Abstract
Dicer is an RNAse III endonuclease that is essential for the biogenesis of microRNAs and small interfering RNAs. These small RNAs post-transcriptionally regulate mRNA gene expression through several mechanisms to affect key cellular events including proliferation, differentiation, and apoptosis. Recently, the role of Dicer function in female reproductive tissues has begun to be elucidated through the use of knock-out mouse models. Loss of Dicer within ovarian granulosa cells, luteal tissue, oocyte, oviduct, and potentially the uterus render females infertile. This review discusses these early studies and other data describing the current understanding of microRNAs and small interfering RNAs in female reproduction.
Dicer mediated post-transcriptional gene regulation
Regulation of fertility in the female is a dynamic and highly regulated process that requires the coordinated actions of multiple tissues and organ systems (e.g. hypothalamus, pituitary, ovary, and reproductive tract) to develop a fertilizable gamete as well as provide a suitable environment for fertilization and subsequent fetal development. To attain this optimal environment, the female reproductive system must be highly malleable to subtle changes in hormones and other external cues. A large body of evidence supports a role for transcriptional regulation in mediating these changes, and recent evidence suggests a hereto underappreciated role for post-transcriptional gene regulation in reproductive tissue and organ function [1]. Post-transcriptional gene regulation encompasses all aspects of messenger RNA (mRNA) turnover, processing, storage and translation, and provides cells with additional mechanisms to regulate protein content following transcription events. Recently, study of post-transcriptional gene regulation has surged due to the discovery of small non-coding RNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs) [2]. Incorporation of miRNA and siRNA into RNA-induced silencing complexes (RISC) allows for the targeting of specific mRNA transcripts, ultimately providing cells with a post-transcriptional regulatory mechanism to either induce or inhibit protein production in response to stimuli, independent of commencement or cessation of mRNA transcription (for reviews, see [3, 4]). This review focuses on the RNA endonuclease III (RNAse III), Dicer, and its enzymatic products, miRNA and siRNA, which elicit post-transcriptional regulatory responses.
Dicer is a cytosolic multi-domain protein comprised of a RNA helicase, domain of unknown function (DUF283), Piwi Argonaute Zwille (PAZ) domain, two RNAse III domains, and a double stranded RNA binding domain (dsRBD) [5, 6]. The RNA helicase unwinds long dsRNA precursors, whereas the dsRBD and PAZ domains are essential for Dicer binding to dsRNA and for determining the length of the siRNA or miRNA products, respectively. Cleavage of miRNA or siRNA precursors is dependent on the two RNAse III domains within the Dicer protein. Dicer is essential in miRNA and siRNA biogenesis (Figure 1), and its function is critical to the cell, as general knock-out of Dicer1 (hereafter referred to as Dicer) in the mouse causes morphologic abnormalities and stunted growth in embryonic day (E) 7.5 embryos and lethality by E11.5 [7]. To further explore Dicer function, multiple groups developed Dicer alleles with the second RNAse III domain flanked by loxP sites (Dicerfl/fl) to facilitate conditional knock-down (cKO) of Dicerfl/fl via tissue specific Cre recombination [8–12]. Although miRNAs are abundant in mammalian somatic tissues and thus most affected by the loss of Dicer, several reports using deep sequencing methods have suggested that siRNAs also might play important role(s) in both somatic tissues and germ cells [13, 14]. Therefore, whereas loss of Dicer within somatic tissues has typically been linked to altered miRNA biogenesis, changes observed in conditional Dicerfl/fl knockout mice might be due to a combined loss of miRNA and siRNA.
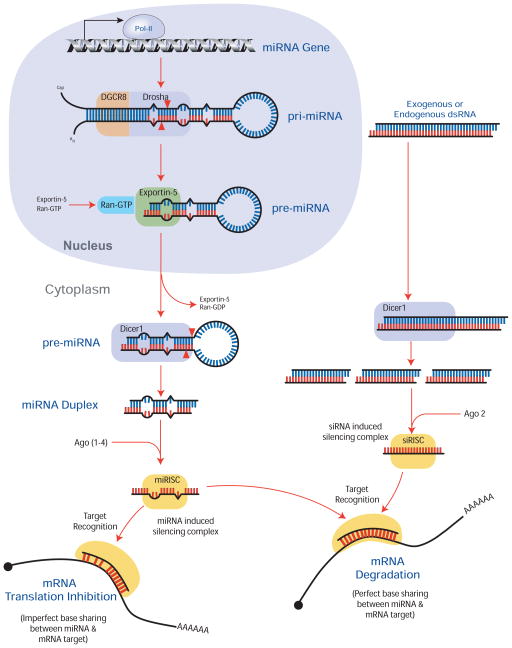
Figure 1. Biogenesis of (a) miRNA and (b) siRNA.
(i) MicroRNAs (miRNAs) are transcribed by RNA polymerase II from intragenic or intergenic regions of the genome. (ii) This transcript, the primary miRNA (pri-miRNA), can range 100–1000 nucleotides in length and shares many properties of mRNA transcripts, including a 7-methylguanasine (m7G) 5′-cap and poly-A tail. Within pri-miRNA, ~70bp stem-loops are recognized and cleaved by the complex of DGCR8 (DiGeorge syndrome critical region gene 8) and the RNAse III endonuclease enzyme, Drosha, to produce the precursor miRNA (pre-miRNA). (iii) Exportin 5 then transports the pre-miRNA from the nucleus to the cytoplasm, where (iv) the RNAse III endonuclease, Dicer, cleaves the stem away from the loop of the pre-miRNA to produce the (v) mature miRNA duplex. (vi) One strand of this ~21nt long duplex is incorporated into the RNA induced silencing complex (RISC) to regulate the translation/degradation of target mRNAs. (b) Whereas exogenous addition of dsRNA (siRNA) to the cell is a common experimental manipulation, recent evidence suggests that endogenous forms of siRNA also exist in animals. (i) Endogenous siRNA forms when long complementary strands of RNA bind to form dsRNA. The individual strands of these dsRNA are products of pseudogenes, transposable elements, or protein-coding genes, and are the result of complementary transcriptional products from cis or trans loci (same or different chromosomes, respectively) or the result of inverted repeat sequences that have folded to form a hairpin structure [9, 10]. (ii) Within the cytoplasm, dsRNA is cleaved into multiple ~21nt fragments by Dicer. (iii) One strand from these duplexes is incorporated into a (siRISC) to bind target mRNA and facilitate siRNA mediated transcript degradation. For a more detailed explanation of miRNA and siRNA biogenesis, see the following [58–60].
MicroRNAs are derived from highly conserved genes that bind partially complementary sequences in the 3′-untranslated region (3′UTR) and/or coding regions of target mRNA transcripts to regulate gene expression (Figure 1, for review [3, 4]). To date, 706 and 547 miRNAs have been identified in the human and mouse, respectively (Version 13 Sanger MirBase) [15]. Because each miRNA is derived from a specific pre-miRNA hairpin loop and represents a specific gene product, a standardized nomenclature for mammalian miRNAs has been established (Box 1). The functions of miRNAs are diverse and play a role in numerous processes including cellular proliferation and differentiation, embryonic development, and apoptosis (for review, see [16–18]).
Box 1. Nomenclature for mammalian miRNA.
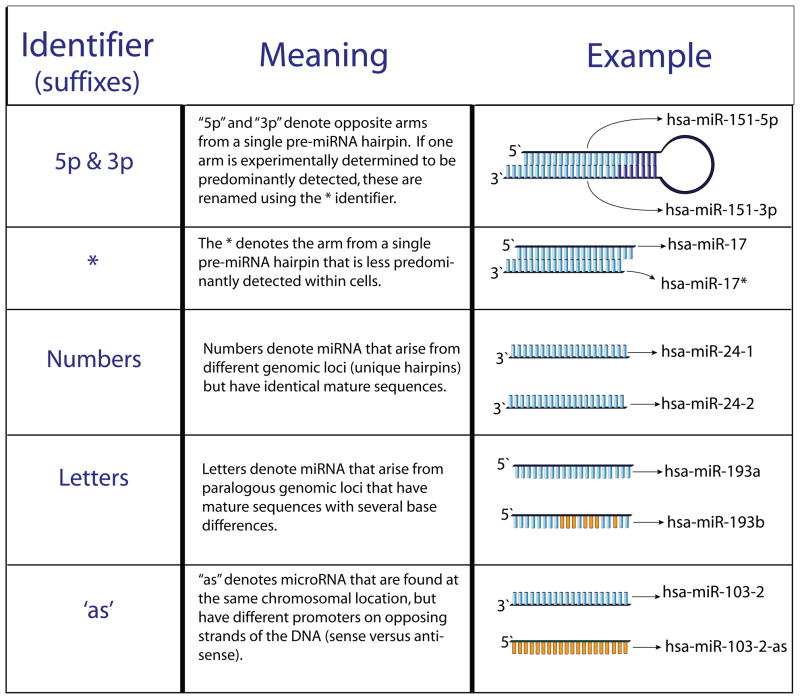
To provide structure in naming and characterizing the thousands of recently identified miRNAs, the Sanger Institute miRNA Registry was established (http://microrna.sanger.ac.uk/; [15]. The current nomenclature for animal miRNAs is described below. The names of miRNAs consist of four components, each conveying a specialized piece of information about the given miRNA: species, form (precursor or mature), identification number, and origin of miRNA (either processing origin or chromosomal origin). Each component is separated by dashes and is represented by the following template: xxx-miR-#-suffix. The xxx signifies the species (i.e., hsa=human, mmu=mouse). To distinguish between the precursor and mature forms of miRNA, a small case ‘r’ (‘mir’) represents the precursor form of the miRNA, whereas an uppercase ‘R’ (‘miR’) represents the mature form of the miRNA. Generally, an identification number is assigned in sequential order of discovery, thus recently identified miRNA have larger numbers. The suffix identifier (which might or might not be separated by a dash depending on the suffix, Figure I) denotes either the processing or chromosomal origin of the miRNA. For the processing origin, opposite arms from a single pre-miRNA are denoted ‘5p’ and ‘3p’. After one arm is experimentally identified as the predominant arm, the less predominant arm is labeled with an asterisk suffix. For genomic origin, miRNAs that arise from different genomic loci but have identical mature sequences are labeled with numbered suffixes, and if they arise from paralogous genomic loci and have highly similar mature sequences, they are labeled with lettered suffixes. In a few cases, miRNA genes have been identified at the same chromosomal location and are found on opposing DNA strands (sense versus antisense) and thus have unique mature miRNA sequences. The miRNA on the antisense chromosome is identified with an ‘as’ suffix and on the sense chromosome an ‘s’ suffix. A given miRNA can have several suffixes. For example, hsa-miR-19b-1, which originates from chromosome 13, is identical to hsa-miR-19b-2 which originates from chromosome X, and is paralogous (shares 70% of mature sequence) to hsa-miR-19a which originates from chromosome 13.
Figure I of Box 1.
Nomenclature of miRNA suffixes
Exogenous siRNAs injected into cells have been widely used to knock down gene expression (reviewed in [19]). Endogenous siRNAs are derived from dsRNA (i.e., pseudogenes, transposable elements, or protein-coding genes). Due to the nature of their biogenesis, siRNAs are typically thought to be fully complementary to their target transcripts, and a standardized nomenclature or estimate of the number of mammalian siRNAs has yet to be established (Figure 1). It remains possible that siRNAs associated with RISC complexes could also act like miRNA and not require full complementation to affect post-transcription gene regulation. Endogenous siRNAs derived from pseudogenes have been shown to target specific mRNA transcripts and regulate transposable elements in the mammalian oocyte [13, 14, 20]. The biological significance of siRNA retrotransposon silencing is thus far unique to the oocyte, and its exact role has yet to be defined [20].
Limited examples of post-transcriptional gene regulation exist within the somatic cells of the reproductive system [21, 22]. However, as the function of Dicer and its products (miRNAs and siRNAs) are studied in the female reproductive tract, vital roles for such post-transcriptional gene regulation in female fertility are becoming evident [1]. In addition, due to the involvement of miRNAs in these crucial biological pathways, it is not surprising that they have been implicated in a number of reproductive diseases and cancers of the reproductive system (for review, see [23–25]). Gaining a better understanding of Dicer generated miRNAs and siRNAs will provide important insight into regulation of the female reproductive system and perhaps the mechanisms that regulate fertility and etiology of reproductive diseases.
Dicer in the oocyte and early embryo
Female germ cells (oocytes) are enclosed in layers of somatic cells (granulosa and theca) form ovarian follicles. The somatic cells support the growth and development of the oocyte and ultimately release (ovulate) a mature transcriptionally quiescent oocyte that can be fertilized by sperm in the oviduct to form the developing zygote. The zygote undergoes a series of reductive cell divisions and eventually becomes an embryonic blastocyst. In the mouse, the embryo is dependent upon maternal transcripts produced during oocyte development until the 2-cell stage when transcription of the embryonic genome begins. Oocytes and fertilized eggs contain 10–15 fold higher levels of Dicer transcripts compared to all other cells/tissues (BioGPS, [26]) and are one of few known mammalian cells/tissues where Dicer expression is regulated [27]. Expression of the Dicer transcript remains steady in the growing mouse oocyte during folliculogenesis [14], through the germinal vesicle (GV) and metaphase II stages [28, 29]. After fertilization, the amount of Dicer mRNA decreases by approximately half and remains low in the 2-cell embryo through the blastocyst stage [28, 29]. Expression of total miRNA during this same period is highest in the mature oocyte and 1-cell zygote before decreasing by half in the 2-cell embryo [30]. Increased miRNA expression observed in the 4-cell embryo [30] could result from the resumption of transcription of the embryonic genome, post-transcriptional regulation of Dicer, or increases in other miRNA processing factors such as Drosha or Exportin 5 (Figure 1).
To examine Dicer function within the oocyte/early embryo, Dicerfl/fl mice were crossed with mice expressing Cre recombinase driven by the oocyte-specific ZP3 (zona pellucida 3) promoter (Dicer ZP3-cKO) or Alpl (alkaline phosphatase, liver/bone/kidney) promoter (Dicer Alpl-cKO [12, 29, 30]. In the Dicer ZP3-cKO model, Cre expression is turned on shortly after initiation of oocyte growth [12, 29, 30], while in the Dicer Alpl-cKO model, Cre expression is turned on as early as the primordial germ cell [12]. Early folliculogenesis and oocyte development in Dicer ZP3-cKO mice appeared normal, evidenced by normal ovulation rates and oocytes indistinguishable from wild-type littermates [29]. The importance of communication between the oocyte and cumulus granulosa cells in follicular and oocyte development is well-documented [31]. Therefore, the lack of a somatic cell phenotype in follicles with oocytes lacking Dicer suggests that oocyte miRNA or siRNA mediated post-transcriptional gene regulation does not play a critical role in regulating somatic cells during folliculogenesis.
While early development and growth of oocytes from Dicer ZP3-cKO females was unaffected, these oocytes were less likely to extrude a polar body following mating, and immunostaining indicated multiple spindles and chromatin condensation defects [12, 29, 30]. Similar spindle defects were observed in oocytes derived from Dicer Alp1-cKO mice [12]. Further analysis of cultured oocytes collected from Dicer ZP3-cKO mice found that meiosis proceeds normally until metaphase II when abnormal spindle formation was observed [12]. Transplantation of wild-type germinal vesicles (nucleus of oocyte) into enucleated oocytes from Dicer ZP3-cKO mice resulted in defective spindle formation, while reciprocal transplantation of mutant germinal vesicles into enucleated wild-type oocytes resulted in normal spindle formation in 74% of oocytes [12]. This data suggests that meiotic defects arise from the ooplasm of the oocyte and not the germinal vesicle [12]. Further, expression analysis of oocytes indicated an overabundance of mRNA transcripts in Dicer ZP3-cKO oocytes, consistent with a loss in miRNA inhibition of translation or mRNA degradation [30].
The roles of individual miRNAs and siRNAs in the oocyte are largely unknown, with a few recent exceptions [29, 30]. Loss of Dicer within oocytes was shown to decrease siRNA levels, whereas corresponding siRNA target transcripts, protein phosphatase 4 regulatory subunit 1 (Ppp4r1) and retrotransposon LTR 10, increased 1.5-fold and 5-fold, respectively [13, 14]. The biological role of siRNA retrotransposon silencing is thus far unique to the oocyte, and its exact role has yet to be defined (see review [19] for more information on RNA silencing in oocytes and early embryos).
Dicer within somatic cells of the ovary
The granulosa and thecal cells of the ovarian follicle support oocyte growth and produce key endocrine hormones (steroids and proteins) that regulate the reproductive system. In response to an ovulatory surge of luteinizing hormone (LH), the mature oocyte is released (ovulation) and with exception of a few cells that exit the ovary with the oocyte, the remaining somatic tissue that comprised the follicle undergoes luteinization (hypertrophy and vascularization) to form the corpus luteum (CL). These LH mediated ovarian events are essential for female fertility. Consistent with other observations showing that Dicer is not dynamically regulated [27], levels of Dicer in granulosa cells before and after the LH surge did not change [32]. Dicer expression in other somatic tissues of the ovary (thecal cells, corpus luteum, interstitium) has not been directly examined, but is not anticipated to change (BioGPS, [26]). In disease states, however, recent studies examining ovarian cancer cells found that Dicer and Drosha levels decrease in tumor cells [33]. Functional deletion studies of Dicer point to a clear and important role for miRNA and/or siRNA in ovarian function and female fertility [34–38].
Otsuka et al. [34] created a general hypomorphic mutation (Dicerhypo; ~75% reduction in Dicer protein) using a gene trap method and observed that Dicerhypo females were infertile due to luteal deficiency. Transplantation of wild-type ovaries into Dicerhypo females restored fertility, indicating that loss of fertility was due to an ovarian defect [34]. Serum progesterone levels in Dicerhypo mice remained low following mating, and histological analyses of ovaries revealed a lack of luteal tissue vascularization. In these mice, the global reduction of Dicer would be predicted to decrease all miRNA; however, selective ovarian bursal replacement of two known angiogenic miRNAs, miR-17-5p and let-7b, was sufficient to partially restore luteal vascular development and progesterone production through day 5.5 post-coitus; however, pregnancy was not maintained [34]. The inability to restore fertility could be due to an insufficient amount of exogenous miRNAs, clearance of exogenous miRNAs, or the need for additional miRNAs in other processes necessary for fertility (e.g. oviductal/uterine function, implantation) [34]. Alternatively, because Dicer was reduced in all cells in this model, the fertility defect could be due to a loss of function of non-reproductive cells (i.e. endothelial cells) within the ovary.
Mice with targeted deletion of Dicer within ovarian granulosa cells of developing follicles were generated by crossing Dicerfl/fl mice with mice expressing Cre recombinase driven by the anti-Mullerian hormone receptor 2 promoter (Dicer Amhr2-cKO; [35–38]). Within the ovary, Amhr2-Cre recombinase is expressed in granulosa cells of preantral and antral follicles [39, 40]. Studies by our group [35] and others [37, 38] crossed the Amhr2-Cre mice to the same Dicer floxed mouse line [9], while Nagaraja et al. [36] used a different Dicer floxed line [11]. Both lines of Dicer mice exhibited defects in ovarian function [35, 36]. The loss of Dicer expression in ovarian granulosa cells reduced natural [35] and equine chorionic gonadotropin (eCG)- and human chorionic gonadotropin stimulated (hCG)-stimulated ovulation rates when compared to wild-type mice [35, 36]. Increased numbers of atretic follicles [36] and trapped oocytes in luteinizing follicles were observed in Dicer Amhr2-cKO mice [35, 36]. Both studies found that loss of Dicer did not impact serum estrogen levels or mating behavior [35, 36]. Collectively, these studies indicate that Dicer affects ovulation rate, likely by influencing the total number of preovulatory follicles that achieve proper development and/or by affecting the ability of follicles to ovulate. Interestingly, evidence for trapped oocytes in luteinized follicles was not reported in the general Dicerhypo mouse model [34]. Discrepancies between the Dicerhypo and Dicer Amhr2-cKO models could be due to the steroidogenic cell specific deletion of Dicer in the Amhr2-cKO model and total cell deletion in the Dicerhypo model. Detailed study of the different stages of follicular development, especially during the peri-ovulatory period, paired with Dicer expression analyses are needed to elucidate how and when loss of Dicer impacts follicular development, ovulation and/or luteinization. It is possible that Dicer is necessary for proper recruitment of follicular waves and loss of Dicer leads to fewer follicles available for growth, maturation, and ovulation.
In contrast to the clear evidence regarding ovulation rate, the two lines of Dicer Amhr2-cKO mice present conflicting evidence regarding whether loss of Dicer in ovarian granulosa cells impacts oocyte function [35, 36]. Nagaraja et al. [36] reported that cultured, fertilized oocytes collected from the oviduct of stimulated Dicer Amhr2-cKO females failed to progress to the 2-cell embryonic stage at the same rate (29.0%) as fertilized oocytes collected from wild-type females (82.8%). Conversely, Hong et al. [35] reported that 97% and 100% of fertilized oocytes collected from Dicer Amhr2-cKO and wild-type females, respectively, one day after mating (no pharmacologic stimulation), progressed to the 2-cell stage. The differences between these studies might have resulted from pharmacologic stimulation versus natural follicular cycle, or because of different Dicer-floxed strains [9, 11].
Functional analyses of Dicer in the ovary implicate miRNA in ovarian function, and the individual miRNA involved in specific functions are now beginning to be determined. Laboratories have identified as many as 177 miRNAs in whole ovaries of newborn, 2 week old and adult mice [41–43]. Several of these miRNA (let-7a, miR-143, miR-21, miR-125, let-7b, and let-7c) have also been described in granulosa cells of primary, secondary, and antral follicles [44]. Human granulosa cell culture studies have implicated a number of miRNAs in the inhibition of steroid production with 36, 51, and 57 specific miRNAs decreasing progesterone, estradiol, and testosterone synthesis, respectively [45]. Conversely, 10, 0, and 1 miRNA were found to stimulate human granulosa cell production of progesterone, estradiol, and testosterone, respectively [45]. Whether these miRNAs are expressed and exhibit similar function within ovarian follicular or luteal tissues remains to be tested. In a cell-specific study, Fiedler et al. [32] identified 212 known miRNAs within granulosa cells isolated from peri-ovulatory follicles, 13 of which were regulated by the LH/hCG surge [32]. Two of these LH-regulated miRNAs (miR-132 and miR-212) were found to post-transcriptionally regulate C-terminal binding protein 1 [32], a protein that was recently shown to repress steroidogenic factor-1 expression [46], a nuclear receptor involved in ovarian, adrenal, and testis function development and function (e.g. steroidogenesis; [47]). In addition to the Otsuka et al. [34] study, these are the only studies linking specific miRNAs to biological endpoints in the ovary, illustrating the need for additional research.
Dicer in the oviduct and uterus
Oocyte fertilization and early pre-implantation embryonic development occurs within the oviduct. Subsequently, the developing embryo passes through the uterotubal junction and enters the uterus where implantation, placentation, and embryonic/fetal development occur. In addition to granulosa cells, Amhr2-Cre is expressed in the oviduct, uterus, cervix, and anterior portion of the vagina [39]. Loss of Dicer in mouse oviducts results in a dramatic phenotype consisting of shortened tubule length, loss of oviductal coils, and large fluid filled sacs [35–38]. Histological analysis of oviducts from Dicer Amhr2-cKO females revealed loss of the smooth muscle layer and disorganization of the epithelium in the oviduct, particularly in the isthmus (region near the uterus) of the oviduct [35–37]. Collection of embryos on day 4 post-mating found all embryos retained in the oviduct of Dicer Amhr2-cKO females, whereas embryos in wild-type females had all traversed the uterotubal junction and resided within the uterus [35]. Similarly, affinity chromatography beads similar in size to preimplantation embryos that were injected into oviducts were unable to enter the uterus in Dicer Amhr2-cKO mice [37]. Since ovulated Dicer Amhr2-cKO oocytes were fertilized following mating [35], this would argue that the block is unidirectional, and/or due to an effect on smooth muscle contractility, cilia, or oocyte size that prevents transport. Nagaraja et al. [36] identified 28 down-regulated miRNAs in oviducts from the Dicer Amhr2-cKO mouse model, 23 of which were predicted to target at least one member of the Wnt or Hox family of genes [36]. In fact, the defect in oviductal transport of the Dicer Amhr2-cKO mouse phenocopies that seen in mice deficient in Wnt/β-catenin signaling [48, 49]. Indeed, β-catenin levels were reduced in oviducts and uteri of Dicer Amhr2-cKO females [35]. In addition to developmental effects, it appears that loss of miRNA in the oviduct might also influence factors secreted into the oviductal lumen, as embryos collected three days after mating from the oviducts of Dicer Amhr2-cKO females were developmentally delayed compared to those collected from wild-type animals [35].
Similar to the oviductal phenotype, the uteri of Dicer Amhr2-cKO animals were also developmentally compromised (with 3 of 4 reports noting severe defects [35–37]). The length and diameter, as well as weights of uteri collected from eCG-stimulated juvenile Dicer Amhr2-cKO females were smaller than wild-type littermates [35, 36]. Histological analysis revealed the presence of all tissue layers within the uterus [35–37], although reduced numbers of uterine glands and a thinner myometrial layer was observed in the one Dicer Amhr2-cKO mouse model [35, 37]. Gonzalez et al [37] further noted that the uterine glands reside in close proximity to the myometrial layer, mimicking a human condition referred to as adenomyosis [50]. The uteri of these mice (n=3 cKO) were not able to sustain pregnancy following embryo transfer. Interestingly, histological analysis failed to observe any defect in other Dicer floxed mouse lines [36], and adenogenesis (i.e., gland formation and location), estrogen responsiveness, and stimulus-induced decidualization reaction all appeared normal [36].
Expression of Dicer mRNA and protein as well as the Argonaute proteins is abundant in the mouse uterus on days 4 through 8 of pregnancy (day 1=presence of vaginal plug), suggesting that miRNA synthesis is ongoing [51]. Furthermore, microarray analysis of miRNA present in the uterus on day 4 of pregnancy (receptive phase) and at implantation sites suggests that miRNAs are important for establishing pregnancy [51, 52]. Comparison of miRNA expression within uterine tissues collected on days 1 and 4 of pregnancy identified 32 miRNA upregulated on day 4 [53]. Expression of miR-101 and miR-199* were stimulated by estradiol and found to post-transcriptionally regulate prostaglandin synthase-2, an enzyme necessary for implantation in mice [53]. Further analysis in the pregnant uterus found 13 miRNAs upregulated in implantation sites compared to inter-implantation sites [53]. Analysis of miR-21, one of the upregulated miRNAs, found that expression was dependent upon the presence of an activated embryo [53]. Uterine miRNAs also appear to be regulated by estrogen, as 49 miRNAs were found to be differentially regulated in response to estradiol [54]. Taken together, these studies suggest a role for uterine miRNAs in implantation and pregnancy.
Future Directions
To date, the study of Dicer function in female reproductive tissues is limited to a few reports (Figure 2). However, these studies all clearly demonstrate that Dicer and its enzymatic products are crucial for female fertility. Conditional deletion of Dicer in the oocyte, ovary, oviduct, and uterus provide convincing evidence that miRNAs/siRNAs are necessary for the overall development and function of the female reproductive system. Recent studies demonstrated that expression of Amhr2-Cre recombinase was leaky, with recombination also occurring in the brain, pituitary, heart, and tail in addition to the known tissues expressing Amhr2 [38, 55]. Additional studies using more refined temporal and cell specific Cre-recombinases are needed to further our understanding of post-transcriptional gene regulation in the female reproductive system (Figure 2). Identification of individual miRNAs or siRNAs in each female reproductive organ will allow researchers to better understand the mechanisms of gene regulation that allow for successful reproduction. Thus far, little is known about the role of siRNAs in somatic cells of the reproductive system, although they appear to be highly abundant [13, 14]. Additional research is needed to determine the functional role siRNA molecules exert on tissue and organ function. Recently, antagomirs and locked nucleic acid oligonucleotides have been used to downregulate specific miRNA expression in a variety of cells in vitro and tissues in vivo [56, 57]. It is anticipated that establishing the spatiotemporal expression patterns of miRNA in the female reproductive tract will provide targets for drug and therapeutic treatments for reproductive diseases such as endometriosis, uterine leiomyomas, and ovarian, uterine, and cervical cancers. Moreover, understanding the role that post-transcriptional gene regulation plays in reproduction will facilitate the elucidation of the etiologies leading to reproductive failure and hopefully provide methods/targets for treatment of infertility and provide new means of contraception.
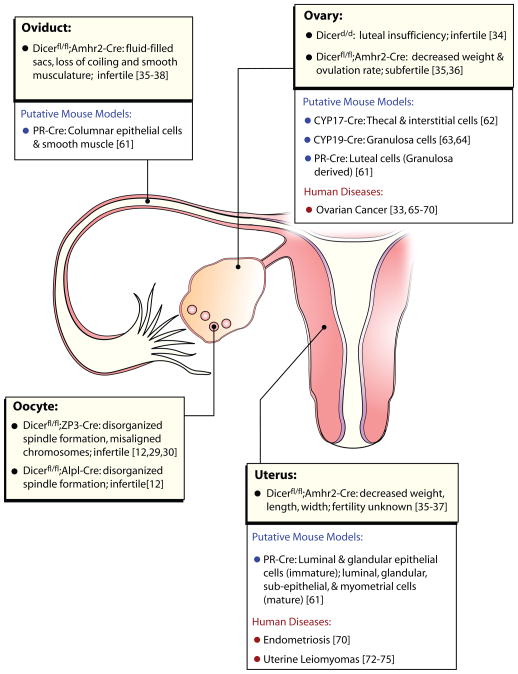
Figure 2. Effects of Dicer deletion on female fertility.
The function of Dicer and its products (siRNA and miRNA) in female fertility have been investigated in Dicer1 floxed mouse lines expressing Cre-recombinase in female reproductive tissues. The descriptions of phenotypes caused by Dicer deletion are listed for each tissue (yellow boxes). Additional female reproductive tissue Cre-recombinases are available. It is anticipated their use will further refine the temporal and cellular importance that Dicer (i.e., miRNA/siRNA) has on female fertility (open boxes). Lastly, a number of expression analysis studies have linked human reproductive diseases to altered miRNA or siRNA profiles they are listed in the (open boxes). Presently, most of these studies are biomarker studies as specific miRNAs/siRNAs have not yet been demonstrated to be causative. Dicerfl/fl;Amhr2-Cre is referred to as Dicer Amhr2-cKO, Dicerfl/fl;ZP3-Cre is referred to as Dicer ZP3-cKO, Dicerfl/fl;Alpl-Cre is referred to as Dicer Alpl-cKO in the text of this review. Gene abbreviations: Alpl; alkaline phosphatase (liver/bone/kidney), Amhr2; anti-mullerian hormone receptor 2, Cre; cre recombinase, CYP17; cytochrome P450 subfamily 17, CYP19; cytochrome P450 subfamily 19, hypo; hypomorph, ZP3; zona pellucida 3, PR; progesterone receptor
Acknowledgments
The authors would like to thank Stephanie Fiedler for help with graphic design, Stan Fernald for illustrations, and Dr. Warren Nothnick for critical reading of the manuscript.
References
- 1.Carletti MZ, Christenson LK. MicroRNA in the ovary and female reproductive tract. J Anim Sci. 2009;87(14 suppl) doi: 10.2527/jas.2008-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruvkun G. The perfect storm of tiny RNAs. Nat Med. 2008;14:1041–1045. doi: 10.1038/nm1008-1041. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, et al. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.van den Berg A, et al. RISC-target interaction: cleavage and translational suppression. Biochim Biophys Acta. 2008;1779:668–677. doi: 10.1016/j.bbagrm.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 6.Provost P, et al. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc Natl Acad Sci U S A. 2002;99:16648–16653. doi: 10.1073/pnas.212633199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 8.Andl T, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harfe BD, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mudhasani R, et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi R, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 12.Mattiske DM, et al. Meiotic maturation failure induced by DICER1 deficiency is derived from primary oocyte ooplasm. Reproduction. 2009;137:625–632. doi: 10.1530/REP-08-0475. [DOI] [PubMed] [Google Scholar]
- 13.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 17.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 18.Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci. 2008;65:545–562. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Svoboda P. RNA silencing in mammalian oocytes and early embryos. Curr Top Microbiol Immunol. 2008;320:225–256. doi: 10.1007/978-3-540-75157-1_11. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dell’Aquila ME, et al. Cumulus expansion, nuclear maturation and connexin 43, cyclooxygenase-2 and FSH receptor mRNA expression in equine cumulus-oocyte complexes cultured in vitro in the presence of FSH and precursors for hyaluronic acid synthesis. Reprod Biol Endocrinol. 2004;2:44. doi: 10.1186/1477-7827-2-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menon KM, et al. A novel post-transcriptional mechanism of regulation of luteinizing hormone receptor expression by an RNA binding protein from the ovary. Mol Cell Endocrinol. 2006;246:135–141. doi: 10.1016/j.mce.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 23.Luo X, Chegini N. The expression and potential regulatory function of microRNAs in the pathogenesis of leiomyoma. Semin Reprod Med. 2008;26:500–514. doi: 10.1055/s-0028-1096130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan Q, Chegini N. MicroRNA signature and regulatory functions in the endometrium during normal and disease states. Semin Reprod Med. 2008;26:479–493. doi: 10.1055/s-0028-1096128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei JJ, Soteropoulos P. MicroRNA: a new tool for biomedical risk assessment and target identification in human uterine leiomyomas. Semin Reprod Med. 2008;26:515–521. doi: 10.1055/s-0028-1096131. [DOI] [PubMed] [Google Scholar]
- 26.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholson RH, Nicholson AW. Molecular characterization of a mouse cDNA encoding Dicer, a ribonuclease III ortholog involved in RNA interference. Mamm Genome. 2002;13:67–73. doi: 10.1007/s00335-001-2119-6. [DOI] [PubMed] [Google Scholar]
- 28.Cui XS, et al. Dicer1 expression in preimplantation mouse embryos: Involvement of Oct3/4 transcription at the blastocyst stage. Biochem Biophys Res Commun. 2007;352:231–236. doi: 10.1016/j.bbrc.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Murchison EP, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang F, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong J, et al. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383:531–535. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- 32.Fiedler SD, et al. Hormonal Regulation of MicroRNA Expression in Periovulatory Mouse Mural Granulosa Cells. Biol Reprod. 2008;79:1030–1037. doi: 10.1095/biolreprod.108.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merritt WM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otsuka M, et al. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest. 2008;118:1944–1954. doi: 10.1172/JCI33680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong X, et al. Dicer1 Is Essential for Female Fertility and Normal Development of the Female Reproductive System. Endocrinology. 2008;149:6207–6212. doi: 10.1210/en.2008-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagaraja AK, et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22:2336–2352. doi: 10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez G, Behringer RR. Dicer is required for female reproductive tract development and fertility in the mouse. Mol Reprod Dev. 2009 doi: 10.1002/mrd.21010. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastorelli LM, et al. Genetic analyses reveal a requirement for Dicer1 in the mouse urogenital tract. Mamm Genome. 2009;20:140–151. doi: 10.1007/s00335-008-9169-y. [DOI] [PubMed] [Google Scholar]
- 39.Jamin SP, et al. Requirement of Bmpr1a for Mullerian duct regression during male sexual development. Nat Genet. 2002;32:408–410. doi: 10.1038/ng1003. [DOI] [PubMed] [Google Scholar]
- 40.Jorgez CJ, et al. Granulosa cell-specific inactivation of follistatin causes female fertility defects. Mol Endocrinol. 2004;18:953–967. doi: 10.1210/me.2003-0301. [DOI] [PubMed] [Google Scholar]
- 41.Choi Y, et al. Microarray analyses of newborn mouse ovaries lacking Nobox. Biol Reprod. 2007;77:312–319. doi: 10.1095/biolreprod.107.060459. [DOI] [PubMed] [Google Scholar]
- 42.Mishima T, et al. MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction. 2008;136:811–822. doi: 10.1530/REP-08-0349. [DOI] [PubMed] [Google Scholar]
- 43.Ro S, et al. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007;13:2366–2380. doi: 10.1261/rna.754207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao N, et al. A network of miRNAs expressed in the ovary are regulated by FSH. Front Biosci. 2009;14:3239–3245. doi: 10.2741/3447. [DOI] [PubMed] [Google Scholar]
- 45.Sirotkin AV, et al. Identification of microRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. J Cell Physiol. 2009;219:415–420. doi: 10.1002/jcp.21689. [DOI] [PubMed] [Google Scholar]
- 46.Dammer EB, Sewer MB. Phosphorylation of CtBP1 by cAMP-dependent protein kinase modulates induction of CYP17 by stimulating partnering of CtBP1 and 2. J Biol Chem. 2008;283:6925–6934. doi: 10.1074/jbc.M708432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lala DS, et al. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol Endocrinol. 1992;6:1249–1258. doi: 10.1210/mend.6.8.1406703. [DOI] [PubMed] [Google Scholar]
- 48.Arango NA, et al. Conditional deletion of beta-catenin in the mesenchyme of the developing mouse uterus results in a switch to adipogenesis in the myometrium. Dev Biol. 2005;288:276–283. doi: 10.1016/j.ydbio.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 49.Deutscher E, Hung-Chang Yao H. Essential roles of mesenchyme-derived beta-catenin in mouse Mullerian duct morphogenesis. Dev Biol. 2007;307:227–236. doi: 10.1016/j.ydbio.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parrott E, et al. Adenomyosis--a result of disordered stromal differentiation. Am J Pathol. 2001;159:623–630. doi: 10.1016/S0002-9440(10)61733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chendrimada TP, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 52.Hu SJ, et al. MicroRNA expression and regulation in mouse uterus during embryo implantation. J Biol Chem. 2008;283:23473–23484. doi: 10.1074/jbc.M800406200. [DOI] [PubMed] [Google Scholar]
- 53.Chakrabarty A, et al. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci U S A. 2007;104:15144–15149. doi: 10.1073/pnas.0705917104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nothnick WB. Regulation of uterine matrix metalloproteinase-9 and the role of microRNAs. Semin Reprod Med. 2008;26:494–499. doi: 10.1055/s-0028-1096129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hernandez Gifford JA, et al. Conditional Deletion of Beta-Catenin Mediated by Amhr2cre in Mice Causes Female Infertility. Biol Reprod. 2009 doi: 10.1095/biolreprod.108.072280. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 57.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 58.Faller M, Guo F. MicroRNA biogenesis: there’s more than one way to skin a cat. Biochim Biophys Acta. 2008;1779:663–667. doi: 10.1016/j.bbagrm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 60.Tay Y, et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 61.Soyal SM, et al. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis. 2005;41:58–66. doi: 10.1002/gene.20098. [DOI] [PubMed] [Google Scholar]
- 62.Bridges PJ, et al. Generation of Cyp17iCre transgenic mice and their application to conditionally delete estrogen receptor alpha (Esr1) from the ovary and testis. Genesis. 2008;46:499–505. doi: 10.1002/dvg.20428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan HY, et al. Targeted disruption of Pten in ovarian granulosa cells enhances ovulation and extends the life span of luteal cells. Mol Endocrinol. 2008;22:2128–2140. doi: 10.1210/me.2008-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan HY, et al. Selective expression of KrasG12D in granulosa cells of the mouse ovary causes defects in follicle development and ovulation. Development. 2008;135:2127–2137. doi: 10.1242/dev.020560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corney DC, et al. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 66.Dahiya N, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iorio MV, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 68.Nam EJ, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 69.Yang H, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 70.Zhang L, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pan Q, et al. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- 72.Pan Q, et al. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med. 2008;12:227–240. doi: 10.1111/j.1582-4934.2007.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Marsh EE, et al. Differential expression of microRNA species in human uterine leiomyoma versus normal myometrium. Fertil Steril. 2008;89:1771–1776. doi: 10.1016/j.fertnstert.2007.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng Y, et al. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008;6:663–673. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- 75.Wang T, et al. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer. 2007;46:336–347. doi: 10.1002/gcc.20415. [DOI] [PubMed] [Google Scholar]