Abstract
Methylmalonate-semialdehyde dehydrogenase (MSDH) belongs to the CoA-dependent aldehyde dehydrogenase subfamily. It catalyzes the NAD-dependent oxidation of methylmalonate semialdehyde (MMSA) to propionyl-CoA via the acylation and deacylation steps. MSDH is the only member of the aldehyde dehydrogenase superfamily that catalyzes a β-decarboxylation process in the deacylation step. Recently, we demonstrated that the β-decarboxylation is rate-limiting and occurs before CoA attack on the thiopropionyl enzyme intermediate. Thus, this prevented determination of the transthioesterification kinetic parameters. Here, we have addressed two key aspects of the mechanism as follows: 1) the molecular basis for recognition of the carboxylate of MMSA; and 2) how CoA binding modulates its reactivity. We substituted two invariant arginines, Arg-124 and Arg-301, by Leu. The second-order rate constant for the acylation step for both mutants was decreased by at least 50-fold, indicating that both arginines are essential for efficient MMSA binding through interactions with the carboxylate group. To gain insight into the transthioesterification, we substituted MMSA with propionaldehyde, as both substrates lead to the same thiopropionyl enzyme intermediate. This allowed us to show the following: 1) the pKapp of CoA decreases by ∼3 units upon binding to MSDH in the deacylation step; and 2) the catalytic efficiency of the transthioesterification is increased by at least 104-fold relative to a chemical model. Moreover, we observed binding of CoA to the acylation complex, supporting a CoA-binding site distinct from that of NAD(H).
Keywords: Coenzyme A, Enzyme Catalysis, Enzyme Kinetics, Fluorescence Resonance Energy Transfer (FRET), Thiol, Aldehyde Dehydrogenase, Coenzyme A Activation, Methylmalonate-semialdehyde Dehydrogenase, Transthioesterification
Introduction
Methylmalonate-semialdehyde dehydrogenases (MSDHs)4 belong to the CoA-dependent aldehyde dehydrogenases (ALDHs), which, together with their hydrolytic counterparts, make up the ALDH superfamily. ALDHs are known to be involved in many essential biological functions such as intermediary metabolism, detoxification, osmotic protection, and cellular differentiation. The enzymes catalyze the NAD(P)-dependent oxidation of a wide variety of aldehydes to their corresponding nonactivated or CoA-activated acids via a common two-step chemical mechanism. The acylation step leads to formation of a thioacyl enzyme intermediate, which then undergoes nucleophilic attack by a water or CoA molecule. Despite mechanistic similarities, previous studies have highlighted major differences in the kinetic mechanism of ALDHs depending on the nature of the deacylation step. In hydrolytic ALDHs, kinetic data support an ordered sequential mechanism in which NAD(P)H dissociates last (1–4). By contrast, CoA-dependent ALDHs exhibit a ping-pong mechanism in which the release of the reduced cofactor occurs before the transthioesterification step (5, 6). Whereas mechanistic and structural aspects have been studied extensively for hydrolytic ALDHs (7–14), considerably less is known about CoA-dependent ALDHs.
MSDHs are present in a wide variety of organisms ranging from bacteria and archaea to plants and mammals where they are described to play a diversity of roles. The MSDHs from Pseudomonas aeruginosa (15, 16) and Streptomyces coelicolor (17) are involved in valine catabolism, and in Bacillus subtilis (18) and Rhizobium leguminosarium bv. viciae (19), MSDH is putatively implicated in myoinositol catabolism. In mammals, MSDH is a mitochondrial enzyme that participates in the distal portions of the valine and pyrimidine catabolic pathways (20). Over the last 2 decades, a possible correlation of organic acidemia with MSDH deficiency has been explored (21, 22). For example, Roe et al. (23) proposed that a psychomotor delay associated with methylmalonyl aciduria is a direct consequence of a lack of MSDH activity. More recently, it was suggested that MSDH may play a role in root development and leaf sheath elongation in rice (24).
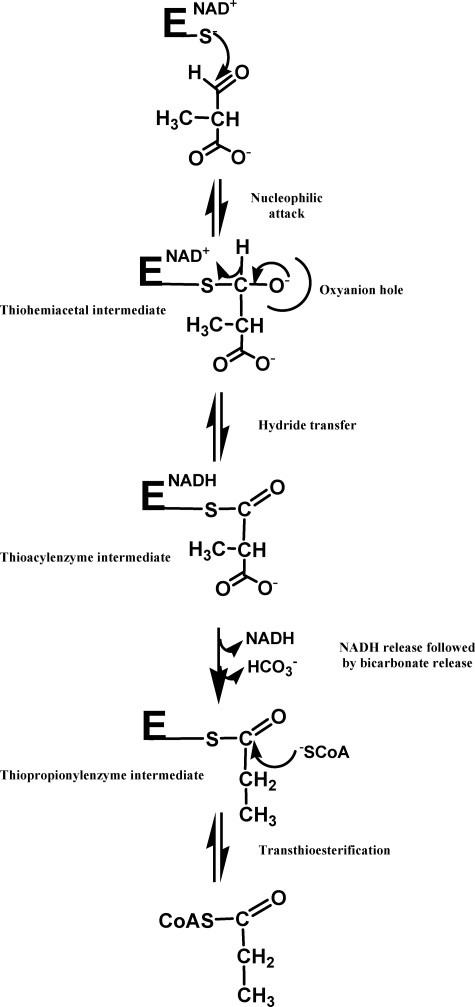
Recently, our group undertook a study of the catalytic mechanism of MSDH from B. subtilis (25). The homotetrameric enzyme catalyzes the NAD+-dependent oxidation of methylmalonate semialdehyde (MMSA) and malonate semialdehyde to propionyl- and acetyl-CoA, respectively. Therefore, MSDH is the only member of the ALDH superfamily known to date to catalyze a β-decarboxylation in addition to the transthioesterification step. From the detailed mechanistic characterization of the MSDH-catalyzed reaction, our studies suggested that NAD binding elicits a structural imprinting of the apoenzyme, which may explain the lag phase observed in the activity assay. Half-site reactivity for MSDH was also demonstrated, with two subunits being active per tetramer. Finally, kinetic studies showed the following: (i) the rate constant associated with the acylation step is high (kac >1000 s−1); (ii) the rate-limiting β-decarboxylation step occurs on the thioacyl enzyme intermediate after NADH release and before transthioesterification; and (iii) bicarbonate is the end product of the thioacyl enzyme decarboxylation (25). Together, these results have led to a proposal for the catalytic mechanism (Scheme 1).
SCHEME 1.
Schematic representation of MSDH-catalyzed reaction with MMSA as substrate (adapted from Ref. 25). When PPA is used as substrate, CoA attacks the same thiopropionyl enzyme intermediate. The acylation step includes all the events up to the hydride transfer, and the deacylation step includes release of NADH up to release of thiopropionyl-CoA.
Nonetheless, several questions about key aspects of the MSDH-catalyzed reaction remain. This is the case for the molecular factors that are responsible for the recognition of a substrate with a carboxylate group. It is not also known whether binding of CoA to MSDH activates its thiol group. Indeed, the rate-limiting step of MSDH occurs before the transthioesterification with MMSA and thus prevented measurement of the pKapp of CoA.
In this study, two invariant Arg residues, expected to stabilize the binding of MMSA via interactions with its carboxylate group, were substituted by Leu. The kinetic data for R124L, R301L, and R124L/R301L MSDHs are described. The results obtained under pre-steady conditions show that both Arg residues participate not only in MMSA binding via stabilizing interactions between the guanidinium groups and the carboxylate but also in the formation of an efficient MSDH·NAD·MMSA ternary complex. When propionaldehyde (PPA), a decarboxylated analog of MMSA that leads to the same thiopropionyl enzyme intermediate, is used as an alternative substrate, the rate-limiting step of the reaction catalyzed by R124L MSDH is then associated with the transthioesterification process. This gave us the opportunity to determine the pKapp of CoA in the deacylation step, which shifts from ∼9.6 to 6.8. When compared with a chemical model, the catalytic efficiency of the transthioesterification for the wild-type MSDH was increased at least 104-fold. Therefore, CoA binding to MSDH optimizes its positioning relative to the thioacyl enzyme intermediate and favors a significant decrease of the pKapp of its thiol group. Moreover, we observed that for the wild-type MSDH (and in a lesser extent for R124L and R301L MSDHs), the catalytic efficiency of the acylation step increased in the presence of CoA, due to an enhancement in the apparent affinity of the enzyme for PPA. These data show for the first time that the CoA-binding site is distinct from that of NAD.
EXPERIMENTAL PROCEDURES
Materials
NAD was purchased from Roche Applied Science. CoA, propionaldehyde, pyruvate, and lactate dehydrogenase (LDH) were from Sigma. Desulfo-CoA was prepared as described by Chase et al. (26). MMSA was synthesized as described by Kupiecki and Coon (27). Its concentration was determined by titration with MSDH.
Site-directed Mutagenesis, Production, and Purification of Wild-type and Mutated MSDHs from B. subtilis
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene). Wild-type, R124L, R301L, and R124L/R301L MSDHs were produced and purified using a procedure described previously (25). Enzyme concentrations were determined spectrophotometrically by using molar absorption coefficients of 2.04 × 105 m−1·cm−1 at 280 nm for wild-type and mutated MSDHs. In the text, enzyme concentrations are expressed per monomer (normality (n)).
Kinetic Parameters for Wild-type and Mutated MSDHs under Steady-state Conditions
Initial rate measurements were carried out at 30 °C on an SAFAS UV mc2® spectrophotometer, by following the appearance of NADH at 340 nm in 50 mm potassium phosphate (pH 8.2). Prior to kinetic measurements in the presence of MMSA, R301L MSDH was preincubated at 30 °C with 2 mm NAD, whereas R124L and R124L/R301L MSDHs were preincubated with 2 mm NAD and 0.5 mm CoA, respectively. When PPA was used as substrate, wild-type and all mutated MSDHs were preincubated at 30 °C with 2 mm NAD. In all the preincubated mixtures, the concentration of MSDHs was 160 μn. The initial rate data were fit to the Michaelis-Menten equation using nonlinear least squares regression analysis to determine the kcat and Km values. All Km values were determined at saturating concentrations of the other substrates as follows: 1 mm MMSA, 1 mm NAD, and 3 mm CoA for R124L and R301L MSDHs; 0.05 m PPA, 1 mm NAD, and 0.5 or 2 mm CoA for wild-type and R124L MSDHs. In the case of R301L MSDH, the substantial increase in the Km value for CoA prevented accurate determination of the Km values for NAD and PPA. For the pH dependence studies, data were collected at 30 °C over a pH range of 5.00–8.75 with a reaction buffer consisting of 22 mm succinic acid, 29 mm imidazole, and 29 mm diethanolamine adjusted with NaCl to a constant ionic strength of 0.06 m (buffer A) and analyzed as described by Marchal et al. (4).
Pre-steady-state Kinetic Measurements
Pre-steady-state kinetic analyses were carried out on an SX18MV-R stopped-flow apparatus (Applied PhotoPhysics), and collected data were analyzed using the SX18MV-R software package.
Kinetics of the Acylation Step and of NADH Dissociation of R124L, R301L, and R124L/R301L MSDHs with MMSA as Substrate
To study the acylation step, progress curves of NADH production were recorded at 340 nm and at 30 °C in 50 mm potassium phosphate (pH 8.2), in the absence of CoA or at subsaturating concentrations of CoA (i.e. 50 μm) for R124L and R124L/R301L MSDHs. One syringe was filled with 16 μn MSDH and 1 mm NAD (after mixing), and the other contained MMSA at various concentrations. No saturation was observed irrespective of the mutated MSDH, and therefore k2 values were estimated from the slope of the linear part of the kobs = f ([MMSA]) profile.
To evaluate the rate of NADH dissociation from the thioacyl enzyme·NADH complex, the coupled pyruvate/LDH assay was used as an NADH-trapping system. One syringe was filled with 16 μn MSDH and 1 mm NAD, and the other contained 0.5 mm MMSA (which is a subsaturating concentration), 16 μn LDH, and 10 mm pyruvate. Experiments were carried out in the absence of CoA (or at a subsaturating concentration of CoA, i.e. 50 μm for R124L and R124L/R301L MSDHs) in 50 mm potassium phosphate (pH 8.2) at 30 °C. Data were fit to a biphasic expression.
Kinetic Properties of Wild-type, R124L, R301L, and R124L/R301L MSDHs with PPA as Substrate
Kinetic analyses of the acylation step were carried out at 30 °C in 50 mm potassium phosphate (pH 8.2). For this, progress curves of NADH production were recorded at 340 nm, in the absence of CoA or in the presence of desulfo-CoA. The k2 values were obtained by nonlinear least square regression of kobs, measured at subsaturating concentrations of substrate PPA, except for the wild-type MSDH for which the second-order rate constant k2 was deduced from the Kapp and the kobs max values. Alternatively, when CoA was added to the reaction mixture, the burst of NADH production associated with acylation was selectively monitored using fluorescence resonance energy transfer (FRET). Following selective excitation of MSDH Trp residues at 295 nm, the fluorescence emission of NADH was measured using a 395-nm cutoff filter. In this experiment, one syringe contained 16 μn wild-type or mutated MSDHs and 1 mm NAD, and the other contained PPA at various concentrations. In the experiments where CoA and desulfo-CoA were used, they were added to the syringe containing the enzyme. Data were fit to Equation 1 to determine kac and Kapp values, where S represents the substrate PPA.
For wild-type and R124L MSDHs, the pH dependence of the acylation rate constant was studied at 30 °C over a pH range 5.00–8.75 in buffer A. One syringe was filled with 16 μn MSDH, 1 mm NAD, and 500 μm CoA or 250 μm desulfo-CoA and the other contained 0.5 m PPA. Formation of NADH was measured by monitoring the FRET signal using a 395-nm cutoff filter. Data were analyzed as described by Marchal et al. (4).
The rate of NADH dissociation from the thioacyl enzyme·NADH complex was determined as described above with MMSA as substrate. One syringe was filled with 16 μn MSDH, 16 μn LDH, and 10 mm pyruvate, and the other contained 1 mm NAD and 0.5 m PPA. Experiments were carried out in the absence of CoA for R124L and R301L MSDHs in 50 mm potassium phosphate (pH 8.2) at 30 °C. For the wild-type MSDH, desulfo-CoA was added to the syringe containing PPA and NAD. Data were fit to a biphasic expression.
RESULTS
Rationale for the Mutations
On the basis of sequence comparison of MSDHs to those of other ALDHs (including hydrolytic and CoA-dependent ALDHs) two invariant Arg residues in MSDHs, i.e. Arg-124 and Arg-301, appear to be only conserved in nonphosphorylating glyceraldehyde-3-phosphate dehydrogenases (GAPNs) (Fig. 1). In GAPN from Streptococcus mutans, these Arg residues were shown to be critical in the binding of d-glyceraldehyde 3-phosphate through stabilizing interactions with the C-3 phosphate (13, 28). Moreover, the x-ray structure of the MSDH from B. subtilis in complex with NAD (Protein Data Bank accession number 1t90) superimposes well on the GAPN structures (Protein Data Bank accession numbers 1qi6 and 2qe0). In particular, Arg-124 and Arg-301 are found on the same secondary structural elements. Therefore, it was reasonable to postulate for MSDH a major role for these residues in the binding of a substrate such as MMSA, which bears a carboxylate group. To validate this assumption, the roles of Arg-124 and Arg-301 have been probed by site-directed mutagenesis. Introduction of a residue such as leucine at positions 124 and 301 would provide experimental support for the role of the electrostatic interactions between the guanidinium groups of Arg-124 and Arg-301 and the carboxylate group of MMSA, while keeping the hydrophobic properties of the aliphatic part of an arginine side chain.
FIGURE 1.
Sequence alignment of the regions around the conserved catalytic Cys-302 and Arg residues in MSDHs and comparison with other ALDHs. The sequence alignment includes some MSDHs whose activity has been demonstrated. Sequence alignment was performed with the BioEdit software, and the numbering of amino acid residues is according to Wang and Weiner (8). The conserved catalytic Cys-302 and the Arg-124 and Arg-301 residues, which are only conserved in MSDH and GAPN sequences, are highlighted. Sequences of 19 GAPNs and 40 hydrolytic and CoA-dependent ALDHs (excluding GAPNs) from the EMBL, Swiss-Prot, GenBankTM, and Pir databases were aligned to determine the consensus sequences.
Kinetic Properties of R124L, R301L, and R124L/R301L MSDHs with MMSA as Substrate
The lag phase exhibited by wild-type MSDH during activity assays with MMSA as substrate was eliminated by preincubating the apoenzyme with NAD, as described previously (25). A similar behavior was observed for R301L MSDH but not for R124L and R124L/R301L MSDHs, for which incubation with both NAD and CoA was required. Therefore, prior to any kinetic studies, R301L MSDH was preincubated with 2 mm NAD, and R124L and R124L/R301L MSDHs with 2 mm NAD and 500 μm CoA. For the wild-type MSDH-catalyzed reaction, the rate-limiting step was previously shown to be associated with the β-decarboxylation process within the deacylation step (25). Therefore, before interpreting the kinetic data of the mutated MSDHs, it was necessary to determine whether the rate-limiting step was the same as the wild-type MSDH. Accordingly, fast kinetic experiments were carried out with the three mutated MSDHs at pH 8.2 and 30 °C in the presence of 2 mm NAD and subsaturating concentrations of MMSA (i.e. 2 mm) and, in the case of R124L and R124L/R301L MSDHs, CoA (50 μm after dilution). A burst magnitude of 2 mol of NADH/mol of tetramer was observed, irrespective of the mutated MSDH. Therefore, these data clearly indicate that all mutated MSDHs also exhibited half-site reactivity, as described for the wild-type MSDH. More importantly, the kobs values determined at 2 mm MMSA were 82-, 36-, and 150-fold higher than the kcat values for R124L, R301L, and R124L/R301L MSDHs, respectively. These data indicate that the rate-limiting step still takes place after hydride transfer.
Steady-state and Pre-steady-state Kinetic Parameters
The kinetic parameters determined at pH 8.2 and at 30 °C under steady-state conditions are summarized in Table 1. Substituting Leu for Arg-301 or Arg-124 resulted in a 5-fold increase in Km for CoA. The Km values for NAD and MMSA decreased from 19- to 38-fold and from 3- to 10-fold for the R301L and R124L MSDHs, respectively, whereas the kcat values decreased from 6- to 18-fold. It was not possible to interpret with confidence the significance of the decrease in Km values for MMSA (and for NAD) observed for the R124L and R301L MSDHs as the Km value cannot be equated with the dissociation constant of the enzyme·substrate complex(es). By contrast, comparison of the apparent affinity constants (Kapp) for MMSA determined under pre-steady-state conditions for the wild-type and the mutated MSDHs provides a more accurate picture of the role(s) of each arginine side chain in MMSA binding (see below). Indeed, the rate constants associated with all the steps following hydride transfer do not contribute to the expression for Kapp. Thus, in this case, the Kapp values are expected to be closer to dissociation constants. For the R124L/R301L MSDH, because of lack of saturation with substrates and very low steady-state initial rates (<10−2 s−1), it was not possible to determine the kcat and Km values.
TABLE 1.
Kinetic parameters of the reactions catalyzed by wild-type and mutated MSDHs under steady-state conditions with MMSA as substrate
Kinetic parameters were deduced from nonlinear least square regression of experimental data sets, according to the Michaelis-Menten equation. All Km values were determined at saturating concentrations of the other substrates, and kcat values are expressed per active subunit (i.e. two active subunits per tetramer). The steady-state initial rates of the reaction of mutated MSDHs were measured at 30 °C in 50 mm potassium phosphate buffer (pH 8.2), under similar conditions as those for the wild-type MSDH. Mutated MSDHs were preincubated with 2 mm NAD or with 2 mm NAD and 500 μm CoA at 30 °C, prior to making the kinetic measurements.
| Km MMSA | Km CoAa | Km NADa | kcatb | |
|---|---|---|---|---|
| mm | μm | mm | s−1 | |
| Wild typec | 0.06 ± 0.01 | 120 ± 20 | 2.30 ± 0.06 | 2.2 ± 0.2 |
| R124L | 0.006 ± 0.001 | 570 ± 70 | 0.06 ± 0.01 | 0.060 ± 0.01 |
| R301L | 0.023 ± 0.005 | 620 ± 80 | 0.12 ± 0.01 | 0.14 ± 0.02 |
| R124L/R301L | <10−2d |
a After dilution of the preincubated enzymes, the remaining NAD (12 μm) and CoA (3 μm) concentrations were taken into account for determination of the Km values.
b For all MSDHs, the rate-limiting step remains associated with the so-called “deacylation” step (i.e. all the steps occurring after hydride transfer).
c Data are from Ref. 25.
d Due to lack of saturation with substrates and very low steady-state initial rates (<10−2 s−1), it was not possible to determine the kcat and Km values.
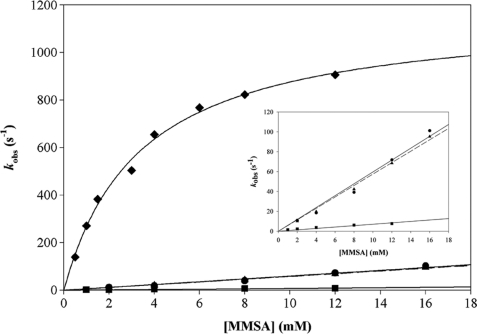
Under pre-steady-state conditions, saturation kinetics were not observed with respect to MMSA in the acylation step up to 12 mm, irrespective of the mutated MSDH (Fig. 2). For R124L and R301L MSDHs, the estimated k2 values of 6000 and 5400 m−1 s−1 are 55- and 62-fold smaller respectively than the k2 value for the wild-type MSDH (Table 2). For R124L/R301L MSDH, the k2 value is 512-fold smaller than that of the wild-type MSDH. The kobs value at 12 mm MMSA is at least 37-fold lower than the kobs (max) of the wild-type MSDH irrespective of the mutated MSDH. Therefore, the 55–512-fold decrease in k2 likely reflects both a decrease of the kobs (max) value and an increase in the Kapp value.
FIGURE 2.
Determination of the catalytic parameters of the acylation step for R124L, R301L, and R124L/R301L MSDH-catalyzed reactions under pre-steady-state conditions with MMSA as substrate. The appearance of NADH was monitored at 340 nm on an SX18MV-R stopped-flow apparatus (Applied PhotoPhysics) at 30 °C. The final concentration of MSDH was 16 μn (expressed in subunit) with the MMSA concentration ranging from 1 to 16 mm. For each substrate concentration, experimental data were analyzed by nonlinear regression using the SX18MV-R software package to obtain kobs. As saturation with MMSA was not observed irrespective of the mutated MSDHs, k2 were extracted from the resulting linear equations. Key: R124L MSDH (●, solid line); R301L MSDH (▴, dashed line), R124L/R301L MSDH (■, solid line), and wild-type MSDH (♦, solid line, from Ref. 25). The inset details the results obtained on the three mutated MSDHs.
TABLE 2.
Determination of the kinetic parameters for the acylation step for R124L, R301L, and R124L/R301L MSDHs under pre-steady-state conditions with MMSA as substrate
Pre-steady-state measurements were performed at 30 °C in 50 mm potassium phosphate buffer (pH 8.2). Mutated MSDHs were preincubated with 2 mm NAD (R301L MSDH) or with 2 mm NAD and 500 μm CoA (R124L and R124L/R301L MSDHs) at 30 °C prior to making the kinetic measurements. The k2 values of the acylation step were obtained by linear regression of kobs determined under subsaturating concentrations of substrate MMSA (up to 16 mm). For the wild-type MSDH, the second-order rate constant k2 was obtained by dividing kobs max by Kapp.
| Kapp MMSA | kobsmax MMSA | k2 MMSA | |
|---|---|---|---|
| mm | s−1 | m−1 s−1 | |
| Wild typea | 3.5 ± 1.0 | 1200 ± 180 | 3.4 × 105 |
| R124L | >15 | 6 × 103 | |
| R301L | >15 | 5.4 × 103 | |
| R124L/R301L | >15 | 6.5 × 102 |
a Data are from Ref. 25.
The lower kobs (max) of the acylation step for all mutated MSDHs, which was measured at pH 8.2, could have been the consequence, in part, of a significant increase in the pKapp of Cys-302. To evaluate this hypothesis, the acylation rate constant was determined for R124L MSDH, over a pH range of 5–8, at 4 mm MMSA, a subsaturating concentration for this mutant. The resulting pH-k2 curve exhibited an increasing monosigmoidal profile with a pKapp value of 6.0, similar to that observed for the wild-type MSDH, which can be assigned to Cys-302, and a k2 max value of 5 × 103 m−1 s−1 at pH 8.0 (curve not shown). Therefore, substituting Arg-124 with Leu does not result in an increase in the pKapp of Cys-302 compared with the wild-type MSDH. Because R124L and R301L MSDHs exhibit similar catalytic parameters, it seems reasonable to postulate a similar pKapp of Cys-302 for R301L MSDH.
NADH Release from the Thioacyl Enzyme·NADH Complex Is Not Rate-limiting for R124L, R301L, and R124L/R301L MSDHs
The rate constant of NADH dissociation from the thioacyl enzyme·NADH complex was determined using LDH as an NADH-trapping system and MMSA as substrate. The experiments were performed using R124L, R301L, and R124L/R301L MSDHs at pH 8.2 and 30 °C under pre-steady-state conditions in the absence of CoA (R301L) or at subsaturating concentrations of CoA (50 μm after dilution) for R124L and R124L/R301L MSDHs. The resulting progress curves were fit to a biphasic expression. Under the experimental conditions used, the rate of NADH oxidation by LDH (130 s−1) was higher than the apparent rate constants of 2.4, 1.5, and 1 s−1, which can be assigned to release of NADH from the thioacyl enzyme·NADH complex for the R124L, R301L, and R124L/R301L MSDHs, respectively (supplemental Fig. S1). These rates, which are likely to underestimate the intrinsic rates of NADH release, are 40-, 11-, and 100-fold higher than the kcat values. Therefore, the rate-limiting step must occur after NADH release from the thioacyl enzyme·NADH complex. Because CoA acts on the same thiopropionyl enzyme intermediate (Scheme 1) regardless of the substrate used (i.e. MMSA or PPA), and the kcat values for R124L and R301L MSDHs are at least 20-fold lower with MMSA compared with those with PPA as substrate (Tables 1 and 3; see also part “Results,” R124L and R301L MSDHs with PPA as substrate), we can assume that the rate-limiting step for the mutants is also associated with the β-decarboxylation process. However, this hypothesis remains to be demonstrated directly.
TABLE 3.
Kinetic parameters for the reactions catalyzed by the wild-type and mutated MSDHs under steady-state conditions with PPA as substrate
Kinetic parameters were deduced from nonlinear least square regression of experimental data sets, according to the Michaelis-Menten equation. All Km values were determined at saturating concentrations of the other substrates, and kcat values are expressed per active subunit (i.e. two active subunits per tetramer). The steady-state initial rates of the reaction of wild-type and mutated MSDHs were measured at 30 °C in 50 mm potassium phosphate buffer (pH 8.2). Wild-type and mutated MSDHs were preincubated with 2 mm NAD at 30 °C before kinetic measurements. ND, not determined.
| Km PPA | Km CoA | Km NADa | kcatb | kcat/Km CoA | |
|---|---|---|---|---|---|
| mm | μm | μm | s−1 | m−1 s−1 | |
| Wild type | 9.8 ± 0.3 | 85 ± 10 | 33 ± 13 | 12.6 ± 0.2 | 1.5 × 105 |
| R124L | 9.3 ± 1.1 | 330 ± 35 | 38 ± 1 | 2.8 ± 0.4 | 8.5 × 103 |
| R301L | ND | >1000 | ND | >3 | 3.0 × 103c |
a After dilution of the preincubated enzyme, the remaining NAD concentration (2 μm) was taken into account for determination of the Km values.
b For all MSDHs, the rate-limiting step remains associated with the so-called deacylation step (i.e. all the steps occurring after hydride transfer).
c This value is only an estimate, since the kinetic parameters cannot be determined with accuracy due to lack of saturation with CoA.
Kinetic Properties of R124L, R301L, and R124L/R301L and Wild-type MSDHs with PPA as Substrate
Acylation with PPA leads to formation of the same thiopropionyl enzyme intermediate that arises following β-decarboxylation of the MMSA substrate (see Scheme 1), and on which CoA attacks. Thus, we hoped that detailed kinetic studies of a reaction lacking the β-decarboxylation step would furnish additional insights into the transthioesterification step, provided that this step became rate-limiting. Accordingly, it was necessary to determine all the kinetic parameters for the wild-type, R124L, and R301L MSDHs.
In contrast to what was observed for R124L and R124L/R301L MSDHs with MMSA as substrate, it was possible to eliminate the lag phase in the activity assay with PPA as substrate by preincubation with 2 mm NAD in the absence of CoA. Thus, in the kinetic experiments detailed below, all MSDHs, including R124L and R124L/R301L mutants, were preincubated with 2 mm NAD only.
Wild-type MSDH
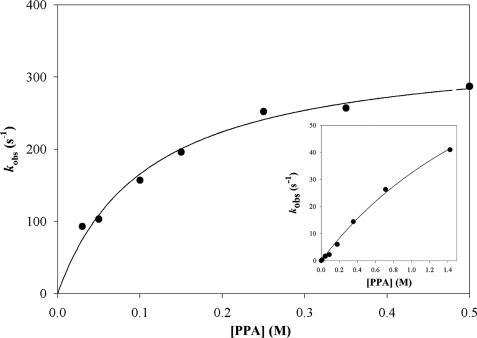
The kinetic parameters determined at pH 8.2 and at 30 °C under steady-state conditions with PPA are summarized in Table 3. The wild-type enzyme displayed Km values of 33 and 85 μm for NAD and CoA, respectively and a kcat value of 12.6 s−1. Under pre-steady-state conditions, fast kinetic experiments were carried out at pH 8.2 and 30 °C in the presence of 1 mm NAD and 0.05–1.40 m PPA. First, the burst magnitude of NADH production showed that only 2 mol of NADH were formed per mol of tetramer irrespective of the PPA concentration used. Therefore, MSDH also exhibits half-site reactivity in the presence of PPA. Second, no saturating kinetic effect with respect to PPA was observed up to 0.6 m PPA (inset of Fig. 3 and Table 4). A second-order rate constant k2 of 36 m−1 s−1 can be deduced from the slope of the linear part of the curve, although only an estimation of the Kapp of PPA (2.6 m) and the kobs (max) (120 s−1) values can be obtained.
FIGURE 3.
Determination of the catalytic parameters for the acylation step of wild-type MSDH in the presence of PPA, under pre-steady-state conditions. The appearance of NADH was selectively monitored using FRET on an SX18MV-R stopped-flow apparatus (Applied PhotoPhysics) at 30 °C. One syringe is filled with 16 μn wild-type MSDH (expressed in subunit), 1 mm NAD, and 500 μm CoA (final concentrations), and the other one contained PPA at various concentrations. Following selective excitation of Trp residues at 295 nm, the fluorescence emission of NADH was measured using a 395-nm cutoff filter. Rate constants kobs were fit to Equation 1, which gave a kobs (max) of 340 ± 20 s−1 and a Kapp for PPA of 0.15 ± 0.02 m. The inset shows the results obtained in the absence of CoA or desulfo-CoA.
TABLE 4.
Determination of the kinetic parameters of the acylation step for the reactions catalyzed by wild-type, R124L, and R301L MSDHs with PPA as substrate
Pre-steady-state measurements were performed at 30 °C in 50 mm potassium phosphate buffer (pH 8.2) in the absence or the presence of CoA. Wild-type and mutated MSDHs were preincubated with 2 mm NAD at 30 °C prior to making the kinetic measurements. In the absence of CoA, the appearance of NADH was monitored by following the change in absorbance at 340 nm. The k2 values were obtained by linear regression of kobs measured at subsaturating concentrations of PPA up to 1.4 m. For the wild-type MSDH, the second-order rate constant k2 was obtained from estimations of the Kapp and the kobs (max) values. In the presence of 500 μm CoA (wild-type) or 250 μm CoA (R124L and R301L MSDHs), formation of NADH was measured by monitoring the FRET signal using a 395-nm cutoff filter, upon selective excitation of Trp residues at 295 nm. The concentration of PPA was varied from 0 to 0.54 and 0 to 0.35 m for R124L and R301L mutants, respectively (see supplemental Fig. S4).
| −CoA |
+CoA |
||||
|---|---|---|---|---|---|
| Kapp PPA | kobs (max) | k2 | Kapp PPA | kobs (max) | |
| m | s−1 | m−1 s−1 | m | s−1 | |
| Wild type | 2.6 ± 0.6 | 120 ± 20 | 36 | 0.15 ± 0.02 | 340 ± 20 |
| R124L | >3 | 110 | 0.18 ± 0.03 | 110 ± 7 | |
| R301L | >3 | 114 | 0.16 ± 0.05 | 117 ± 16 | |
More importantly, we determined a kobs value of 5 s−1 at 0.05 m PPA (i.e. a saturating concentration under steady-state conditions), which was 2.5-fold lower than the kcat value of 12.6 s−1. This value showed that the kinetics of the acylation step under steady-state conditions are CoA-dependent in contrast to what was observed with MMSA. To determine the contribution of the CoA binding to the acylation kinetics, NADH production was monitored by following change in absorbance or fluorescence. Unfortunately, addition of 500 μm CoA did not allow an accurate determination of the rate constant associated with acylation because the acylation and deacylation steps could not be kinetically resolved due to their relative magnitudes (i.e. a higher rate constant for the acylation step but a much smaller signal amplitude compared with that observed under steady-state conditions). Therefore, we took advantage of the fact that one (or several) of the five tryptophan residues of wild-type MSDH could participate as donors in FRET with NADH acceptor, to selectively monitor the burst of NADH production associated with acylation. The advantage of this experimental approach is that it eliminates a contribution of the steady-state phase to the fluorescence signal, as NADH is released before the transesterification step.
Following selective excitation of Trp residues at 295 nm, the fluorescence emission of NADH was measured using a 395-nm cutoff filter. Fast kinetic experiments were then carried out at pH 8.2 and 30 °C in the presence of 500 μm CoA, 1 mm NAD, and 0.05–0.5 m PPA. At 0.05 m PPA, the kobs value is already 8-fold higher than the kcat value (i.e. 103 versus 12.6 s−1). A substrate-saturating effect was observed, allowing determination of kobs (max) and Kapp values of 340 s−1 and 0.15 m, respectively (Fig. 3 and Table 4). Thus, addition of saturating concentrations of CoA led to a 2.8-fold increase in kobs (max) and a 17-fold decrease in Kapp for PPA. Similar experiments were performed in the presence of 1 mm NAD and 0.5 m PPA, with CoA concentrations ranging from 0 to 500 μm, resulting in determination of a Kapp of 35 μm for CoA and a kobs (max) of 330 s−1 (supplemental Fig. S2). The CoA dependence of the acylation step was further supported by single turnover experiments carried out in the presence of 1 mm NAD, 0.05 m PPA, and 50 or 250 μm desulfo-CoA, which is a nonactive CoA analog. Under these experimental conditions, accumulation of the thioacyl enzyme was observed due to the very low steady-state rate, which is now associated with the hydrolysis of the thioacyl enzyme intermediate (i.e. 10−3 s−1). The kobs values of 45 and 96 s−1 are ∼9 and 19-fold higher than the kobs determined in the absence of CoA, and 3.5- and 7.6 higher than the kcat value. Clearly, a desulfo-CoA dependence of the acylation step was also observed. Moreover, all the results showed that the rate-limiting step takes place after hydride transfer and could be associated with NADH dissociation, transthioesterification, product release, or any potential isomerization steps.
As described earlier, the rate constant for NADH dissociation from the thioacyl enzyme·NADH complex was determined using LDH as an NADH-trapping system. The experiment was performed at pH 8.2 and 30 °C under pre-steady-state conditions in the presence of 1 mm NAD, 0.5 m PPA, and 50 or 100 μm desulfo-CoA. This yielded an apparent rate constant of 13 s−1, which is similar to the kcat value, irrespective of the desulfo-CoA concentration used (supplemental Fig. S3). This result, together with the kinetic data obtained under steady-state conditions, showed the following: (i) NADH dissociation occurs prior to transthioesterification, as already demonstrated for the MSDH-catalyzed reaction in the presence of MMSA; (ii) NADH dissociation is likely rate-limiting for the overall reaction; and (iii) the catalytic efficiency (kcat/Km) of the thioester exchange with CoA is high, i.e. at least 1.5 × 105 m−1 s−1.
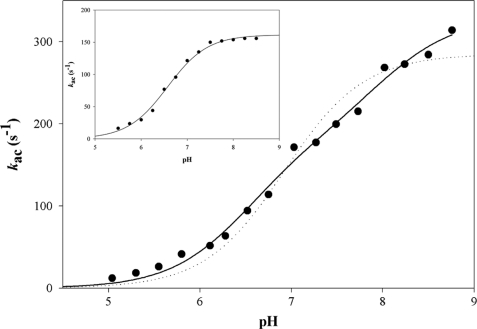
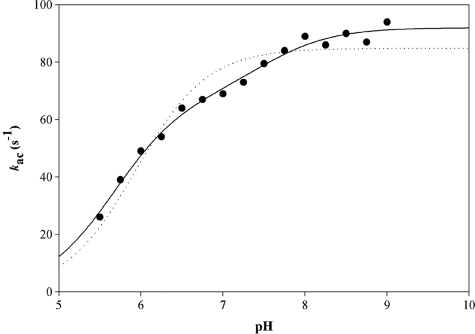
The pH dependence of the acylation rate constant was determined in the presence of 1 mm NAD and saturating concentrations of 500 μm CoA. To avoid any kac/K contribution in the kac-pH profile, experiments were carried out at 0.5 m PPA, which was shown to be mostly saturating (3-fold the Kapp value) over the full pH range (5.00–8.75). Formation of NADH was measured by monitoring the FRET signal using a 395-nm cutoff filter. The pH-kac curve exhibited an increasing bisigmoidal profile that can be related to the contribution of two ionizable groups, one with a pKapp of 6.5 that must be deprotonated for acylation and one with a pKapp of 7.9 whose deprotonation increased the acylation rate by 1.9-fold with rate constants kac (i) of 176 s−1 and kac (max) of 330 s−1, respectively (Fig. 4). When desulfo-CoA was used instead of CoA, the pH-kac curve exhibited an increasing monosigmoidal profile that can be related to the contribution of one ionizable group with a pKapp of 6.5 and a rate constant kac of 160 s−1 (Fig. 4, inset).
FIGURE 4.
pH dependence of the acylation rate kac for the wild-type MSDH-catalyzed reaction with PPA as substrate. Pre-steady-state data were collected at 30 °C on an SX18MV-R stopped-flow apparatus (Applied PhotoPhysics) using the 20-μl cell, by rapidly mixing 16 μn wild-type MSDH, 1 mm NAD, 0.5 m PPA, and 500 μm CoA (final concentrations) over a pH range of 5.00–8.75 in buffer A. At pH values lower than 5, MSDH was not stable. Following selective excitation of Trp residues at 295 nm, formation of NADH was measured by monitoring the FRET signal using a 395-nm cutoff filter. Each individual point is the average of at least six measurements with an error of <5%. Experimental data (●) were fit by nonlinear least square regression analysis against a two-pKa model which gave pKapp values of 6.5 ± 0.1 and 7.9 ± 0.3 with rate constants kac (i) of 176 ± 28 and kac (max) of 330 ± 16 s−1, respectively (solid line). The data do not fit to a one-pKa model (dotted line). The inset shows the results obtained in the presence of 250 μm desulfo-CoA. Experimental data were fit by nonlinear least square regression analysis against a one-pKa model, which gave a pKapp of 6.5 ± 0.1 with rate constant kac of 162 ± 2 s−1 (solid line).
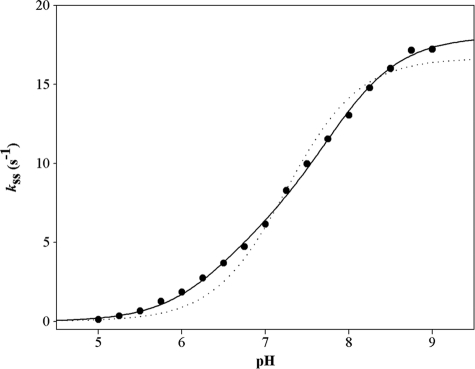
Additionally, the pH dependence of the steady-state rate constant was determined at 30 °C in the presence of 1 mm NAD, 50 mm PPA (5-fold the Km value), and 500 μm CoA. Analysis of the pH-kss curve provided similar pKapp values to those extracted from the pH-kac curve (i.e. 6.5 and 7.8), as well as a kss (i) of 6 s−1 and kss (max) of 18 s−1 (Fig. 5). Differences in buffer composition and ionic strength (0.15 m versus 0.06 m) likely explain the slightly higher value of kss (max) (18 s−1) relative to kcat (12.6 s−1).
FIGURE 5.
pH dependence of the steady-state rate constant kss for wild-type MSDH with PPA as substrate. The pH dependence of kss was studied at 30 °C over the pH range 5.0–9.0 in buffer A. The remaining experimental conditions were as follows: 1 mm NAD, 0.05 m PPA, and 500 μm CoA. Experiments were done twice with experimental errors on the individual points ≤ 5%. Data (●) were fit by nonlinear least square regression analysis against a two-pKapp model that yielded pKapp values of 6.5 ± 0.1 and 7.8 ± 0.1, and rate constants kss(i) of 6.0 ± 0.7 s−1 and kss (max) of 18 ± 0.2 s−1, respectively (solid line). The data do not fit to a one-pKa model (dotted line).
R124L and R301L MSDHs
The kinetic parameters determined at pH 8.2 and at 30 °C under steady-state conditions are summarized in Table 3. For R124L MSDH, the Km values for NAD and PPA are similar to those of the wild-type enzyme, whereas the Km for CoA is 3.9-fold higher and the kcat is 4.5-fold smaller. Substituting Leu for Arg-301 led to a significant increase in Km for CoA (at least 12-fold) that prevented accurate determination of Km values for NAD and PPA. However, the likely underestimated kcat value of 3 s−1 is in the same range as that determined for R124L MSDH, and smaller by only 4-fold relative to the wild-type MSDH.
Under pre-steady-state conditions, fast kinetic experiments were carried out at pH 8.2 and 30 °C in the presence of 1 mm NAD and 0.05–1.4 m PPA. A linear dependence of the acylation rate versus PPA concentration was observed up to 1.4 m, irrespective of the mutated MSDH. For R124L and R301L MSDHs, k2 values of 110 and 114 m−1 s−1 were estimated that are ∼2.5-fold higher than the k2 value of the wild-type MSDH (Table 4). Moreover, the kobs value determined at 0.05 m PPA for R124L MSDH is 2-fold higher than the kcat value (i.e. 5.5 versus 2.8 s−1). The kobs value for R124L MSDH supports a rate-limiting step that occurs after the hydride transfer, although no definitive conclusion can be made for R301L MSDH due to the fact that the kcat value of 3 s−1 is likely to be an underestimate.
The fact that in the absence of CoA, the kobs was higher than the kcat (at least for R124L MSDH) did not exclude CoA binding to the mutated MSDHs and therefore a kinetic effect of CoA on the acylation step. To evaluate this assumption, fast kinetic experiments were carried out on R124L MSDH in the presence of 1 mm NAD and 250 μm CoA, with PPA concentrations ranging from 0 to 0.54 m. Under these experimental conditions, a Kapp of 0.18 m for PPA and a kobs (max) of 110 s−1 were determined (supplemental Fig. S4). Therefore, binding of CoA leads to a 5.5-fold increase of the second-order rate constant kobs (max)/Kapp when compared with the k2 value determined in the absence of CoA. Similar experiments were carried out in the presence of 1 mm NAD and 0.54 m PPA with CoA concentrations ranging from 0 to 400 μm. A Kapp of 90 μm for CoA was determined, which is in the range of that found for the wild-type MSDH (35 μm).
The kobs value determined at 0.05 m of PPA is ∼5-fold higher than the kobs determined in the absence of CoA and 10-fold higher than the kcat value (i.e. 29 versus ∼3 s−1). This latter result confirmed that the rate-limiting step takes place after hydride transfer as already shown for the wild-type MSDH. For the R301L MSDH, similar experimental conditions were used except the concentration range of PPA was 0–0.35 m, yielding a Kapp of 0.16 m for PPA and a kobs max of 117 s−1 (supplemental Fig. S4). As observed for R124L MSDH, binding of CoA led to a 6.5-fold increase in the second-order rate constant kobs (max)/Kapp relative to the k2 value determined in the absence of CoA.
Additionally, the rate constants for NADH dissociation from the thioacyl enzyme·NADH complex were determined for R124L and R301L MSDHs. The experiments were performed at pH 8.2 and 30 °C under pre-steady-state conditions and in the presence of 1 mm NAD and 0.5 m PPA but in the absence of CoA. Apparent rate constants of 12 and 8.4 s−1 were assigned to NADH release for R124L and R301L MSDHs, respectively (curves not shown), in the range of that determined for the wild-type MSDH. These rates are higher than the kcat values at least for R124L MSDH (Table 3). Thus, it can be concluded that the rate-limiting step for R124L MSDH and probably also for R301L MSDH is associated with the transthioesterification process. This finding is in accord with the fact that the kcat values for R124L and R301L MSDHs with MMSA as substrate, for which the rate-limiting step is likely associated with the β-decarboxylation process, are at least 20-fold lower compared with those determined with PPA. The fact that the rate-limiting step for wild-type MSDH with PPA is associated with NADH release (12.6 s−1) supports an intrinsic rate of transthioesterification that is at least 4-fold higher than that of R124L MSDH.
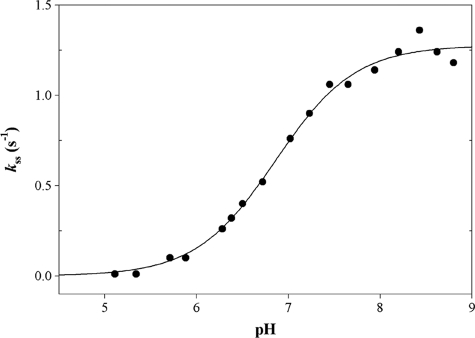
Finally, for the R124L MSDH, the pH dependence of the acylation rate constant was determined in the presence of 1 mm NAD, 0.5 m PPA, and 500 μm CoA. As already described for the wild-type MSDH, the formation of NADH was measured by monitoring the FRET signal using a 395-nm cutoff filter. The pH-kac curve exhibited an increasing bisigmoidal profile that can be related to the contribution of two ionizable groups, one of pKapp 5.7 that must be deprotonated for acylation and one of pKapp 7.5 whose deprotonation increased the acylation rate by 1.5-fold with rate constants kac (i) of 67 s−1 and kac (max) of 92 s−1, respectively (Fig. 6). The pH dependence of the steady-state rate constant was determined at 30 °C in the presence of 1 mm NAD, 0.05 m PPA, and 500 μm CoA. The pH- kss curve exhibited a monosigmoidal profile showing dependence on the contribution of one ionizable group of pKapp 6.8, and a kss (max) of 1.3 s−1 (Fig. 7). Differences in buffer composition and ionic strength likely explain why the kss (max) is half of the kcat value, i.e. 2.8 s−1.
FIGURE 6.
pH dependence of the acylation rate constant kac for the R124L MSDH-catalyzed reaction with PPA as substrate. Pre-steady-state data were collected at 30 °C on an SX18MV-R stopped-flow apparatus (Applied PhotoPhysics) using the 20-μl cell, by rapidly mixing 16 μn R124L MSDH (expressed in subunits), 1 mm NAD, 0.5 m PPA, and 500 μm CoA (final concentrations) over a pH-range of 5.50–8.75 in buffer A. Following excitation at 295 nm, formation of NADH was measured by monitoring the fluorescence emission using a 395-nm cutoff filter. Each individual point is the average of at least six measurements with an error of <5%. Experimental data (●) were fit by nonlinear least square regression analysis against a two-pKa model, which gave pKapp values of 5.7 ± 0.1 and 7.5 ± 0.2 and rate constants kac (i) of 67 ± 4 and kac (max) of 92 ± 2 s−1, respectively (solid line). The data do not fit to a one-pKa model (dotted line).
FIGURE 7.
pH dependence of the steady-state rate constant kss for the R124L MSDH-catalyzed reaction with PPA as substrate. The pH dependence of kss was studied at 30 °C over the pH range 5.00–8.75 in buffer A. The remaining experimental conditions were as follows: 1 mm NAD, 0.05 m PPA, and 500 μm CoA. Experiments were done twice with experimental errors on the individual points ≤5%. Data (●) were fit by nonlinear least square regression analysis against a single pKapp model that yielded a pKapp of 6.8 ± 0.1 and a kss of 1.3 ± 0.1 s−1 (solid line).
DISCUSSION
The kinetic mechanism of the CoA-dependent ALDH family to which MSDHs belong was shown to be of ping-pong type with NADH release preceding transthioesterification (5, 6). However, the MSDH-catalyzed reaction also includes a supplementary β-decarboxylation step that occurs before transthioesterification. Our recent mechanistic characterization of the wild-type MSDH from B. subtilis showed that the β-decarboxylation process is rate-limiting and occurs with formation of bicarbonate. As discussed earlier, sequence alignments and three-dimensional structure comparisons suggested that the two invariant residues Arg-124 and Arg-301 in MSDHs, contributed to the binding of MMSA through electrostatic interaction between the carboxylate and the guanidinium groups. This assumption is supported by the kinetic parameters of the acylation step for R301L, R124L, and R124L/R301L MSDHs. Indeed, no saturating kinetic effect with respect to MMSA was observed up to 12–16 mm, in contrast to the wild-type MSDH. The second-order rate constant k2 is decreased by a factor of 55, 62, and at least 512 for R124L, R301L, and R124L/R301L MSDHs, respectively. The magnitude of these decreases is 3–30-fold higher than that described for R124L GAPN (28). In the case of MSDH, the decrease of k2 likely includes contributions from both Kapp and kobs (max). Therefore, Arg-124 and Arg-301 participate not only in MMSA binding via a stabilizing electrostatic interaction with the carboxylate but also likely in positioning MMSA efficiently relative to Cys-302 within the MSDH·NAD·MMSA ternary complex. From an evolutionary point of view, it is interesting to note that such a stabilization mode is not unique within the ALDH superfamily. Indeed, in the succinate semialdehyde dehydrogenase, a member of the hydrolytic ALDH family, the stabilization of the carboxylate group of the substrate is mainly achieved through hydrogen bonds with two arginine (Arg-213 and Arg-334) residues (29). Although differing in position in primary structures, superimposition of the GAPN, MSDH, and succinate semialdehyde dehydrogenase active sites shows that Arg-213 and Arg-334 are structurally and functionally equivalents to Arg-124 and Arg-301, respectively (not shown).
With the PPA substrate, no kinetic role of Arg-124 and Arg-301 in the acylation process was expected due to the absence of the carboxylate group. This assumption is supported by the values of the second-order rate constant k2 of the acylation step determined in the absence of CoA, which are similar for the wild-type and the mutated MSDHs (i.e. 36, 110, and 114 m−1 s−1, respectively) and which are ∼104-fold lower compared with that determined for the wild-type MSDH with MMSA as substrate. Much more importantly, the kobs value determined at 50 mm PPA (i.e. a saturating concentration under steady-state conditions) was 2.5-fold lower than the kcat value. This result suggested that CoA binds to the wild-type MSDH prior to the hydride transfer. Favoring this assumption is the fact that the k2 value for the acylation step determined in the presence of saturating concentration of CoA is 72-fold higher than in the absence of CoA. The increase of the k2 value is mainly due to a Kapp effect for PPA, as it is decreased by a factor of at least 25. This result suggests a higher affinity of the wild-type MSDH for PPA because of CoA binding. When CoA is bound, the Kapp of PPA is only 40-fold higher than that for MMSA in the acylation step. The stronger affinity with MMSA is likely due to stabilizing interactions between the carboxylate of MMSA and both Arg-124 and Arg-301.
The R124L and R301L MSDHs also exhibited an increase in k2 by factors of 5.5- and 6.5-fold, respectively, in the presence of 250 μm CoA. Again, as for the wild-type MSDH, the increase of the k2 value is mainly due to a Kapp effect for PPA, which suggests a higher affinity of the mutated MSDHs for PPA. These results, taken together, raise interesting questions as to the nature of the molecular/structural factors that favor the increased affinity of the wild-type, R124L, and R301L MSDHs·NAD complexes for PPA in the presence of CoA. The only information currently available on this issue is that the thiol group of CoA plays no role because the kinetics of the acylation step are similar when desulfo-CoA is used instead of CoA. Substituting Leu for Arg-124 led to an increase of the Kapp and Km values for CoA by factors of 3 and 4, respectively. Considering the 4.5-fold decrease of the kcat value, the catalytic efficiency of the thioester exchange with CoA (∼8.5 × 103 m−1 s−1) is at least 17-fold decreased (∼1.5 × 105 m−1 s−1 for the wild-type MSDH). The Arg-124 probably favors an efficient binding of CoA within the deacylation complex. This is likely to also be the case for Arg-301 whose substitution by Leu leads to an even more significant increase of the Km value for CoA.
In contrast to PPA, when MMSA was used as substrate, no kinetic effect on the acylation step was observed for the wild-type and R301L MSDHs when CoA was added. Therefore, two alternatives are possible, either CoA binds but has no kinetic effect on the rate of the acylation step or CoA does not bind. Further information on CoA binding to the MSDH·NAD binary complex comes from our earlier studies of the lag phase in enzymatic turnover (25). Indeed, we have shown that formation of the MSDH·NAD binary complex elicits a slow conformational reorganization within the active site of the wild-type MSDH. The lag phase, which is eliminated when the wild-type and R301L MSDHs are preincubated with NAD, also disappears for R124L and R124L/R301L MSDHs but only when CoA is also added. This supports CoA binding at a minimum to the R124L and R124L/R301L MSDH·NAD complexes. Thus, for mutants where Arg-124 is substituted by Leu, the lag phase is also a good probe of the CoA binding to the enzyme·NAD complex. The reasons why R124L mutants behave differently from R301L and wild-type MSDHs remain to be explained.
Together, the kinetic data obtained in this study favor the existence of two distinct binding sites for CoA and NAD in contrast to the suggestion made in our previous paper that was based on the hypothesis that the early release of NADH (i.e. before transthioesterification) occurs because the NADH-binding site partially overlaps the CoA-binding site.
The fact that with PPA as substrate CoA binds to wild-type MSDH in the acylation step gave us the opportunity to measure the pKapp of the thiol group of CoA in the acylation step. In the absence of CoA, the obtained kac versus pH curve with PPA fits to a monosigmoidal profile, yielding a pKapp of ∼6. This value is similar to that previously found for the wild-type MSDH and the quintuple Cys → Ala mutant, in which Ala was substituted for all the Cys residues, except for Cys-302, with MMSA as substrate (25). Therefore, as already interpreted, the pKapp of ∼6 corresponds to the pKapp of Cys-302. In the presence of saturating concentrations of CoA, the pH-kac curve exhibits an increasing double sigmoidal profile with two pKapp values of 6.5 and 7.9 (Fig. 4). The double sigmoidal profile is therefore representative of two species of Cys-302 that participate in the acylation process, i.e. Cys-302-S−/CoA-SH with a kac of 100 s−1 and Cys-302-S−/CoA-S− with a kac of 330 s−1. Consequently, the pKapp of 6.5 corresponds to Cys-302 and that of 7.9 can be assigned to the thiol group of CoA. Indeed, when desulfo-CoA was used instead of CoA (i.e. no CoA-SH species was present), a monosigmoidal profile is observed with a pKapp of 6.5 (Fig. 4).
As NADH dissociation from the acylation complex was shown to be rate-limiting, it was not possible to determine the pKapp of the CoA within the deacylation complex. The picture is clearly different for at least R124L MSDH for which the rate-limiting step is now associated with transthioesterification. From the pH-kac curve obtained for R124L MSDH in the presence of CoA (Fig. 6), two pKapp values of 5.7 and 7.5 were extracted, which can be assigned to Cys-302 and CoA, as already proposed for the wild-type MSDH. The fact that a monosigmoidal profile was obtained for the pH-kss curve (Fig. 7) with a pKapp of 6.8 suggests a similar pKapp for both Cys-302 and CoA. Alternatively, the pKapp of 6.8 corresponds only to that of CoA. In any case, the pKapp values show that the CoA is more activated in the deacylation than in the acylation complex, consistent with the requirement for it to efficiently attack the thioacyl enzyme intermediate. Presumably, the change in pKapp between the two complexes arises from differences in the local environment or precise positioning of the thiol of CoA. In addition, the k2 (kcat/Km) values for the MSDH thioester exchange with CoA are ≥104- and ≈103-fold higher for the wild-type and R124L MSDHs, respectively, when compared with the second-order rate constant k2 for the attack of the mercaptoethanol anion (which has a pKapp value of 9.6 (30) similar to that of CoA), on a nonactivated thiol ester, i.e. 12 m−1 s−1 (31). Among the different factors that can explain such differences, a major one should be CoA binding to MSDH that likely optimizes the positioning of CoA relative to the thioacyl enzyme intermediate and thus favors a significant decrease of its pKapp.
In conclusion, Arg-301 and Arg-124 are essential in the binding of MMSA via stabilizing interactions with the carboxylate group. The use of PPA has permitted us to show that the CoA also binds to the acylation complex and therefore to a site distinct from that of NAD. The catalytic efficiency of the MSDH thioester exchange with CoA is at least 104-fold increased compared with the second order-rate constant k2 for the attack of the mercaptoethanol anion on a nonactivated thiol ester. This difference in reactivity is the consequence of the binding of CoA to MSDH, which likely optimizes the positioning of CoA relative to the thioacyl enzyme intermediate and consequently favors a significant decrease of ∼3 units of the pKapp of the thiol of CoA. The results obtained in this study leave some questions opened. For example, as the NAD- and CoA-binding sites do not overlap, the CoA-binding site remains to be characterized. In this context, the resolution of the three-dimensional structure of a ternary MSDH·NAD·CoA complex would be informative.
Supplementary Material
Acknowledgments
We gratefully thank S. Boutserin for the very efficient technical help and Prof. C. Gerardin for helping to synthesize desulfo-CoA. We also thank Prof. K. Weissman for careful reading of the manuscript.
This work was supported in part by the CNRS, the University of Nancy I, the Institut Fédératif de Recherche 111 Bioingénierie, and local funds from the Région Lorraine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- MSDH
- methylmalonate semialdehyde dehydrogenase
- ALDH
- aldehyde dehydrogenase
- GAPN
- nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase
- LDH
- lactate dehydrogenase
- MMSA
- methylmalonate semialdehyde
- PPA
- propionaldehyde.
REFERENCES
- 1. Bradbury S. L., Jakoby W. B. (1971) J. Biol. Chem. 246, 1834–1840 [PubMed] [Google Scholar]
- 2. MacGibbon A. K., Blackwell L. F., Buckley P. D. (1977) Biochem. J. 167, 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vallari R. C., Pietruszko R. (1981) Arch. Biochem. Biophys. 212, 9–19 [DOI] [PubMed] [Google Scholar]
- 4. Marchal S., Rahuel-Clermont S., Branlant G. (2000) Biochemistry 39, 3327–3335 [DOI] [PubMed] [Google Scholar]
- 5. Shone C. C., Fromm H. J. (1981) Biochemistry 20, 7494–7501 [DOI] [PubMed] [Google Scholar]
- 6. Söhling B., Gottschalk G. (1993) Eur. J. Biochem. 212, 121–127 [DOI] [PubMed] [Google Scholar]
- 7. Farrés J., Wang T. T., Cunningham S. J., Weiner H. (1995) Biochemistry 34, 2592–2598 [DOI] [PubMed] [Google Scholar]
- 8. Wang X., Weiner H. (1995) Biochemistry 34, 237–243 [DOI] [PubMed] [Google Scholar]
- 9. Vedadi M., Meighen E. (1997) Eur. J. Biochem. 246, 698–704 [DOI] [PubMed] [Google Scholar]
- 10. Liu Z. J., Sun Y. J., Rose J., Chung Y. J., Hsiao C. D., Chang W. R., Kuo I., Perozich J., Lindahl R., Hempel J., Wang B. C. (1997) Nat. Struct. Biol. 4, 317–326 [DOI] [PubMed] [Google Scholar]
- 11. Steinmetz C. G., Xie P., Weiner H., Hurley T. D. (1997) Structure 5, 701–711 [DOI] [PubMed] [Google Scholar]
- 12. Cobessi D., Tête-Favier F., Marchal S., Branlant G., Aubry A. (2000) J. Mol. Biol. 300, 141–152 [DOI] [PubMed] [Google Scholar]
- 13. D'Ambrosio K., Pailot A., Talfournier F., Didierjean C., Benedetti E., Aubry A., Branlant G., Corbier C. (2006) Biochemistry 45, 2978–2986 [DOI] [PubMed] [Google Scholar]
- 14. Talfournier F., Pailot A., Stinès-Chaumeil C., Branlant G. (2009) Chem. Biol. Interact. 178, 79–83 [DOI] [PubMed] [Google Scholar]
- 15. Bannerjee D., Sanders L. E., Sokatch J. R. (1970) J. Biol. Chem. 245, 1828–1835 [PubMed] [Google Scholar]
- 16. Steele M. I., Lorenz D., Hatter K., Park A., Sokatch J. R. (1992) J. Biol. Chem. 267, 13585–13592 [PubMed] [Google Scholar]
- 17. Zhang Y. X., Tang L., Hutchinson C. R. (1996) J. Bacteriol. 178, 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoshida K. I., Aoyama D., Ishio I., Shibayama T., Fujita Y. (1997) J. Bacteriol. 179, 4591–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fry J., Wood M., Poole P. S. (2001) Mol. Plant-Microbe Interact. 14, 1016–1025 [DOI] [PubMed] [Google Scholar]
- 20. Goodwin G. W., Rougraff P. M., Davis E. J., Harris R. A. (1989) J. Biol. Chem. 264, 14965–14971 [PubMed] [Google Scholar]
- 21. Gray R. G., Pollitt R. J., Webley J. (1987) Biochem. Med. Metab. Biol. 38, 121–124 [DOI] [PubMed] [Google Scholar]
- 22. Chambliss K. L., Gray R. G., Rylance G., Pollitt R. J., Gibson K. M. (2000) J. Inherit. Metab. Dis. 23, 497–504 [DOI] [PubMed] [Google Scholar]
- 23. Roe C. R., Struys E., Kok R. M., Roe D. S., Harris R. A., Jakobs C. (1998) Mol. Genet. Metab. 65, 35–43 [DOI] [PubMed] [Google Scholar]
- 24. Tanaka N., Takahashi H., Kitano H., Matsuoka M., Akao S., Uchimiya H., Komatsu S. (2005) J. Proteome Res. 4, 1575–1582 [DOI] [PubMed] [Google Scholar]
- 25. Stines-Chaumeil C., Talfournier F., Branlant G. (2006) Biochem. J. 395, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chase J. F., Middleton B., Tubbs P. K. (1966) Biochem. Biophys. Res. Commun. 23, 208–213 [DOI] [PubMed] [Google Scholar]
- 27. Kupiecki F. P., Coon M. J. (1960) Biochem. Prep. 7, 69–71 [Google Scholar]
- 28. Marchal S., Branlant G. (2002) J. Biol. Chem. 277, 39235–39242 [DOI] [PubMed] [Google Scholar]
- 29. Kim Y. G., Lee S., Kwon O. S., Park S. Y., Lee S. J., Park B. J., Kim K. J. (2009) EMBO J. 28, 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jocelyn P. C. (1972) Biochemistry of the SH Group, Academic Press, New York [Google Scholar]
- 31. Hupe D. J., Jencks W. P. (1977) J. Am. Chem. Soc. 99, 451–464 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.