 It was ascribed to the low level of molecules recruited to the damage sites. To circumvent this, laser micro-irradiation has been previously adopted to generate more concentrated DSBs, thus permitting focus detection by fluorescent microscopy.16 We used a multiphoton laser irradiation system (Ti:Sapphire laser, 800 nm) similar to the previous reports,17,18 which is superior to other single-photon based systems because of reduced damages to non-focal area along the light path. Interestingly, upon laser irradiation, Kif4A rapidly became enriched at the radiated nuclear site (a “bar” region) in less than 5 minutes and colocalized with the DSB responsive maker γ-H2AX (Fig. 2). Kif4A was retained in the irradiated bar area for up to 1–2 hours and then dissolve, suggesting a role in early stages of DNA damage response and/or repair (data not shown). Together, the results suggested that Kif4A is an early responsive molecule during DNA DSB damage.
Figure 2.
Kif4A co-localizes with γ-H2AX upon laser micro-irradiation in HeLa cells. Cells were miro-irradiated within a bar area of nucleus as described in the Methods section, fixed at various time points and processed for immunofluorescent staining to detect Kif4A and γ-H2AX (an early responsive DNA double strand break marker). DAPI staining shows the DNA/nucleus. Scale bar, 10 µm.
Depletion of Kif4A leads to defective Rad51 IRIF formation
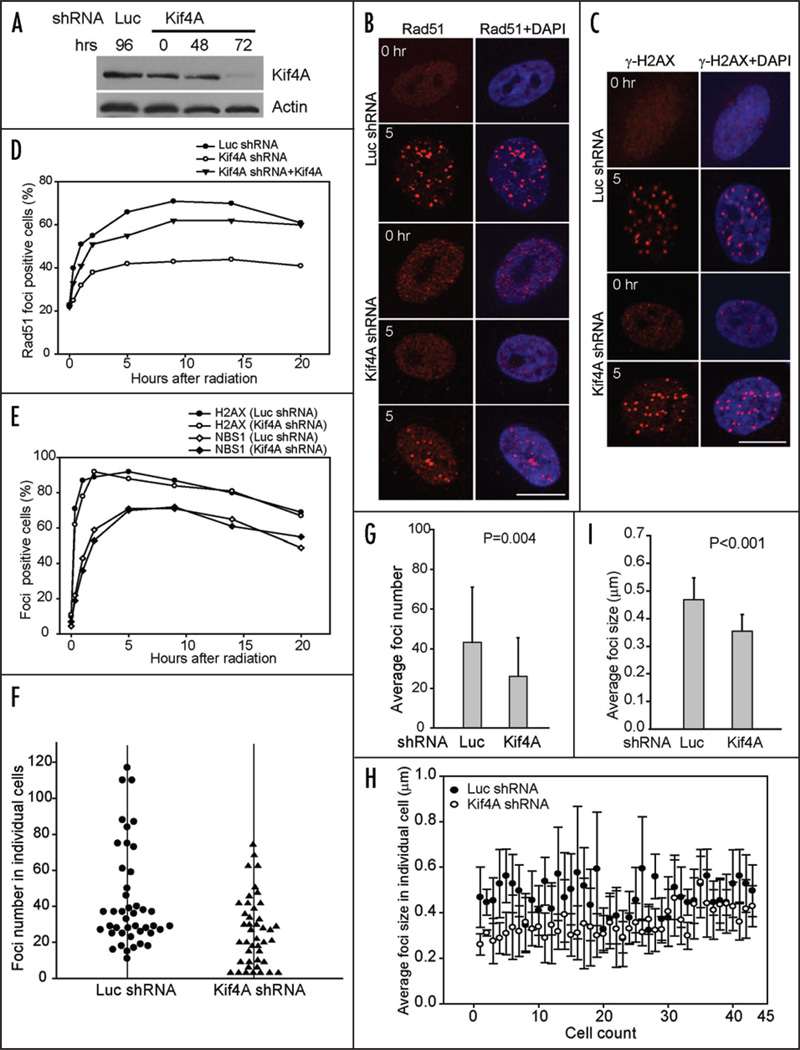
Biochemical studies have indicated that BRCA2 functions to promote Rad51-ssDNA nucleofilament formation and Rad51-mediated recombination, which requires both the BRC repeats and the OB folds at the C-terminal region of BRCA2.12,19–22 In cells, BRCA2 is required for the nuclear foci formation of Rad51 upon ionizing radiation.23,24 Since Kif4A binds to the OB motif 1 of BRCA2, we speculate that Kif4A might regulate the action of BRCA2 in controlling Rad51 and consequently facilitate the repair process. Alternatively, Kif4A might help establish proper chromatin structure to allow the efficient local processing of chromatin lesions. Because the cell biological measurement of Rad51 IRIF is much more straightforward than BRCA2, we thus chose to test whether Kif4A affects Rad51 IRIF formation in a dynamic and quantitative manner. To test this possibility, we employed transcription-based RNAi (shRNA) to knockdown Kif4A in HeLa cells. As shown in Figure 3A, Kif4A was efficiently depleted 72 hours post shRNA transfection (85–90% protein reduction) (Fig. 3A). The cell cycle profile was not apparently changed at this time point (Fig. 5D), thus permitting us to examine the direct effect of Kif4A depletion on Rad51 IRIF, the formation of which occurs preferentially in S and G2 phases when sister chromatids are available.
Figure 3.
Kif4A is required for efficient Rad51 ionizing radiation induced foci formation. (A) Downregulation of Kif4A by short-hairpin (sh) RNA. HeLa cells expressing luciferase or Kif4A shRNA were harvested at indicated time points and used for Western blot. P84, loading control. (B and C) HeLa cells co-expressing luciferase or Kif4A shRNA were subjected to ionizing radiation (4 gray) 72 hours after RNA interference and then processed for Rad51 or γ-H2AX immunostaining. Scale bar, 10 µm. (D) Kinetics of Rad51 IRIF upon luciferase or Kif4A shRNA treatment (with or without RNAi-resistant Kif4A cDNA). Cells are exposed to 4 gray IR at 72-hour post RNAi expression, then fixed at various time points and processed for staining to detect Rad51 nuclear foci. Cells with at least 3 nuclear foci (diameter ≥ 0.2 µm) were considered foci-positive. (E) Kinetics of γ-H2AX and NBS1 IRIF. Cells were identical as in (D). (F and G) The nuclear foci in foci-positive cells were enumerated in individual cells. Each dot (Luciferase control) or triangle (Kif4A shRNA) represents a single cell in the histogram to show the distribution of foci numbers, the average of each is shown in (G). (H and I) The size (diameter) of individual nuclear foci was measured by ImageJ software in each cell. In the histogram, each solid dot (Luciferase control) or open circle (Kif4A shRNA) represents the mean size of all nulcear foci in a given cell, the average of which is shown in (I). The t-test was performed to obtain p value.
Figure 5.
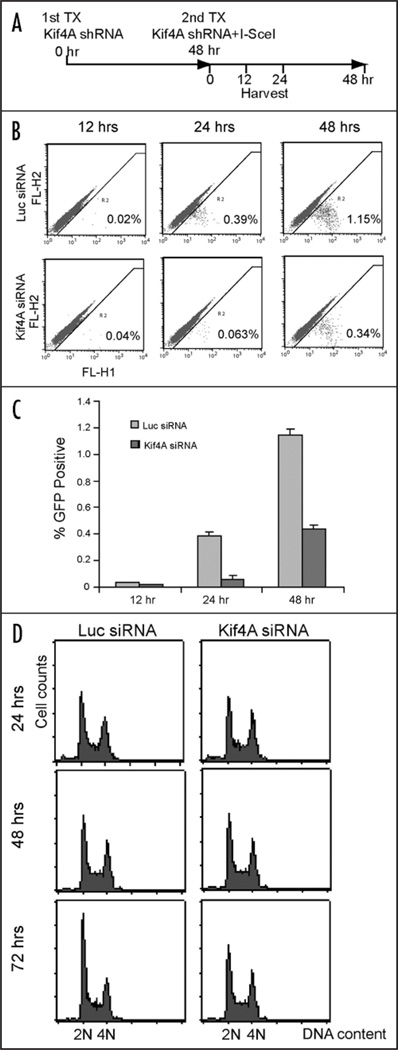
Kif4A depletion led to reduced homologous recombination in cells. (A) Time scheme for the transfection of Kif4A shRNA and I-SceI expression plasmids. (B) Percentage of GFP positive population detected by FACS analysis with samples collected at 12, 24 or 48 hours after 2nd Kif4A shRNA/I-SceI expression. The results were showed separately in a histogram (C). (D) cell cycle profiles of cells harvested at 24, 48 or 72 hours after Kif4A shRNA transfection (with I-SceI co-transfected at 48 hrs) as in (A).
We examined the Rad51 foci formation before and after IR treatment. In untreated cells, ~22% of the cells contain Rad51 foci that are S-phase specific and IR independent.25,26 A comparable percentage of foci-positive cells were detected in luciferase control or Kif4A shRNA targeted cells in the absence of IR treatment, suggesting that S phase specific Rad51 foci were not affected by Kif4A depletion (Fig. 3B and C). However, when cells were treated with IR, Rad51 foci were rapidly and efficiently induced in control treated cells, where a maximum of 71% foci-positive cells were readily observed 9 hours post radiation (Fig. 3D). The foci-positive populations then decreased gradually. In contrast, Rad51 IRIF were markedly reduced, though not abolished, in Kif4A shRNA targeted cells, where only 43% cells were foci positive at similar time point (9 hrs) (Fig. 3D). The ratio of foci positive cells remained, however, unchanged at later time points, suggesting continuous presence of unrepaired DNA lesions. Moreover, ectopic expression of RNAi-resistant Kif4A with wild type coding capacity significantly restored the normal Rad51 IRIF formation kinetics (Fig. 3E). In contrast, the kinetics of IR-induced foci assembly of γ-H2AX and NBS1 were not influenced by Kif4A depletion (Fig. 3E). Taken together, these results suggest that Kif4A is specifically required for efficient and proper Rad51 foci formation upon ionizing radiation. Thus, Kif4A is a likely upstream regulator of Rad51, similar to and most likely in cooperation with BRCA2.
We further examined the average nuclear foci number and size in individual cells within the IRIF-positive population. We found that the remaining foci-positive cells among the Kif4A defective population generally showed a reduced foci number and smaller foci size in comparison to control cells (Fig. 3F–I). The mean foci number is 43 and 26 for luciferase control and Kif4A shRNA targeted population, respectively (p = 0.004; N = 43 cells for each group) (Fig. 3F and G), whereas the averaged focus size (diameter) is 0.47 µm for the control group and 0.35 µm for Kif4A-depleted group (p < 0.001; N = 43 cells for each group) (Fig. 3H and I). Therefore, Kif4A downregulation led to attenuated Rad51 foci formation quantitatively and qualitatively.
Knockdown of Kif4A renders cells hypersensitive to ionizing radiation
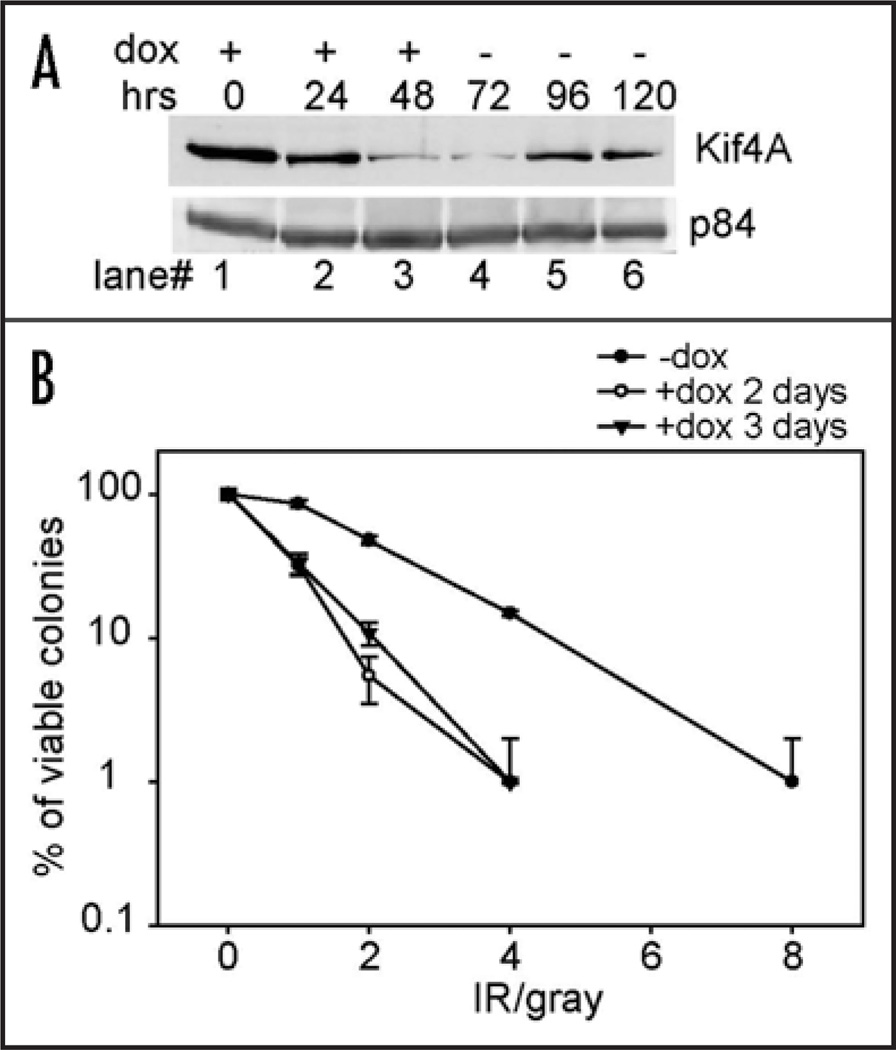
Since BRCA2 deficient cells are hypersensitive to ionizing radiation and Kif4A is required for efficient Rad51 IRIF formation, we asked whether Kif4A depletion might lead to a similar phenotype. To test this, we employed a colonogenic survival assay by using two different Kif4A shRNA to deplete endogenous Kif4A via transfection. We found that Kif4A-deficient cells exhibited a markedly reduced viability of colonies upon ionizing radiation of varying dosages (data not shown). Since the long-term depletion of Kif4A may induce mitotic defects and introduce experimental biases, we additionally established a tet-on inducible system for stable and yet transient expression of Kif4A shRNA in HeLa cells (designated as HeLa-tetK4Ai). In HeLa-tetK4Ai cells, Kif4A was downregulated by ~80% at 48 or 72 hours post Kif4A shRNA induction by doxy-cycline (Fig. 4A). The protein level recovered to 40–50% of the normal level at 96-hr time point when doxycycline was withdraw at 72-hour (Fig. 4A). No significant cell death was observed using this procedure, suggesting that mitotic catastrophe was minimal (data not shown). Thus, there was a 24~48 hour period when the Kif4A level was fairly low, allowing us to conveniently examine the cellular viability upon DNA damage treatment without the complication of long-term or excessive Kif4A shRNA exposure. By following a similar colony formation assaying procedure, we then treated the cells with various doses of IR at 48 or 72 hrs post Kif4A shRNA induction. The cells were then replated in doxycycline-free medium and subjected to a colonogenic survival assay. As shown in Figure 4B, HeLa-tetK4Ai cells transiently depleted of Kif4A were increasingly sensitive to ionizing radiation compared to control. These results are consistent with the observation that Rad51 IRIF formation was impaired in Kif4A-depleted cells and further supported a critical cellular role for Kif4A during ionizing radiation induced DNA damage response.
Figure 4.
Kif4A depletion renders cell hypersensitive to ionizing radiation. (A) Doxycycline (dox) was added into HeLa-tetK4Ai cell culture (see materials and methods) to induce Kif4A shRNA expression and then withdrawn 72 hours later to allow Kif4A expression recovery. Western blot showed Kif4A level changes during the course. (B) HeLa-tetK4Ai cells were exposed to Kif4A shRNA induced transiently for 48 or 72 hrs before the treatment of ionizing radiation and then replated for colony formation in doxycycline-free medium. Viable colonies were scored from triplicate experiments and the compiled survival curves were presented in logarithmic scale. Colonogenic survival assay was performed as described as previously.32
Kif4A depletion resulted in reduced homologous recombination in cells
Since BRCA2/Rad51 are directly involved in DNA DSB repair processes, it is possible that Kif4A plays a similar role in association with BRCA2/Rad51. To test this hypothesis, we adopted an in vivo recombination assay based on restoring the expression of a fluorescent GFP from two homologous yet non-functional GFP sequences via homologous recombination in cells. This HR system has been successfully used previously for in vivo HR assays.27 First, we established a HeLa subline (HeLa-pDR-GFP) that stably incorporates the GFP HR substrate sequence in the genome (HR sequence copy number not determined). Then the cells were transfected with Kif4A shRNA twice (time point 0 and 48 hours respectively). At the second transfection of Kif4A shRNA (48 hr point), I-SceI expression plasmids were con-transfected. At various time points (12, 24 and 48 hours, etc) post I-SceI transfection, cells were harvested for GFP detection by FACS analysis. Notably, at 12 hour point, both control and Kif4A shRNA targeted cells showed minimal level of GFP signal presumably because of lack of HR events. However, at time points 24 and 48 hours, we detected significant reduction of GFP-positive populations in Kif4A shRNA treated cells (6.2 and 3.4 fold difference respectively). Because HR occurs preferentially in late S and G2 phase cells, we next test whether the above described HR reduction is attributed to cell cycle change or not. By FACS analysis, we found that the cell cycle distribution is not apparently changed in kif4A or control treated cells at time point 24, 48 or even 72 hours, suggesting that reduction of HR activity in Kif4A depleted cells is due to loss of Kif4A but not indirect effect of cell cycle changes (Fig. 5D). Thus, our results indicated that Kif4A plays a role in the homologous recombination pathway, presumably in part through modulating the BRCA2/Rad51 pathway.
Discussion
In this study, we uncovered a functional and physical link between BRCA2 and the chromokinesin member Kif4A. Our work presents a novel nuclear role for a conventional chromokinesin. BRCA2 plays a prominent role in the DNA damage response and Rad51-mediated DNA DSB repair via homologous recombination. Our results support a direct or indirect role of Kif4A in DNA damage response, consistent with a previous report.8 Kif4A specifically regulates the Rad51 pathway but not the γ-H2AX and NBS1. It is possible that Kif4A regulates BRCA2 during certain steps of recombination in a transient and conditional manner. Thus, the cellular deficiency of Kif4A might in part mimic the inactivation of BRCA2 and manifest itself through the deficient Rad51 IRIF formation and cellular hypersensitivity to IR treatment. How is Kif4A involved in the DNA damage response/repair processes? Given the relatively large size of BRCA2 and Kif4A, precedent problems with proteolytic degradation during protein preparation has precluded direct biochemical test in vitro. At present, we surmise that there are several possibilities: (1) Kif4A regulates or competes against the ssDNA binding activity of BRCA2 since Kif4A binds to the OB1 motif of BRCA2. Remarkably, the OB1 motif harbors several breast cancer-associated point mutations. Whether these mutations are interrelated to Kif4A function is not clear at present; (2) Kif4A binds to and processes structured DNAs that arise as recombination intermediates, together with BRCA2 and/or Rad51, and possibly other cofactors; (3) Kif4A may act as a DNA binding factor to help establish a proper local chromatin structure surrounding the DNA lesions possibly in an ATP-dependent manner, and therefore allows efficient repair to occur. On this note, many non-histone chromosome components are known to contain ATP-binding domains. Kif4A is probably a true chromosome component since its Xenopous homologue xKlp1 is more resistant to salt extraction than the bona fide chromosome component Topoisomerase II.11 Clearly, a systematic biochemical study is warranted in order to distinguish these different possibilities.
The nuclear function of Kif4A has an important implication since at least one additional chromokinesin, Kid, is also localized to the nucleus during interphase. We propose that chromokinesins possess unique biochemical activities towards the interphase chromatin, which are required for the maintenance of genomic stability. Careful dissection of the underlying mechanisms in human cells might provide novel insights into the etiology of tumorigenesis.
Materials and methods
Cloning and antibodies
Kif4A fragments were N-terminally tagged with GFP in the pEGFP-C system (Clontech, CA). One of the mutants (K4-1) was additionally tagged with a nuclear localization signal (NLS) derived from SV-40 large T-antigen. The BRCA2 mammalian expression constructs were engineered in a pEGFPC2-NLS vector to allow nuclear distribution. Flag-tagged Kif4A was engineered in the p3XFlag-CMV-10 vector (Sigma). Antibodies used for detecting BRCA2 (BBA: purified rabbit polyclonal antibody; p400: mouse monoclonal) and Kif4A have been described previously.28,29 Rabbit polyclonal anti-Rad51 and γ-H2AX (Calbiochem, CA) were purchased commercially. Rabbit polyclonal anti-NBS1, mouse monoclonal anti-Rad51 and p84 antibodies were purchased from Genetex (San Antonio, TX).
Cell culture and immunoprecipitation assay
HeLa (human cervical adenoma) and U2-OS (human osteosarcoma) cells were cultured in regular Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, 2 mM L-glutamine, and penicillin/streptomycin (DMEM) (Invitrogen, CA). Capan-1 (human pancreatic cancer) cells were cultured in DMEM/F12 medium supplemented with 10% FBS. For immunoprecipitation assay, cells were lysed in Lysis-250 buffer (50 mM Tris-HCl pH 7.5, 250 mM NaCl, 0.3% NP-40, and 10 mM NaF) supplemented with protease inhibitors. Cell extract was incubated with ethidium bromide (150 µg/ml) for 30 min on ice followed by centrifugation to remove debris. The resulted supernatant was further clarified by incubation with protein A-Sepharose beads (Amersham Biosciences) at 4°C for 30 min. The clarified extract was then used for immunoprecipitation by primary antibodies and subsequent binding by protein A-Sepharose beads. Finally, the beads were washed in ice-cold PBS buffer, and the precipitates were dissolved in SDS sample buffer for SDS-PAGE separation.
Immunofluorescent staining and microscopy
Immunofluorescent study was performed essentially as described previously.30 Briefly, cells grown on cover slips were washed with PBS and fixed for 20 minutes in 3.2% formaldehyde plus 0.1% Triton X-100. Cells were permeated by 0.05% Saponin or 0.5% Triton X-100 at room temperature for 20 minutes and blocked with diluted goat serum (10% in PBS) for 30 minutes. Cells were then incubated with primary antibodies at RT for 1–3 ours. For secondary antibody labeling, FITC or Texas Red-conjugated secondary antibodies (Southern Biotechnology Associates; Birmingham, AL) were used for 45-minute incubation. Cells were then washed extensively in PBS plus 0.5% NP-40, further stained with DAPI and mounted in ProLong Gold anti-fade (Molecular Probes, OR). Immunofluorescent microscopy was performed with a Zeiss Axioskop plus microscope equipped with a deconvolution module. Images captured were further processed with Adobe Photoshop. The foci number and size (diameter no less than 0.2 µm) was measured by using the Image J software for histogram presentation and statistical analysis by SimgaPlot/SigmaStat.
RNA interference
Short hairpin (sh) RNAs were expressed in the pSuperior (Oligoengine, Seattle, WA) or the pBS/U6 vector system.31 Two sequences were designed to target Kif4A (1): 5' GGG AGG TTG CAG ATA AGC GGA A 3'; (2): 5' GGG ACT CAC TGA GAA GAC TGT T 3'. To achieve maximal expression of shRNA, four expression-cassettes, each of which contains a H1 or U6 promoter, the RNAi targeting sequence, and an RNA pol III stop site (TTTTT), were inserted tandemly into the pcDNA vector to create pcDNA-4Kif4Ai. The knockdown of Kif4A was achieved by transfection of pcDNA-4Kif4Ai with FUGENE 6 (Roche Scientific). To establish the inducible shRNA-expressing cell line, we first established a HeLa sub-line (named HeLa-tet) stably expressing the Tet repressor. Kif4A shRNA-targeting sequence was cloned into the pSuperior vector to generate pSuperior-4Kif4Ai, which was then stably integrated into HeLa-tet cells. The obtained inducible line is designated as HeLa-tetK4Ai. To induce the constitutive expression of Kif4A shRNA, doxycycline was added into the medium (2 µg/ml). A shRNA targeting firefly luciferase was used as control: 5'-AAG ATT CAA AGT GCG CTG CTG-3'.
Irradiation treatment and colonogenic survival assays
For laser micro-irradiation assay, cells were grown on grid glass bottom dishes (MakTek), and irradiated with the Sapphire multiphoton laser (Chameleon Ultra model; Coherent Inc., Santa Clara, CA) integrated with a Zeiss LSM 510meta confocal system at 800 nm (140 fs pulse width, 80 MHz, 7% output) for 3 seconds. Cells were fixed in 3% PFA and processed for immunofluorescent staining. For IRIF formation analysis, pcDNA-4Kif4Ai was expressed in HeLa cells by transfection with FUGENE 6 (Roche). The IRIF formation kinetics was obtained by analyzing the time-dependent foci formation profile of cells with or without ionizing radiation (4 gray). To generate RNAi resistant Kif4A versions (pcDNA-Kif4A) for mammalian expression, three silent point mutations were introduced by site-directed mutagenesis into the wobble positions of the RNAi targeting region of Kif4A. For complementation assay, RNAi-resistant Kif4A expression plasmid was co-expressed with pcDNA-4Kif4Ai. The colony formation assay is performed similarly as described previously.32
Homologous recombination assay
To analyze the chromosomal homologous recombination, the GFP HR substrate plasmid (pDR-GFP, courtesy of Dr. Andrew Pierce)) was transfected and stably integrated into HeLa cells via selection against puromycin, which was further confirmed by FACS to examine the GFP-positive cells upon I-SceI ectopic expression.27 For HR assays, HeLa-pDR-GFP stable cells were co-transfected (FuGENE 6) with I-SceI expression construct pCBASce and siRNAs against luciferase or Kif4A. At 12, 24 and 48 hours, cells were harvested for flow cytometric analysis. Two-color fluorescence analysis revealed the percentage of GFP-positive cells was scored out of a total of 50,000 viable events.
Acknowledgements
We thank Randy Wei, Chi-Fen Chen and Ling Wang for technical assistance, W.H. Lee for his support. We thank Dr. Andrew Pierce for the HR substrate plasmids. GW is supported by a postdoctoral award from Susan G. Komen Foundation; LZ by a DOD postdoctoral fellowship, LK by a NIH cancer research training grant to UCI cancer research program, and EG by a DOD predoctoral fellowship. This study was in part supported by the laser microbeam and medical program (LAMMP) at UCI.
Abbreviations
- IRIF
ionizing radiation induced focus
- DSB
double-strand break
- dsDNA
double stranded DNA
- ssDNA
single-stranded DNA
- HR
homologous recombination
- NHEJ
non-homologous end joining
- GFP
green fluorescence protein
- IP
immunoprecipitation
- RNAi
RNA interference
- FACS
fluorescence activated cell sorting
References
- 1.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 2.Heald R. Motor function in the mitotic spindle. Cell. 2000;102:399–402. doi: 10.1016/s0092-8674(00)00044-1. [DOI] [PubMed] [Google Scholar]
- 3.Mazumdar M, Misteli T. Chromokinesins: multitalented players in mitosis. Trends Cell Biol. 2005;15:349–355. doi: 10.1016/j.tcb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. Embo J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazumdar M, Sundareshan S, Misteli T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J Cell Biol. 2004;166:613–620. doi: 10.1083/jcb.200401142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu C, Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci USA. 2005;102:343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Midorikawa R, Takei Y, Hirokawa N. KIF4 motor regulates activity-dependent neuronal survival by suppressing PARP-1 enzymatic activity. Cell. 2006;125:371–383. doi: 10.1016/j.cell.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 8.Mazumdar M, Lee JH, Sengupta K, Ried T, Rane S, Misteli T. Tumor formation via loss of a molecular motor protein. Curr Biol. 2006;16:1559–1564. doi: 10.1016/j.cub.2006.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang SZ, Adler R. Chromokinesin: a DNA-binding, kinesin-like nuclear protein. J Cell Biol. 1995;128:761–768. doi: 10.1083/jcb.128.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C. Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell. 1995;81:117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- 12.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 13.Shivji MK, Venkitaraman AR. DNA recombination, chromosomal stability and carcinogenesis: insights into the role of BRCA2. DNA Repair (Amst) 2004;3:835–843. doi: 10.1016/j.dnarep.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 15.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Lukas C, Bartek J, Lukas J. Imaging of protein movement induced by chromosomal breakage: tiny ‘local’ lesions pose great ‘global’ challenges. Chromosoma. 2005;114:146–154. doi: 10.1007/s00412-005-0011-y. [DOI] [PubMed] [Google Scholar]
- 17.Meldrum RA, Botchway SW, Wharton CW, Hirst GJ. Nanoscale spatial induction of ultraviolet photoproducts in cellular DNA by three-photon near-infrared absorption. EMBO Rep. 2003;4:1144–1149. doi: 10.1038/sj.embor.7400028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, Houtsmuller AB, van Gent DC. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 20.Petalcorin MI, Sandall J, Wigley DB, Boulton SJ. CeBRC-2 stimulates D-loop formation by RAD-51 and promotes DNA single-strand annealing. J Mol Biol. 2006;361:231–242. doi: 10.1016/j.jmb.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 21.San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 23.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 24.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 25.Tashiro S, Kotomura N, Shinohara A, Tanaka K, Ueda K, Kamada N. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene. 1996;12:2165–2170. [PubMed] [Google Scholar]
- 26.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 27.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YM, Lee S, Lee E, Shin H, Hahn H, Choi W, Kim W. Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochem J. 2001;360:549–556. doi: 10.1042/0264-6021:3600549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu G, Lee WH, Chen PL. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. J Biol Chem. 2000;275:30618–30622. doi: 10.1074/jbc.C000390200. [DOI] [PubMed] [Google Scholar]
- 31.Sui G, Soohoo C, Affar EB, Gay F, Shi Y, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CF, Chen PL, Zhong Q, Sharp ZD. W.H.L. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]