Abstract
Recent studies on the molecular mechanisms responsible for cell cycle deregulation in cancer have puzzled out the role of oncogenes in mediating unscheduled cellular proliferation. This is reminiscence of their activity as proto-oncogenes that drives scheduled cell cycle progression under physiological conditions. Working on the cell cycle regulatory activity of proto-oncogene, we observed that c-ETS1 transcriptionally up-regulated both cyclin E and CDK2 genes, the master regulators of G1/S-phase transition. The process was mediated by kinetic coherence of c-ETS1 expression and its recruitment to both promoters during G1/S-phase transition. Furthermore, enforced expression of c-ETS1 helped G0-arrested cells to progress into G1/S-phases apparently due to the activation of cyclin E/CDK2 genes. Physiological induction of c-ETS1 by EGF showed the remodeling of mononucleosomes bound to the c-ETS1 binding site on both promoters during their activation. The exchange of HDAC1 with histone acetyltransferase-p300 was contemporaneous to the chromatin remodeling with consequent increase in histone H3K9 acetylation. Furthermore, the ATP-dependent chromatin remodeler hBRM1 recruitment was also associated with nucleosome remodeling and promoter occupancy of phospho-Ser5 RNA polymerase II. Intriguingly, the activity of the HBx viral oncoprotein was dependent on c-ETS1 in a hepatotropic manner, which led to the activation of cyclin E/CDK2 genes. Thus, cyclin E and CDK2 genes are key physiological effectors of the c-ETS1 proto-oncogene. Furthermore, c-ETS1 is indispensable for the hepatotropic action of HBx in cell cycle deregulation.
Keywords: Cell Cycle, Chromatin Remodeling, Cyclins, Ets Family Transcription Factor, Transcription Regulation, EGF, HBx, Cdk2
Introduction
Mechanisms that coordinate cell cycle progression are driven by sequential activation and inactivation of a family of cyclin-dependent kinases (CDKs).4 The activation occurs predominately by the periodic expression of its regulatory subunit cyclin and activating phosphorylation of the kinase subunit (1). Redundant nature of cyclins and CDKs implies that the functional importance lies in their temporal expression rather than their effector molecules (2).
G1-specific cyclin E expression periodically oscillates in every cycle of proliferating cells, ensuring its ordered progression (3). Periodicity is mainly controlled at the transcriptional level leading to its peak expression during G1/S-phase transition (4, 5). The cyclin E/CDK2 activity assumes special significance as it is the rate-limiting regulator of the G1/S-phase transition and act as a switch for various cellular processes including initiation of DNA replication (6, 7). Perturbation of this rate-limiting step by viral oncoproteins is a common theme that causally relates to the plethora of cancers (8).
The regulation of the cyclin E promoter by mitogens, various transactivators, and growth factors has been analyzed extensively in the recent past (9–12). In contrast, CDK2 regulation has been studied mainly at post translational level (13), and only a few reports have discussed the oscillation of CDK2 levels before S-phase entry (14, 15). This suggests that the regulation of CDK2 at the transcriptional level could be as important as its post-translational control. Although the cyclin E-CDK2 complex is assembled and active during the same window of cell cycle, their transcriptional regulation by common transactivators remains obscure.
c-ETS1 (transcription factor E-26 transforming sequence-1) is a classic example of the proto-oncogene and the founding member of ETS family proteins. The unique cis element “GGA(A/T),” known as the ETS binding site (EBS), is among the eight most important DNA motifs in minimal responsive synthetic promoters and identified in the promoter/enhancer regions of >200 genes (16). c-ETS1 is known to be associated with different aspects of cancer, including extracellular matrix remodeling, invasion, angiogenesis (17), and also has an important role in proliferation and differentiation of hematopoietic cells (18). Moreover, inappropriate expression of c-ETS1 is an early event in a wide variety of cancers, and its overexpression results in a transformed phenotype (19). Furthermore, the serum-inducible nature of the c-ETS1 promoter (20) suggested its plausible role in cell cycle regulation. However, the physiological effectors of c-ETS1 involved in the regulation of cell cycle remains enigmatic.
In the present study, we have shown that cyclin E and CDK2 genes are the physiological effectors of c-ETS1. The stimulation of c-ETS1 by EGF leads to up-regulation of both genes, which involves chromatin remodeling and cross-talk of associated co-factors. Furthermore, the up-regulation of cyclin E and CDK2 genes by viral oncoprotein HBx is dependent on a c-ETS1-responsive element in a tissue-specific manner.
EXPERIMENTAL PROCEDURES
Expression Vectors and Reporter DNA Constructs
The human cyclin E promoter reporter construct pCycECAT (pE-WT) (−1195/+79) was kindly provided by Professor J. R. Nevins (Duke University Medical Center) (9), whereas the CDK2 promoter reporter construct −2400CDK2/LUC (DSC37) (pCDK2-WT) was from Gary Stein (University of Massachusetts Medical School) (21). The c-ETS1 expression construct was from Hiroyuki Sugimoto (22), and c-ETS1 dominant negative (c-ETS1 DN) construct (pAPrEts-Z) was kindly provided by Arthur Gutierrez-Hartmann (23). The β-galactosidase expression plasmid pCH110 (Amersham Biosciences) and the EGFP expression plasmid pEGFP-C1 (BD bioscience) were used as transfection control.
Site-directed Mutagenesis of c-ETS1 Elements
The c-ETS1 response element in the cyclin E promoter (pE-WT) reporter construct was mutated by PCR using the QuikChangeTM site-directed mutagenesis kit (Stratagene, La Jolla, CA) to get pE-mut. Likewise, the CDK2 promoter (pCDK2-WT) was also mutated for its proximal (pCDK2-prox.mut), distal (pCDK2-dis.mut), or both c-ETS1 sites (pCDK2-mut). The following set of primers were used: forward, pE-mut F, 5′-actcagggcccctcgagcggcgtctc-3′; pE-mut R, 5′-gagacgccgctcgaggggccctgagt-3′; pCDK2-prox.mut F, 5′-agggaaacgctcgaggcaggggcggg-3′; pCDK2-prox.mut R, 5′-cccgcccctgcctcgagcgtttccct-3′; pCDK2-dis.mut F, 5′-agattcccggctcgagggtttccaaa-3′; and pCDK2-dis.mut R, 5′-tttggaaaccctcgagccgggaatct-3′. The mutated bases are boldfaced and underlined.
Cell Culture, Reagents, and Antibodies
Maintanance of human hepatoma Huh7 and HepG2 cells, human embryonic kidney HEK293 (ATCC CRL-1573), and human epithelial cervical HeLa (ATCC CCL2) cell lines were described elsewhere (24). Transfection was carried out in a 60-mm culture dish (0.6 × 106 cells) with 2.0 μg of indicated plasmids by Lipofectamine (Invitrogen) according to the manufacturer's instructions. EGFP-C1 (0.5 μg) was used in each experiment as a transfection control (supplemental Fig. 1). For reporter assays, 0.25 μg of CAT or luciferase reporter plasmids were cotransfected with 0.5 μg of indicated expression plasmids. siRNA against c-ETS1 was procured from Santa Cruz Biotechnology and used for transfection according to manufacturer's instructions. Wherever mentioned, 24-h post-starved cells were treated with EGF (10 ng/ml, Calbiochem) for 18 h. Antibodies were obtained from the following sources: Santa Cruz Biotechnology for RNA polymerase II (pol II; CTD4H8), phospho-Ser5 pol II, HDAC1, p300, cyclin E, CDK2, GAPDH, c-ETS1 and hBRM1; and Upstate Biotechnology for H3K9Ac. mAb (monoclonal antibody) (B-8/2/8) against HBx has been reported previously (25).
CAT and Luciferase Assay
The chloramphenicol acetyltransferase (CAT) assay was performed as described previously (24). Luciferase assay was performed according to the manufacturer's instructions (Promega). The relative CAT and luciferase activities were measured after normalizing each sample with β-galactosidase activity. Wherever HBx expression vector was used in a reporter assay, mean EGFP fluorescence was used for normalization of reporter activity.
Flow Cytometry (FACS)
Huh7 cells were starved for 48 h then stimulated with serum for the indicated time periods. Wherever indicated, 24 h post-transfected cells were subjected to serum starvation for another 24 h. Flow cytometry of cells was done as described in Ref. 26.
RNA Isolation and Quantitative RT-PCR Assay
Total RNA was isolated from cells using TRIzol reagent as per the supplier's instructions (Invitrogen). RT-PCR was performed with M-MuLV reverse transcriptase (Fermentas) according to the manufacturer's guidelines. The real-time quantitative PCR (qPCR) was done using specific primers (supplemental Table 1) as described previously (24).
Western Blotting
Protocol for Western blotting of protein samples can be found elsewhere (26).
EMSA
In vitro binding of c-ETS1 to putative EBS elements was performed by EMSA as described previously (24). The end labeling of oligonucleotides (supplemental Table 2) was done using [γ-32P]ATP and T4 Polynucleotide Kinase (Fermentas) per the manufacturer's instructions.
MNase Southern Hybridization Assay
Nuclei isolation and MNase digestion was done as described earlier (24) with minor modifications. Complete digestion of nuclei with MNase (Fermentas) was performed for 20 min at 37 °C (50 units MNase/2 × 106 nuclei), whereas incomplete digestion was performed for 5 min at 37 °C (5 units MNase/2 × 106 nuclei). For southern hybridization, 10 μg of DNA yielded from either complete or incomplete digestion of MNase was electrophoresed and transferred to nylon membrane for hybridization with end-labeled DNA probes (supplemental Table 3).
MNase CHART-PCR Assay
Mononucleosomes obtained by complete digestion of MNase as mentioned above were resolved on agarose gel (1.8%) and eluted using QIAquick Gel extraction kit (Qiagen). The genomic DNA obtained by gel elution was used to perform SYBR green real-time PCR in triplicates with cyclin E and CDK2 primers (supplemental Table 3). The Ct values generated by MNase CHART-PCR were converted to DNA concentrations as described previously (27) using the standard curve of corresponding primer set and normalized for input variations. The standard curve for each primer set was generated using serial dilutions of genomic DNA (28). The results were expressed as relative nucleosome occupancy. Because MNase introduces double-stranded breaks in the nucleosome linker regions, the level of product generated by CHART-PCR assay is inversely proportional to the amount of nucleosomal DNA digested. This was expressed as nucleosome occupancy.
ChIP-qPCR Assay
ChIP assay was carried out as described previously (24). Chromatin obtained was purified using QIAquick PCR purification kit (Qiagen). The eluted genomic DNA was subjected to either semi-quantitative PCR or SYBR green real-time qPCR with indicated primer sets (supplemental Table 3) as described in CHART-PCR assay. The results were expressed as fold enrichment over mock.
X15-Myc Transgenic Mouse Model
Processing of liver sample for Western blotting and RNA isolation from X15-Myc transgenic mouse model has been reported earlier (29).
Statistical Analysis
Data are expressed as mean ± S.D. Means were compared with one-factor analysis of variance followed by Fisher protected least significant difference to assess specific group differences. Differences were considered significant at p < 0.05.
RESULTS
Cyclin E and CDK2 Genes Are Up-regulated at Promoter Level by c-ETS1
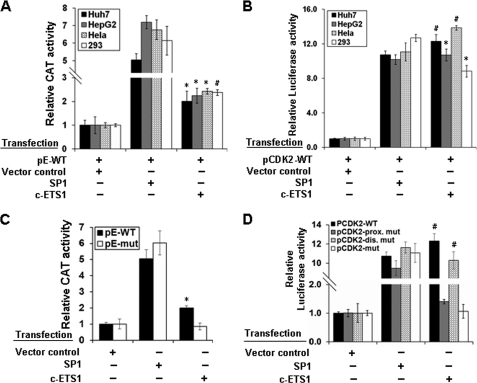
Despite the same window of expression (14), the transcriptional regulation of cyclin E and CDK2 genes involving common regulatory cis elements is poorly understood. Our bioinformatics analysis revealed the presence of single c-ETS1 element at the −526 position on the cyclin E promoter, whereas two putative sites on the CDK2 promoter at positions −67 (proximal) and −384 (distal) (supplemental Fig. 2, A and B). Because c-ETS1 is a serum-inducible transcription factor (20), we performed reporter assay in the presence of c-ETS1 using cyclin E-CAT (pE-WT) and CDK2-luciferase (pCDK2-WT) reporter constructs in hepatic (Huh7 and HepG2) and non-hepatic (HEK293 and HeLa) cell lines. SP1 expression was used as positive control for both cyclin E (30) and CDK2 (15) reporters. Irrespective of the cell lines used, both cyclin E and CDK2 promoters were up-regulated nearly 2- and 12-fold, respectively, by c-ETS1 (Fig. 1, A and B). Furthermore, a dose-dependent activation of both promoters was observed with increasing amount of c-ETS1 (supplemental Fig. 3, A and B). The specificity of promoter stimulation was evident from their competitive inhibition by c-ETS1 DN (supplemental Fig. 3, C and D).
FIGURE 1.
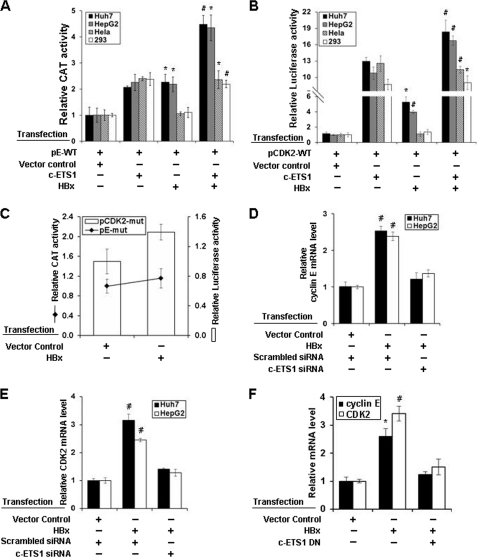
Functional characterization and localization of c-ETS1 element in cyclin E and CDK2 promoters. A and B, Huh7, HepG2, HeLa, and HEK293 cells were transfected with either pE-WT (A) or pCDK2-WT (B) reporter construct along with the expression vectors of SP1, and c-ETS1 and the relative CAT and luciferase activities were measured. C, pE-WT and pE-mut reporters were transfected in Huh7 cells along with SP1 and c-ETS1 expression vectors, and the relative CAT activity was measured. D, pCDK2-WT, pCDK2-prox.mut, pCDK2-dis.mut, and pCDK2-mut luciferase reporters were transfected in Huh7 cells along with the expression constructs of SP1 and c-ETS1, and the relative reporter activity was measured. Data shown in A–D are the mean ± S.D. of three independent experiments. The asterisk and number sign indicate statistically significant difference at p < 0.05 and p < 0.01, respectively.
The involvement of the c-ETS1 element in transactivation was confirmed using the mutated reporter constructs: pE-mut, pCDK2-prox.mut, pCDK2-dis.mut, and pCDK2-mut (supplemental Fig. 1, A and B). The c-ETS1-mediated transactivation of the pE-mut reporter was significantly reduced in the presence of c-ETS1, whereas the SP1-mediated promoter stimulation remained unchanged (Fig. 1C). Likewise, the transactivation of pCDK2-prox.mut and pCDK2-mut was also abolished (Fig. 1D). However, no appreciable reduction was observed with pCDK2-dis.mut (Fig. 1D), indicating the functional importance of the proximal c-ETS1 element. As a reason, all subsequent studies were performed in relation to the c-ETS1 proximal element of the CDK2 promoter. Nevertheless, c-ETS1 appeared to be a modulator of both cyclin E and CDK2 promoters.
c-ETS1-mediated Stimulation of Cyclin E and CDK2 Genes Accelerate G1/S-phase Progression
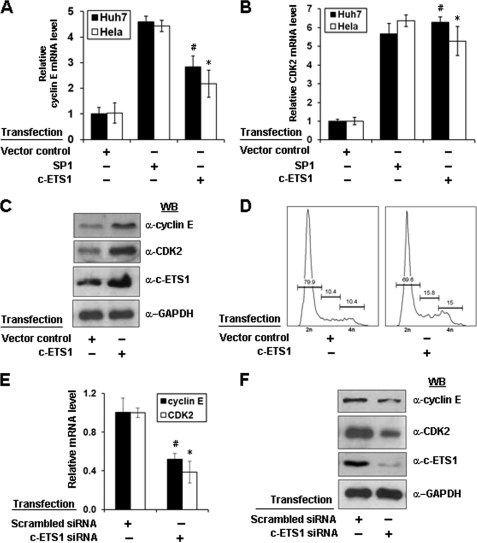
Because c-ETS1 stimulated both cyclin E and CDK2 promoters, we next directly monitored the expression of these genes at mRNA and protein levels after enforced expression of c-ETS1 or by RNA interference. We observed that c-ETS1 expression led to a significant increase in the mRNA levels of both cyclin E and CDK2 genes independent of cell lines used (Fig. 2, A and B). Furthermore, it was specific because mRNA levels were down-regulated in the presence of siRNA against c-ETS1 (Fig. 2E). The up-regulation of both cyclin E and CDK2 genes were further confirmed at protein levels by Western blot analysis. As expected, overexpression of c-ETS1 showed a significant increase in the level of both cyclin E and CDK2 protein (Fig. 2C), which was abrogated by c-ETS1 siRNA (Fig. 2F and supplemental Fig. 4A). Because cyclin E-CDK2 complexes play a key role in G1/S-phase transition, we also studied the effect of c-ETS1 overexpression on cell cycle progression. The FACS analysis clearly indicated that even starved cells had progressed into S-phase in the presence of c-ETS1 (Fig. 2D). Thus, c-ETS1 has a modulatory role in the expression of cyclin E and CDK2 genes and cell cycle progression.
FIGURE 2.
Effect of c-ETS1 on cyclin E and CDK2 expression and cell cycle progression. A and B, Huh7 and HeLa cells were transfected with SP1 and c-ETS1 expression vectors and the mRNA levels of cyclin E (A) and CDK2 (B) were measured by qRT-PCR. The sequences of primers are given in supplemental Table 1. C, Huh7 cells were transfected with either control or c-ETS1 expression vector and 48 h post-transfection, the cell extracts were processed for Western blotting with cyclin E, CDK2, c-ETS1, and GAPDH antibodies. D, 24 h post-transfected Huh7 cells were subjected to serum starvation for another 24 h and analyzed by flow cytometry. E and F, Huh7 cells transfected with indicated siRNA were analyzed for cyclin E and CDK2 transcripts level by qRT-PCR (E) or Western blotted (WB) for protein levels of cyclin E, CDK2, c-ETS1, and GAPDH (F). Data shown in A, B, and E are the mean ± S.D. of three independent experiments. The asterisk and number sign indicate statistically significant difference at p < 0.05 and p < 0.01, respectively.
c-ETS1 Binds to Its Cognate “cis” Elements in Cyclin E and CDK2 Promoters
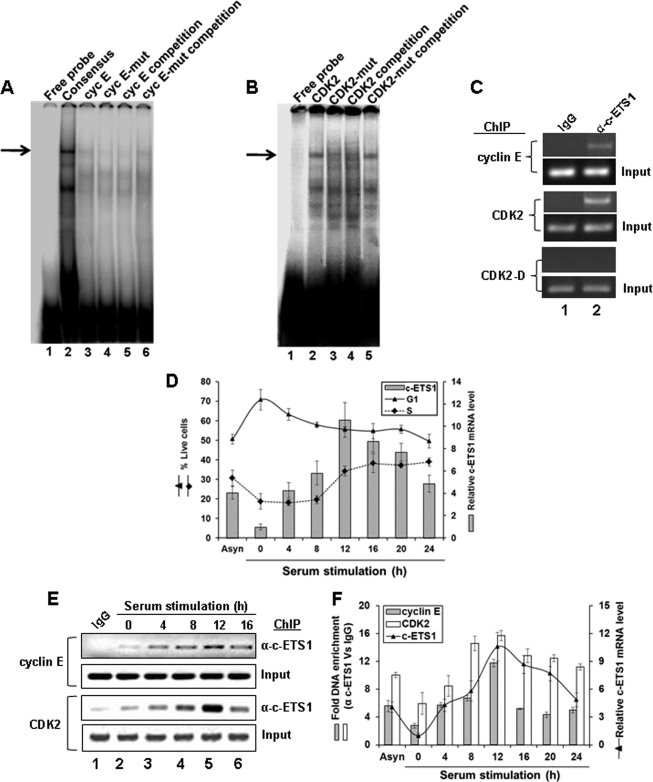
The binding of c-ETS1 to its putative sites on cyclin E and CDK2 promoters was examined by EMSA using nuclear extracts of Huh7 cells. As shown in Fig. 3, a typical protein-DNA complex was observed with the consensus c-ETS1 element as well as those derived from cyclin E (Fig. 3A, lanes 2 and 3) and CDK2 promoters (Fig. 3B, lane 2). As expected, the mutant c-ETS1 element for cyclin E (cyc E mut) and CDK2 proximal region (CDK2 mut) did not exhibit such complexes (Fig. 3A, lane 4, and Fig. 3B, lane 3, respectively). Furthermore, the wild type c-ETS1-DNA complexes could be competitively displaced by excessive unlabeled probes but not by unlabeled mutant probes (Fig. 3A, lanes 5 and 6, and Fig. 3B, lanes 4 and 5, respectively).
FIGURE 3.
Kinetics of c-ETS1 expression and recruitment to cyclin E and CDK2 promoters during cell cycle. A and B, EMSA to show the binding of a γ-32P-labeled Consensus (A, lane 2), WT c-ETS1 element of cyclin E (Cyc E; A, lane 3), and CDK2 (B, lane 2) to the nuclear extracts of Huh7 cells. Cold competition with 100-fold molar excess of unlabeled WT and mut c-ETS1 elements of cyclin E (A, lanes 5 and 6) and CDK2 (B, lane 4 and 5) were used to show the binding specificity. Impaired binding by γ-32P-labeled mut c-ETS1 oligonucleotides of cyclin E (A, lane 4) and CDK2 (B, lane 3) further ensured the specific binding of WT c-ETS1 oligonucleotides. The EMSA primers are given in supplemental Table 2. Arrows indicate the positions of the c-ETS1-DNA complex. C, ChIP analysis showing the recruitment of c-ETS1 on cyclin E and CDK2 promoters. IgG indicates the control antibody. D, the relative c-ETS1 mRNA level (bar) measured by qRT-PCR was plotted against percent G1 and S-phase Huh7 cells that were serum-starved for 48 h and stimulated for indicated time points. E, semiquantitative ChIP-PCR analysis showing the recruitment kinetics of c-ETS1 on cyclin E and CDK2 promoters. Huh7 cells were subjected to ChIP-PCR analysis upon serum stimulation for the indicated time periods. IgG indicates the control antibody. F, ChIP-qPCR showing fold DNA enrichment over mock due to c-ETS1 occupancy on cyclin E and CDK2 promoters (bars) was plotted against the relative c-ETS1 mRNA level (line) measured by qRT-PCR. Data shown in D and F are the mean ± S.D. of three independent experiments.
Further evidence of c-ETS1 interaction to its responsive elements on cyclin E and CDK2 promoters came from ChIP analysis on asynchronously growing Huh7 cells (Fig. 3C). Thus, c-ETS1 appears to directly interact with cyclin E and CDK2 promoters both in vitro as well as in vivo.
Recruitment of c-ETS1 on Cyclin E and CDK2 Promoters Parallels G1/S-phase Transition
As cyclin E and CDK2 appeared to be the effectors of c-ETS1 and cyclin E/CDK2 activity is essential for G1/S-phase progression (3), we next analyzed the expression profile of c-ETS1 along the cell cycle. The G0-arrested Huh7 cells were released by serum stimulation and harvested every 4 h window until 24 h. Percent live cells were plotted against the endogenous mRNA level of c-ETS1 measured during the same time frame. The results (Fig. 3D) clearly indicated that the G1/S-phase transition during 8–12 h coincided with the peak of c-ETS1 expression. This is consistent with the fact that the c-ETS1 promoter is serum-inducible and exhibits maximum activity during 8 h post-serum stimulation (20). The c-ETS1 expression profile and its recruitment on cyclin E and CDK2 promoters during cell cycle were also analyzed by semi-quantitative and quantitative ChIP-PCR assay (Fig. 3, E and F). As shown in Fig. 3F, the c-ETS1 expression apparently coincided with its occupancy on both promoters. Thus, c-ETS1 expression and its recruitment on cyclin E and CDK2 promoters follow the similar kinetics and parallel the G1/S-phase transition.
EGF-mediated Expression of c-ETS1 Leads to Cyclin E and CDK2 Induction
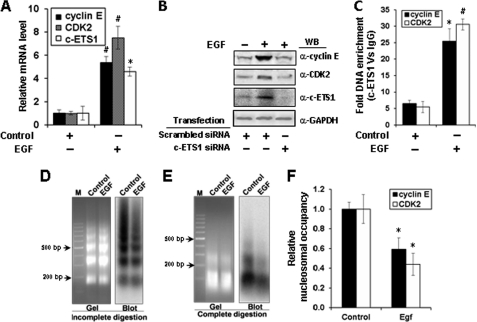
EGF is reported as a physiological inducer of c-ETS1 (31), we wondered whether cyclin E and CDK2 genes can also be stimulated by EGF. c-ETS1 expression was measured in the presence of increasing concentrations of EGF treatment (1–10 ng/ml), and optimal expression of c-ETS1 was observed with 5–10 ng/ml (supplemental Fig. 4B). Quantitative analysis of mRNA expression showed nearly 5-fold increase in the levels of cyclin E and almost 7-fold increase in CDK2 transcripts (Fig. 4A). Furthermore, as expected, the EGF-mediated induction of c-ETS1 and its downstream genes cyclin E and CDK2 could be down-regulated in the presence of c-ETS1 siRNA (Fig. 4B). Moreover, there was a marked (5–6-fold) increase in the recruitment of c-ETS1 on both cyclin E and CDK2 promoters (Fig. 4C). These results confirmed that both cyclin E and CDK2 genes are up-regulated by EGF-mediated c-ETS1 stimulation.
FIGURE 4.
Regulation of cyclin E, CDK2, and c-ETS1 genes by EGF and the nuclesosomal status of c-ETS1 elements of cyclin E and CDK2 promoters post-EGF induction. Huh7 cells were treated with EGF (10 ng/ml) for 18 h and processed for the following experiments. A, relative mRNA levels of cyclin E, CDK2, and c-ETS1 measured by qRT-PCR. B, Western blot (WB) showing the expression of cyclin E, CDK2, and c-ETS1 protein in cells treated with EGF and transfected with indicated siRNAs. C, ChIP-qPCR to show the occupancy of c-ETS1 on cyclin E and CDK2 promoters. D and E, MNase Southern assay showing the ethidium bromide-stained gel (left) and the corresponding hybridized blot (right) with a probe bearing the proximal c-ETS1 element of the CDK2 promoter to show nucleosome array generated by partial MNase digestion (D) or mononucleosomal DNA after complete MNase digestion (E). M, DNA marker lane. F, MNase CHART-PCR analysis to show the relative nucleosome occupancy of c-ETS1 element on both cyclin E and CDK2 promoters. Data shown in A, C, and F are the mean ± S.D. of three independent experiments. The asterisk and number sign indicate statistically significant difference at p < 0.05 and p < 0.01, respectively.
c-ETS1 Elements on Both Cyclin E and CDK2 Promoters Are Nucleosomal and Remodeled after EGF-mediated Induction of c-ETS1
We showed earlier that −448 to −595 region of the cyclin E promoter comprising the c-ETS1 element (−526 to −535) is nucleosomal (24). The present study, using MNase CHART-PCR, revealed the remodeling of this mononucleosome upon EGF induction (Fig. 4F). However, the nucleosomal status of c-ETS1 element on the CDK2 promoter is largely unknown. To address this issue, we employed the MNase Southern assay to the nuclei isolated from the EGF-treated Huh7 cells. Partial MNase digestion revealed a physiologically spaced nucleosomal ladder of DNA fragments of ∼160 bp periodicity (Fig. 4D), whereas the complete MNase digestion shown positioned mononucleosome (Fig. 4E) at c-ETS1 element on the CDK2 promoter. Upon induction with EGF, the nucleosomal status was altered albeit with differences in the input levels. This caveat was overcome by the MNase CHART-PCR assay that allows the quantitative measurement of chromatin remodeling (27). Just as cyclin E, the mononucleosome assembled on the proximal c-ETS1 element of the CDK2 promoter also remodeled after EGF treatment (Fig. 4F). Thus, induction of c-ETS1 leads to remodeling of mononucleosomes positioned on c-ETS1 responsive elements of both cyclin E and CDK2 promoters.
Recruitment of c-ETS1 Facilitates Cross-talk of Cofactors over Cyclin E and CDK2 Promoters
Next, we studied the interplay of co-factors associated with chromatin remodeling and transcription initiation cycle upon EGF-mediated c-ETS1 induction. We carried out ChIP-qPCR with HDAC1 and p300 antibodies following EGF treatment. HDAC1 occupancy on cyclin E and CDK2 promoters declined upon c-ETS1 induction with a concomitant increase in the occupancy of histone acetyltransferase-p300 (Fig. 5, A and B). This is consistent with the nucleosome remodeling of both promoters after EGF treatment (Fig. 4E). The increase in the p300 occupancy prompted us to look at the levels of acetylation of lysine 9 residue of histone H3 (H3K9Ac), which is considered as a hallmark of active transcription (32). As expected, there was a 4-fold increase in the acetylation of cyclin E promoter and a nearly 2-fold increase was observed in the acetylation of the CDK2 promoter (Fig. 5, A and B). Interestingly, the histone H3K9 acetylation correlated well with the increased occupancy of phospho-Ser5 pol II transcription initiation marker on both promoters (Fig. 5, A and B). Furthermore, increased hBRM1 occupancy confirmed the involvement of not only the histone modifiers but also the chromatin remodelers after EGF treatment (Fig. 5, A and B). Thus, c-ETS1-mediated expression of cyclin E and CDK2 genes involves the cross-talk of co-factors associated with chromatin remodeling.
FIGURE 5.
Co-factors interplay over c-ETS1 element of cyclin E and CDK2 promoter after EGF induction. Occupancy of cofactors pol II, phospho-Ser5 pol II, H3K9Ac, p300, HDAC1, and hBRM1 on cyclin E (A) and CDK2 (B) promoters measured by ChIP-qPCR. Data shown in A and B are the mean ± S.D. of three independent experiments. An asterisk and number sign indicate statistically significant difference at p < 0.05 and p < 0.01, respectively.
HBx Protein Cooperates with c-ETS1 in Up-regulation of Cyclin E and CDK2
The viral oncoprotein HBx can overcome the G0 and G1/S checkpoints even in the absence of serum (26). Because cyclin E and CDK2 levels were up-regulated in the presence of c-ETS1, we wondered whether HBx can aid this process. We performed reporter transactivation assay using cyclin E and CDK2 reporter constructs in hepatic (Huh7 and HepG2) and non-hepatic (HEK293 and HeLa) cell lines after co-expressing c-ETS1 and HBx. HBx stimulated both promoters and had an additive effect with c-ETS1 in hepatic cell lines but not in non-hepatic cell lines (Fig. 6, A and B). Stimulation of both promoters by HBx was significantly reduced with mutated c-ETS1 reporters in the hepatic cell line (Fig. 6C). Furthermore, the increased level of cyclin E and CDK2 transcripts in presence of HBx was significantly reduced by siRNA against c-ETS1 and c-ETS1 dominant negative construct (Fig. 6, D–F). Next, to show c-ETS1 was critical for cell cycle progression through induction of cyclin E and CDK2 genes, we performed FACS analysis after c-ETS1 silencing. Presence of HBx under c-ETS1 silenced condition did not lead to S-phase progression, substantiating our earlier observation on c-ETS1 dependence of HBx activity for G1/S-phase transition. Interestingly, enforced co-expression of cyclin E and CDK2 bypassed the requirement of c-ETS1 (by knockdown) for G1/S-phase progression (supplemental Fig. 5).
FIGURE 6.
Induction of cyclin E and CDK2 transcription by HBx. A and B, Huh7, HepG2, HeLa, and HEK293 cells were transfected with either pE-WT (A) or pCDK2-WT (B) reporter constructs along with the expression vectors of c-ETS1 and HBx and the relative reporter activity was measured. C, Huh7 cells were transfected with either pCDK2-mut (bar) or pE-mut (line) along with the expression construct of HBx, and the relative reporter activity was measured. D–F, the relative mRNA levels of cyclin E (D) and CDK2 (E) genes were measured by qRT-PCR in Huh7 cells transfected with HBx along with either indicated siRNAs or c-ETS1 dominant negative. Data shown in A–F are the mean ± S.D. of three independent experiments. The asterisk and number sign indicate statistically significant difference at p < 0.05 and p < 0.01, respectively.
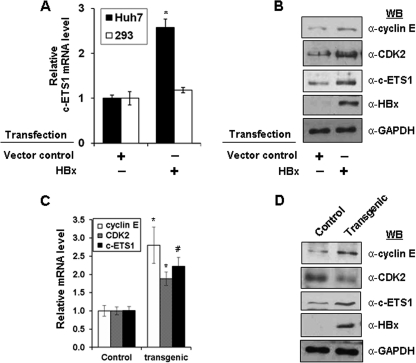
We also investigated the direct regulation of c-ETS1 by HBx. A marked increase in c-ETS1 mRNA levels was observed in the presence of HBx in Huh7 cells but not in HEK293 cells (Fig. 7A). Furthermore, this was also confirmed at protein levels in Huh7 cells (Fig. 7B). These results, together with HBx-mediated up-regulation of cyclin E and CDK2 protein levels confirmed the modulation of both genes by HBx via c-ETS1 responsive elements. Interestingly, analysis of c-ETS1 transcript and protein levels in the liver of X15-Myc transgenic mice also confirmed a significant increase in c-ETS1 mRNA and protein levels (Fig. 7, C and D). Besides c-ETS1, we also observed increased cyclin E and CDK2 mRNA and protein levels in transgenic environment (Fig. 7, C and D). Thus, HBx-mediated direct targeting of c-ETS1 seems to require a hepatotropic environment for modulating cyclin E and Cdk2 expression.
FIGURE 7.
Deregulation of c-ETS1 proto-oncogene by HBx. A, the relative mRNA levels of c-ETS1 were measured by qRT-PCR in both Huh7 and HEK293 cells transfected with HBx. C, the relative mRNA levels of cyclin E, CDK2, and c-ETS1 were measured by qRT-PCR in the liver samples of control and X15-Myc transgenic mice. B and D, immunoblots to show the levels of cyclin E, CDK2, c-ETS1, HBx, and GAPDH in the Huh7 cells transfected with HBx (B) or in the liver samples of 1-month-old X15-Myc transgenic mice (D). Data shown in A and C are the mean ± S.D. of three independent experiments. An asterisk and number sign indicate statistically significant difference at p < 0.05 and p < 0.01, respectively.
DISCUSSION
Considering the timing of their expression and broader range of substrates, coregulation of cyclin E and CDK2 is of paramount importance in the orchestration of S-phase progression. In the current study, we investigated the activation of both cyclin E and CDK2 promoters by the c-ETS1 proto-oncogene. c-ETS1 is well known to be involved in diverse cellular processes such as proliferation, differentiation, development, transformation, and apoptosis (33). Intriguingly, c-ETS1 null animals do not show any proliferative phenotype (34). Nevertheless, the c-ETS1 targeting-mediated cell cycle progression has been reported only for the cyclin D1 gene (35). Furthermore, c-ETS1 allows rat embryo fibroblasts to grow in serum-free medium (36). Thus, it was noteworthy to investigate the role of c-ETS1 in the regulation of serum-inducible promoters such as cyclin E and CDK2.
Our reporter gene studies showed a significant increase in the cyclin E and CDK2 promoter activity in presence of c-ETS1 independent of cell lineage (Fig. 1, A and B) and is of no surprise, due to ubiquitous nature of c-ETS1 expression (37). There was a built-in specificity in this mechanism as confirmed by competitive inhibition using c-ETS1 dominant negative or mutation of c-ETS1 elements (supplemental Fig. 3, C and D and Fig. 1, C and D). Interestingly for the CDK2 promoter, only the proximal c-ETS1 element appeared to be functionally important because mutation in the distal element did not impair the reporter gene activity (Fig. 1D). Furthermore, the occupancy of c-ETS1 to its responsive elements on both promoters was evident from our EMSA and ChIP studies (Fig. 3, A–C). Moreover, siRNA-mediated silencing of c-ETS1 recapitulated its role in targeting both cyclin E and CDK2 genes (Fig. 2, E and F).
The regulation of cell cycle progression by c-ETS1 was evident from enforced expression of c-ETS1 that helped overcoming the quiescence state (G0) imposed by deprivation of serum. The accelerated G1/S-phase progression was apparently due to the activation of cyclin E and CDK2 genes (Fig. 2D). Interestingly, a similar effect has been reported for viral oncoprotein HBx in the induction of cyclin A promoter (38). Moreover, c-MYC is also reported to play a critical role in sustaining an E2F-independent G1/S-promoting mechanism by regulating cyclin E-CDK2 function (39). Thus, coalescence of our results with earlier reports unveiled that the c-ETS1 proto-oncogene has the same potential as c-MYC and HBx oncoproteins in mediating proliferation function of cells and that its deregulation could create a microenvironment conducive for cancerous growth.
Our kinetic studies on c-ETS1 expression clearly showed its peak during G1/S transition (Fig. 3D), which is in accordance to the earlier report (20). Furthermore, the expression kinetics of c-ETS1 was well correlated with its occupancy on both cyclin E and CDK2 promoters (Fig. 3E) that would be important for the consequent events in transcription cycle (40). Elucidation of a “transcriptional clock” that directs sequential and combinatorial assembly of transcriptionally productive complexes would be achieved by induction of promoter elements (41). In this light, we used EGF as a physiological inducer of c-ETS1 as it is known to stimulate c-ETS1 transcription and protein expression (31). Corroborating with ectopic expression of c-ETS1, the induction of endogenous c-ETS1 by EGF also recapitulated a similar event in the activation of cyclin E and CDK2 genes (Fig. 4, A and B).
The chromatin structure and remodeling plays a pivotal role in the control of eukaryotic gene regulation by influencing all stages of transcription (42). Analysis of the nucleosomal status of EBS indicated that similar to cyclin E (24), the CDK2 promoter EBS also assembled into mononucleoesome (Fig. 4, D and E) and remodeled upon c-ETS1 induction by EGF (Fig. 4F). The SWItch/Sucrose Non-Fermentable-ATPases, including BRG1 and hBRM complexes (43), are known to increase the binding of transcription factors to mononucleosomes as well as nucleosomal arrays (44, 45). Consistently, we also observed the increased recruitment of hBRM1 during activation of cyclin E and CDK2 promoters by c-ETS1 (Fig. 5, A and B). The acetylation of N-terminal tails of histones is correlated with disruption of higher order chromatin structure and activation of transcription, whereas its deacetylation relates to its reversal and repression of transcription (46). Among the histone acetyltransferases that catalyze acetylation of histones, p300/CBP is well established for its role in the regulation of plethora of genes, including cell cycle regulators (47). Moreover, HDAC1 is reported to be the negative regulator of cyclin E by RB protein (48). In this line, we observed the exchange of HDAC1 with histone acetyltransferase p300 on both promoters during induction of c-ETS1 by EGF treatment (Fig. 5, A and B). Earlier reports suggested that p300 forms co-activator complex with PCAF (49), and this complex mediates the histone H3K9 acetylation (50). Consistently, the increased p300 occupancy was observed with the contemporaneous raise in the acetylation of histone H3K9 levels (Fig. 5, A and B). Moreover, nucleosomes with histone H3-K4me3 and H3K9, K14 acetylation modifications, together with pol II occupy the promoters of most protein-coding genes and serve as the hallmarks of transcription initiation (32). We also observed increased pol II occupancy on both cyclin E and CDK2 promoters with the concomitant rise in histone H3K9 acetylation. Furthermore, the elevation of serine 5-phosphorylated pol II occupancy confirmed the activation of both genes upon induction of c-ETS1 (Fig. 5, A and B). Collectively, our results suggested that chromatin remodeling by both ATP-dependent remodelers and histone modifiers could be the underlying mechanism of co-factors interplay in the regulation of c-ETS1-mediated activation of cyclin E and CDK2 genes.
Overwhelming evidence suggests that HBx protein of mammalian hepadnavirus (Hepatitis B virus) with transactivator and mitogenic signaling functions has a definitive role in the development of hepatocellular carcinoma (51). Cells expressing HBx show increased rate of entry to S-phase, breakdown of the G1/S-phase checkpoint, and accelerated G2/M progression due to activation of cyclins and cell division cycle 2 kinases (52, 26). Because c-ETS1 regulates G1/S-transition by activation of cyclin E and CDK2 genes, we investigated whether HBx can deregulate the cell cycle by cooperating with c-ETS1. We indeed observed a synergy between HBx and c-ETS1 exclusively in a hepatotropic environment (Fig. 6, A and B), which mimicked the oncogenic nature of HBx in a transgenic environment (53). Because HBx is unable to bind directly to any defined DNA-binding sequences (54, 55), the responsive elements of other transcription factors, including EBS (56), could mediate the transactivation function of HBx. Consistently, mutation studies and RNA interference against c-ETS1 impaired the up-regulation of both cyclin E and CDK2 promoters in presence of HBx (Fig. 6, C–E). The synergism between HBx and c-ETS1 is likely due to up-regulation of c-ETS1 by HBx as evident from both cell culture studies and in vivo studies in X15-Myc transgenic animals (Fig. 7, A–D).
Collectively, based on these results, it may be concluded that cyclin E and CDK2 genes are the physiological effector molecules of c-ETS1. Thus, direct targeting of c-ETS1 by HBx could be associated with the cell cycle regulatory process and poses fresh challenges in understanding of hepatocellular carcinoma.
Supplementary Material
Acknowledgments
We are grateful to the following scientists for the generous gift of the following recombinant constructs: Dr. J. R. Nevins (Duke University Medical Center) for pCycECAT reporter, Dr. Gary Stein (University of Massachusetts Medical School) for −2400CDK2/LUC (DSC37) reporter, Dr. Hiroyuki Sugimoto for the c-ETS1 expression construct, Dr. Arthur Gutierrez-Hartmann for the c-ETS1 DN construct, Dr. Michael G. Brattain (Roswell Park Cancer Institute) for SP1 expression vectors, Dr. J. M. Roberts (Fred Hutchinson Cancer Research Center) for cyclin E expression constructs, and Dr. E. Harlow (Massachusetts General Hospital Cancer Center) for CDK2 expression constructs. Ravinder Kumar helped in cell culture work.
This work was supported by a core grant of the International Centre for Genetic Engineering and Biotechnology, New Delhi.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–5.
- CDK
- cyclin-dependent kinase
- c-ETS1
- E-26 transforming sequence 1
- CHART-PCR
- chromatin accessibility assay based on real-time PCR
- HDAC
- histone deacetylase
- MNase
- micrococcal nuclease
- qRT-PCR
- quantitative RT-PCR
- EBS
- Ets binding site
- CAT
- chloramphenicol acetyltransferase
- pol II
- RNA polymerase II.
REFERENCES
- 1. McGowan C. H. (2003) Prog. Cell. Cycle. Res. 5, 1–4 [PubMed] [Google Scholar]
- 2. Santamaria D., Ortega S. (2006) Front. Biosci. 11, 1164–1188 [DOI] [PubMed] [Google Scholar]
- 3. Le Cam L., Polanowska J., Fabbrizio E., Olivier M., Philips A., Ng Eaton E., Classon M., Geng Y., Sardet C. (1999) EMBO J. 18, 1878–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dulić V., Lees E., Reed S. I. (1992) Science 257, 1958–1961 [DOI] [PubMed] [Google Scholar]
- 5. Koff A., Giordano A., Desai D., Yamashita K., Harper J. W., Elledge S., Nishimoto T., Morgan D. O., Franza B. R., Roberts J. M. (1992) Science 257, 1689–1694 [DOI] [PubMed] [Google Scholar]
- 6. Donnellan R., Chetty R. (1999) FASEB J. 13, 773–780 [DOI] [PubMed] [Google Scholar]
- 7. Krude T. (2000) J. Biol. Chem. 275, 13699–13707 [DOI] [PubMed] [Google Scholar]
- 8. Schang L. M. (2003) Prog. Cell. Cycle. Res. 5, 103–124 [PubMed] [Google Scholar]
- 9. Ohtani K., DeGregori J., Nevins J. R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 12146–12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Botz J., Zerfass-Thome K., Spitkovsky D., Delius H., Vogt B., Eilers M., Hatzigeorgiou A., Jansen-Dürr P. (1996) Mol. Cell. Biol. 16, 3401–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Geng Y., Eaton E. N., Picón M., Roberts J. M., Lundberg A. S., Gifford A., Sardet C., Weinberg R. A. (1996) Oncogene 12, 1173–1180 [PubMed] [Google Scholar]
- 12. Pérez-Roger I., Solomon D. L., Sewing A., Land H. (1997) Oncogene 14, 2373–2381 [DOI] [PubMed] [Google Scholar]
- 13. Morgan D. O. (1995) Nature 374, 131–134 [DOI] [PubMed] [Google Scholar]
- 14. Geng Y., Weinberg R. A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10315–10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shiffman D., Brooks E. E., Brooks A. R., Chan C. S., Milner P. G. (1996) J. Biol. Chem. 271, 12199–12204 [DOI] [PubMed] [Google Scholar]
- 16. Sementchenko V. I., Watson D. K. (2000) Oncogene 19, 6533–6548 [DOI] [PubMed] [Google Scholar]
- 17. Oda N., Abe M., Sato Y. (1999) J. Cell. Physiol. 178, 121–132 [DOI] [PubMed] [Google Scholar]
- 18. Takai N., Miyazaki T., Nishida M., Nasu K., Miyakawa I. (2002) Int. J. Mol. Med. 9, 287–292 [DOI] [PubMed] [Google Scholar]
- 19. Seth A., Watson D. K. (2005) Eur. J. Cancer. 41, 2462–2478 [DOI] [PubMed] [Google Scholar]
- 20. Majérus M. A., Bibollet-Ruche F., Telliez J. B., Wasylyk B., Bailleul B. (1992) Nucleic Acids Res. 20, 2699–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie R. L., Gupta S., Miele A., Shiffman D., Stein J. L., Stein G. S., van Wijnen A. J. (2003) J. Biol. Chem. 278, 26589–26596 [DOI] [PubMed] [Google Scholar]
- 22. Sugimoto H., Sugimoto S., Tatei K., Obinata H., Bakovic M., Izumi T., Vance D. E. (2003) J. Biol. Chem. 278, 19716–19722 [DOI] [PubMed] [Google Scholar]
- 23. Bradford A. P., Conrad K. E., Wasylyk C., Wasylyk B., Gutierrez-Hartmann A. (1995) Mol. Cell. Biol. 15, 2849–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Janbandhu V. C., Singh A. K., Mukherji A., Kumar V. (2010) J. Biol. Chem. 285, 17453–17464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar V., Jayasuryan N., Reddi H., Sahal D., Panda S. K. (1998) Hybridoma 17, 157–164 [DOI] [PubMed] [Google Scholar]
- 26. Mukherji A., Janbandhu V. C., Kumar V. (2007) Biochem. J. 401, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rao S., Procko E., Shannon M. F. (2001) J. Immunol. 167, 4494–4503 [DOI] [PubMed] [Google Scholar]
- 28. Overbergh L., Valckx D., Waer M., Mathieu C. (1999) Cytokine 11, 305–312 [DOI] [PubMed] [Google Scholar]
- 29. Lakhtakia R., Kumar V., Reddi H., Mathur M., Dattagupta S., Panda S. K. (2003) J. Gastroenterol. Hepatol. 18, 80–91 [DOI] [PubMed] [Google Scholar]
- 30. Kim S., Kang J. K., Kim Y. K., Seo D. W., Ahn S. H., Lee J. C., Lee C. H., You J. S., Cho E. J., Lee H. W., Han J. W. (2006) Biochem. Biophys. Res. Commun. 342, 1168–1173 [DOI] [PubMed] [Google Scholar]
- 31. Gilles F., Raes M. B., Stéhelin D., Vandenbunder B., Fafeur V. (1996) Exp. Cell Res. 222, 370–378 [DOI] [PubMed] [Google Scholar]
- 32. Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. (2007) Cell. 130, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dittmer J. (2003) Mol. Cancer. 2, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Muthusamy N., Barton K., Leiden J. M. (1995) Nature 377, 639–642 [DOI] [PubMed] [Google Scholar]
- 35. Tanaka H., Terada Y., Kobayashi T., Okado T., Inoshita S., Kuwahara M., Seth A., Sato Y., Sasaki S. (2004) J. Am Soc. Nephrol. 15, 3083–3092 [DOI] [PubMed] [Google Scholar]
- 36. Topol L. Z., Tatosyan A. G., Ascione R., Thompson D. M., Blair D. G., Kola I., Seth A. (1992) Cancer Lett. 67, 71–78 [DOI] [PubMed] [Google Scholar]
- 37. Hollenhorst P. C., Jones D. A., Graves B. J. (2004) Nucleic Acids Res. 32, 5693–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bouchard M., Giannakopoulos S., Wang E. H., Tanese N., Schneider R. J. (2001) J. Virol. 75, 4247–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Santoni-Rugiu E., Falck J., Mailand N., Bartek J., Lukas J. (2000) Mol. Cell. Biol. 20, 3497–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kodadek T., Sikder D., Nalley K. (2006) Cell 127, 261–264 [DOI] [PubMed] [Google Scholar]
- 41. Métivier R., Penot G., Hübner M. R., Reid G., Brand H., Kos M., Gannon F. (2003) Cell 115, 751–763 [DOI] [PubMed] [Google Scholar]
- 42. Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 43. Becker P. B., Hörz W. (2002) Annu. Rev. Biochem. 71, 247–273 [DOI] [PubMed] [Google Scholar]
- 44. Imbalzano A. N., Kwon H., Green M. R., Kingston R. E. (1994) Nature 370, 481–485 [DOI] [PubMed] [Google Scholar]
- 45. Kwon H., Imbalzano A. N., Khavari P. A., Kingston R. E., Green M. R. (1994) Nature. 370, 477–481 [DOI] [PubMed] [Google Scholar]
- 46. Davie J. R., Spencer V. A. (1999) J. Cell. Biochem. 32, 141–148 [DOI] [PubMed] [Google Scholar]
- 47. Goodman R. H., Smolik S. (2000) Genes Dev. 14, 1553–1577 [PubMed] [Google Scholar]
- 48. Brehm A., Miska E. A., McCance D. J., Reid J. L., Bannister A. J., Kouzarides T. (1998) Nature 391, 597–601 [DOI] [PubMed] [Google Scholar]
- 49. Glass C. K., Rosenfeld M. G. (2000) Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 50. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 51. Benhenda S., Cougot D., Buendia M. A., Neuveut C. (2009) Adv. Cancer Res. 103, 75–109 [DOI] [PubMed] [Google Scholar]
- 52. Benn J., Schneider R. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11215–11219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kumar V. (2008) in The Pleiotropic Functions of the Viral Protein HBx and the Development of Liver Cancer (Kobarg J. ed) pp. 221–240, Research Signpost, Trivandrum, India [Google Scholar]
- 54. Maguire H. F., Hoeffler J. P., Siddiqui A. (1991) Science 252, 842–844 [DOI] [PubMed] [Google Scholar]
- 55. Avantaggiati M. L., Natoli G., Balsano C., Chirillo P., Artini M., De Marzio E., Collepardo D., Levrero M. (1993) Oncogene 8, 1567–1574 [PubMed] [Google Scholar]
- 56. Yoo Y. D., Ueda H., Park K., Flanders K. C., Lee Y. I., Jay G., Kim S. J. (1996) J. Clin. Invest. 97, 388–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.