Abstract
Here we describe a straightforward, efficient, and reliable way to clone an insert of choice into a plasmid of choice without restriction endonucleases or T4 DNA ligase. Chimeric primers containing plasmid sequence at the 5′ ends and insert sequence at the 3′ ends were used to PCR-amplify insertion sequences of various sizes, namely the genes for GFP (gfp), β-D-glucuronidase (gusA), and β-galactosidase (lacZ), as well as the entire luxABCDE operon. These inserts were employed as mega-primers in a second PCR with a circular plasmid template. The original plasmid templates were then destroyed in restriction digests with DpnI, and the overlap extension PCR products were used to transform competent Escherichia coli cells. Phusion DNA polymerase was used for the amplification and fusion reactions, so both reactions were easy to monitor and optimize.
Keywords: overlap extension PCR cloning, recombinant vector, Phusion, restriction enzyme ligation independent
Numerous alternative approaches to PCR cloning (1) have been developed, including TA cloning (2), ligation independent cloning (LIC) (3–4), recombinase-dependent cloning (5–7), and PCR-mediated cloning (8–10). The practical utility of any cloning method is predicated upon its reliability, rather than its convenience, price, or efficiency under optimum conditions. The methods that are easiest to monitor and optimize ultimately prove the most reliable. TA cloning and LIC require end modifications that cannot be monitored by gel electrophoresis. Recombinases are generally sold as proprietary components of cloning kits, so few consumers optimize the in vitro recombination reactions. Overlap extension PCR cloning, described here, is not the first form of PCR-mediated cloning (8–10). It is, however, relatively straightforward, efficient, and reliable.
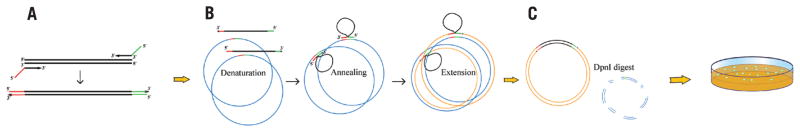
The first of two PCRs (Figure 1A) creates a linear insert with plasmid sequences at both ends (see Supplementary Materials for methods and instructions for primer design). These extensions subsequently allow the strands of the PCR product (Figure 1A) to act as a pair of oversized primers on the vector fragment (Figure 1B). After denaturation and annealing, the insert strands hybridize to the vector and extend to form new double-stranded plasmid. Phusion DNA polymerase (Cat. no. F-530; New England BioLabs, Ipswich, MA, USA), crucial for performance of the technique, does not possess strand displacement activity. Therefore, the final product of the reaction is a double-stranded fusion plasmid with two nicks (one on each strand). This relaxed double-stranded plasmid is then transformed into competent Escherichia coli cells, which seal the nicks with DNA repair enzymes (Figure 1C). We employ the same thermostable polymerase for both PCRs, so inexperienced users can clone efficiently without mastering the idiosyncrasies of multiple restriction enzymes, polymerases, glycosylases, recombinases, and ligases.
Figure 1. An outline of the overlap extension PCR cloning.
(A) First, the insert is PCR-amplified with the chimeric primers so that the final PCR product has overlapping regions with the vector. (B) Then, vector and insert are mixed, denatured and annealed; the hybridized insert then is extended by Phusion DNA polymerase using vector as a template until polymerase reaches 5′ end of the insert. After several PCR cycles, the new plasmid with two nicks (one on each strand) gets accumulated as a product. (C) The new plasmid can be transformed into E. coli after the parental plasmid is destroyed by DpnI digest.
We first used gfp for proof-of-principle experiments. The gfp gene was PCR-amplified (Figure 1A) with the chimeric primers (5′ ends complementary to the pQE30 plasmid; 3′-end complementary to gfp). Overlap extension PCR (Figure 1B) was performed with five different DNA polymerases (Supplementary Table S1). High concentrations of the insert and relatively low annealing temperatures in the reaction (5–10°C below the calculated melting temperature of the primer/plasmid complex) are important for efficient overlap extension. Some of the PCR products correspond to the relaxed form of the desirable vector, as revealed by agarose gel analysis (Figure 2A). The original pQE30 vector was destroyed in reactions with DpnI restriction endonuclease (Figure 1C). A small aliquot from each reaction was used to transform E. coli cells.
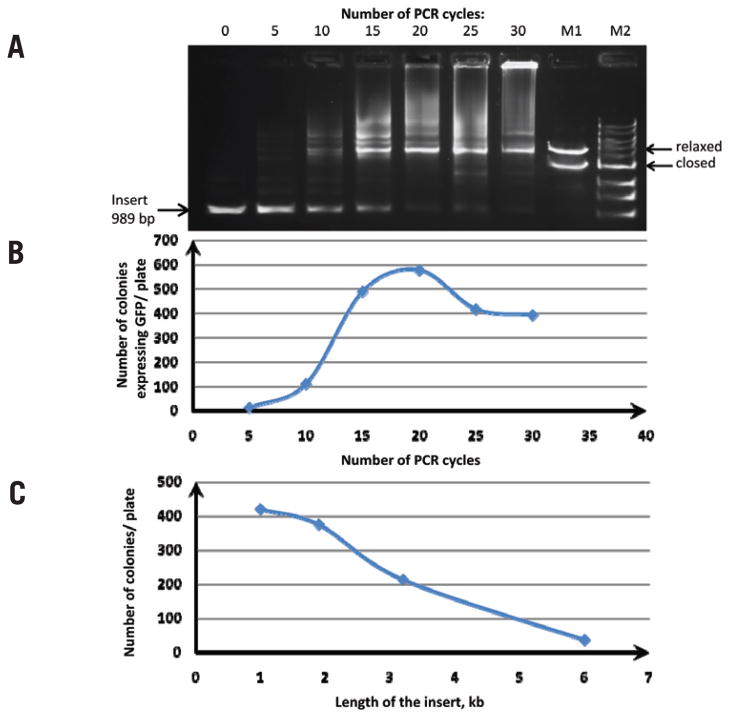
Figure 2. Analysis of the overlap extension PCR cloning reaction.
(A) Products of the overlap extension PCR cloning reaction after 0, 5, 10, 15, 20, 25, and 30 cycles by agarose gel electrophoresis. Three nanograms of pQE30 vector were mixed with 175 ng insert (250 molar excess) in 10 μL total volume; a 4-μL aliquot of reaction was separated on a 0.8% agarose gel. M2, 1 kb DNA ladder; M1, assembled plasmid in closed circular and relaxed circular forms. (B) Overlap extension PCR cloning efficiency of a gfp gene as a function of the number of PCR cycles. Twenty microliters of competent E. coli cells were transformed with 1 μL pQE30/insert overlap extension PCR. The number of green colonies was plotted against the number of PCR cycles for each plate. (C) Overlap extension PCR cloning efficiency as a function of the insert length. Phusion DNA polymerase was used to PCR-amplify products of various sizes: GFP (gfp) gene, β-D-glucuronidase (gusA) gene, β-galactosidase (lacZ) gene, and the luxABCDE operon from the carrying pIMBB plasmid. These products were gel-purified and used in the overlap extension PCR reaction with pQE30 vector. Three nanograms of pQE30 vector were mixed with 175–500 ng insert in a total reaction volume of 10 μL and subjected to 18 cycles of PCR. After DpnI treatment, the overlap extension PCR products were used to transform competent E. coli cells. The number of colonies per plate was plotted against the size of the insert.
Phusion DNA polymerase was better suited for overlap extension PCR cloning than the competitors we tested (Supplementary Table S1), perhaps due to its superior processivity and fidelity (11–12). Phusion DNA polymerase is 10× more processive than the native Pfu polymerase (Cat. no. 600135; Stratagene, La Jolla, CA, USA), and produced 46× more colonies (Supplementary Table S1). It also produced 35× more colonies than Expand Long Template DNA polymerase mix (Cat. No. 11681834001; Roche, Basel, Switzerland), which is mostly Taq DNA polymerase. Zuo and Rabie developed a similar method with Taq DNA polymerase alone, and reported similarly modest cloning efficiencies (8). Taq DNA polymerase is relatively processive (60% that of Phusion), but the overall fidelity of the mixture is only 3.8% that of Phusion. More than 98% of the colonies transformed with DNA produced by Phusion DNA polymerase were visibly green, indicating minimal cloning error or carryover of the original vector.
We measured the efficiency of the overlap extension PCR cloning as a function of temperature cycles; the number of recombinant clones increased geometrically during the first 15 cycles and peaked at 17–18 cycles (Figure 2B). Further cycles resulted in a slight (~30%) decrease in the quantity of clones produced, associated with the accumulation of the high–molecular weight DNA products observed in agarose gels (Figure 2A). The insert/plasmid ratio can also have a pronounced effect on the outcome of the reaction. We compared three different vector:insert ratios (1:5; 1:50 and 1:250) in overlap extension PCR cloning reaction with Phusion DNA polymerase. All three ratios resulted in the appearance of the nicked form of the plasmid (as judged by agarose gel electrophoresis, not shown) and recombinant clones (as judged by transformation and growth of green colonies). The 1:250 ratio produced the most recombinant clones.
We then applied overlap extension PCR cloning to clone the genes for GFP (gfp, 1 kb), β-D-glucuronidase (gusA, 1.9 kb), and β-galactosidase (lacZ, 3.2 kb), as well as the entire luxABCDE operon (6 kb). The correct structure of all the recombinant vectors was confirmed by restriction analysis and reporter protein function. The apparent error rate associated with our method, as judged by the fraction of colonies that did not exhibit full reporter activity, was <3% regardless of the size of the insert (data not shown). At the same time, the number of colonies that we observed on plates after transformation decreased considerably with the increase in insert length (Figure 2C). The graph is almost linear, which suggests that 6.7 kb is the upper limit for inserts with this technique.
The outcome of any cloning project is largely dependent upon the worker’s effort and attention to detail. The overlap extension PCR cloning reaction described here is as easy to monitor and optimize as any other long PCR protocol (13). In general, PCR yields are poor when the reaction conditions are too stringent (primers fail to anneal) or too relaxed (non-specific priming). Both are manifested by empty lanes in agarose gels, although the latter can also result in smears or undesired bands (See Supplementary Materials for details on primer design and PCR reaction optimization). The stringency of PCR can be controlled by altering reactant concentrations (primers, template), annealing temperature, buffer ingredients (magnesium, pH, DMSO) or the number of temperature cycles. Overall, this cloning approach proved to be insensitive to the presence of the internal repeated elements (see Supplementary Materials for details). Phusion DNA polymerase can be used to catalyze both the PCR amplification of the insert and overlap extension reactions, so practitioners will only need to familiarize themselves with the idiosyncrasies of a single enzyme.
Supplementary Material
Acknowledgments
We thank Natasha Degtereva and Joseph Kramer of Emory University for critical reviews of this manuscript. This work was supported by the National Institutes of Health (NIH; grant nos. 1 R01 GM074264 and 1 R01 GM086824, to I.M.). This paper is subject to the NIH Public Access Policy.
Footnotes
Supplementary material for this article is available at www.BioTechniques.com/article/113418.
Competing interests
The authors declare no competing interests.
References
- 1.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. CSH Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 2.Marchuk D, Drumm M, Saulino A, Collins FS. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rashtchian A, Thornton CG, Heidecker G. A novel method for site-directed mutagenesis using PCR and uracil DNA glycosylase. PCR Methods Appl. 1992;2:124–130. doi: 10.1101/gr.2.2.124. [DOI] [PubMed] [Google Scholar]
- 4.Weeks SD, Drinker M, Loll PJ. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr Purif. 2007;53:40–50. doi: 10.1016/j.pep.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 6.Cheo DL, Titus SA, Byrd DR, Hartley JL, Temple GF, Brasch MA. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res. 2004;14:2111–2120. doi: 10.1101/gr.2512204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 8.Zuo P, Rabie BM. One-step DNA fragment assembly and circularization for gene cloning. Curr Issues Mol Biol. 2009;12:11–16. [PubMed] [Google Scholar]
- 9.Shuldiner AR, Scott LA, Roth J. PCR-induced (ligase-free) subcloning: a rapid reliable method to subclone polymerase chain reaction (PCR) products. Nucleic Acids Res. 1990;18:1920. doi: 10.1093/nar/18.7.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuldiner AR, Tanner K, Scott LA, Moore CA, Roth J. Ligase-free subcloning: a versatile method to subclone polymerase chain reaction (PCR) products in a single day. Anal Biochem. 1991;194:9–15. doi: 10.1016/0003-2697(91)90144-i. [DOI] [PubMed] [Google Scholar]
- 11.Benson LM, Null AP, Muddiman DC. Advantages of Thermococcus kodakaraenis (KOD) DNA polymerase for PCR-mass spectrometry based analyses. J. Am. Soc. Mass Spectrom. 2003;14:601–604. doi: 10.1016/S1044-0305(03)00148-X. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Prosen DE, Mei L, Sullivan JC, Finney M, Vander Horn PB. A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res. 2004;32:1197–1207. doi: 10.1093/nar/gkh271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 2004;32:e19. doi: 10.1093/nar/gnh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.