Abstract
Protein-disulfide isomerase (PDI) has been proposed to exhibit an “unfoldase” activity against the catalytic A1 subunit of cholera toxin (CT). Unfolding of the CTA1 subunit is thought to displace it from the CT holotoxin and to prepare it for translocation to the cytosol. To date, the unfoldase activity of PDI has not been demonstrated for any substrate other than CTA1. An alternative explanation for the putative unfoldase activity of PDI has been suggested by recent structural studies demonstrating that CTA1 will unfold spontaneously upon its separation from the holotoxin at physiological temperature. Thus, PDI may simply dislodge CTA1 from the CT holotoxin without unfolding the CTA1 subunit. To evaluate the role of PDI in CT disassembly and CTA1 unfolding, we utilized a real-time assay to monitor the PDI-mediated separation of CTA1 from the CT holotoxin and directly examined the impact of PDI binding on CTA1 structure by isotope-edited Fourier transform infrared spectroscopy. Our collective data demonstrate that PDI is required for disassembly of the CT holotoxin but does not unfold the CTA1 subunit, thus uncovering a new mechanism for CTA1 dissociation from its holotoxin.
Keywords: Cholera Toxin, Endoplasmic Reticulum(ER), Fourier Transform IR (FTIR), Protein Conformation, Protein Folding, Protein Isomerase, Protein Stability, Protein Translocation, Surface Plasmon Resonance (SPR)
Introduction
Cholera toxin (CT)2 is an AB5 protein toxin that consists of a catalytic A moiety and a cell-binding B moiety (1, 2). The B subunit is pentameric ring-like structure that adheres to GM1 gangliosides on the plasma membrane of a target cell. The A subunit is initially synthesized as a 26 kDa protein that undergoes proteolytic nicking to generate a disulfide-linked A1/A2 heterodimer. The 21 kDa CTA1 polypeptide is an ADP-ribosyltransferase that modifies and activates Gsα in the host cell cytosol. CTA1 can be divided into three subdomains: the A11 subdomain contains the catalytic core of the toxin; the A12 subdomain is a short extended linker that connects the A11 and A13 subdomains; and the A13 subdomain is a globular structure with many hydrophobic residues as well as a cysteine residue involved with the single disulfide bridge between CTA1 and CTA2 (3). The 5 kDa CTA2 polypeptide maintains numerous non-covalent interactions with the central pore of the B pentamer and thereby anchors CTA1 to CTB5. A ribbon diagram of the CT holotoxin which highlights the subdomain structure of CTA1 is provided in supplemental Fig. S1.
To reach its cytosolic Gsα target, CT moves from the cell surface to the endoplasmic reticulum (ER) by retrograde vesicular traffic (4). A C-terminal KDEL sequence in the CTA2 subunit is thought to target and/or retain CT in the ER (4, 5). Conditions in the ER lead to reductive cleavage of the CTA1/CTA2 disulfide bond and chaperone-assisted dissociation of CTA1 from CTA2/CTB5 (6–9). Unfolding of the free A1 subunit then activates the quality control system of ER-associated degradation (ERAD), thereby promoting CTA1 translocation to the cytosol (10, 11). Most exported ERAD substrates are degraded by the ubiquitin-proteasome system, but it was hypothesized that CTA1 and the A chains of other ER-translocating toxins avoid this fate because they lack substantial numbers of lysine residues for ubiquitin conjugation (12). Subsequent experimental studies verified the paucity of lysine residues protects CTA1 and other toxin A chains from ubiquitin-dependent proteasomal degradation (13–15). The translocated pool of CTA1 instead interacts with ADP-ribosylation factors and possibly other host factors to regain an active, folded conformation in the cytoplasm (16–18).
The reduced form of protein-disulfide isomerase (PDI), an ER-localized oxidoreductase and molecular chaperone (19, 20), was originally proposed to unfold the holotoxin-associated CTA1 subunit and to thereby promote the separation of CTA1 from CTA2/CTB5 (9). It was further posited that unfolding of the CTA1 polypeptide was directly coupled with toxin delivery to the Derlin-1 pore (21, 22). Release of CTA1 from PDI in preparation for translocation through the Derlin-1 pore was thought to involve the oxidation of PDI by Ero1p (23). Thus, PDI is viewed as a redox-dependent chaperone that binds to holotoxin-associated CTA1 in a reduced state, actively unfolds the toxin, and then releases the dissociated CTA1 subunit upon its oxidation by Ero1p.
Many aspects of PDI-CTA1 interactions remain controversial. Lumb and Bulleid demonstrated that PDI does not act in a redox-dependent fashion when assisting the folding of other polypeptide chains (24). Recent experiments using zebrafish and mammalian cells have questioned the role of Derlin-1 in CTA1 translocation (25). Our structural studies have indicated that CTA1 will unfold spontaneously after dissociation from the holotoxin at physiological temperature (17, 26, 27). It is therefore possible that, in terms of cholera intoxication, the primary function of PDI is to simply dislodge CTA1 from CTA2/CTB5. The dissociated CTA1 polypeptide would then spontaneously unfold and consequently trigger the ERAD translocation mechanism.
In this work we evaluated the role of PDI in disassembly of the CT holotoxin and unfolding of the CTA1 subunit. The prevailing model of PDI-toxin interactions is largely based upon the results of a protease sensitivity assay which demonstrated that CTA1 shifts from a protease-resistant conformation to a protease-sensitive conformation in the presence of reduced PDI (9, 13, 23, 28, 29). As folded proteins are generally more resistant to proteolysis than unfolded variants of the same protein, this shift was interpreted to represent the PDI-induced unfolding of CTA1 and the PDI-induced displacement of CTA1 from CTA2/CTB5. Yet this assay, which was typically performed at 30 °C, only provides an indirect measure of protein folding. Here, we developed a surface plasmon resonance (SPR) assay to directly monitor in real time the disassembly of the CT holotoxin. Biophysical methods were used to directly examine the conformation of CTA1 in the presence or absence of PDI. Cell-based assays further probed the role of PDI in CT intoxication and CTA1 translocation. Our collective data indicate that PDI does not unfold the CTA1 polypeptide and that PDI can displace CTA1 from CTA2/CTB5 in a process which does not involve substantial alterations to the structure of CTA1. In further contrast with the current model of PDI-toxin interactions where PDI acts as an unfoldase, we found that the PDI-induced shift of CTA1 to a protease-sensitive conformation does not correlate to the disassembly of the CT holotoxin. Moreover, the release of PDI from CTA1 does not require Ero1p but instead results from the spontaneous unfolding of CTA1 which occurs after its dissociation from the holotoxin. The unstable nature of the isolated CTA1 polypeptide at physiological temperature thus plays a central role in toxin translocation and leads to a new model of CT disassembly in the ER.
EXPERIMENTAL PROCEDURES
Materials
His-tagged CTA1, CTA11–168, and CTA11–133 constructs were purified as previously described (26). CT and the CTA1/CTA2 heterodimer were purchased from List Biologicals (Campbell, CA), while PDI, chemicals, and other reagents were purchased from Sigma-Aldrich. Anti-CTA and anti-CTB antibodies were purchased from Sigma-Aldrich; anti-PDI and anti-KDEL antibodies were purchased from Enzo Life Sciences (Plymouth Meeting, PA); and the anti-CTA monoclonal antibody 35C2 (30) was kindly provided by Dr. Randall K. Holmes (University of Colorado Denver School of Medicine).
SPR
Experiments were performed with a Reichert (Depew, NY) SR7000 SPR Refractometer as previously described (17). Antibodies were perfused over the plate at the following dilutions in phosphate-buffered saline containing 0.05% Tween 20: anti-PDI antibody, 1:10,000; anti-CTB antibody, 1:15,000; anti-CTA monoclonal antibody, 1:500; anti-CTA polyclonal antibody, 1:5,000; and anti-KDEL antibody, 1:10,000. The flow rate for all steps was 41 μl/min.
13C Labeling and Purification of CTA1-His6
Escherichia coli strain BL21 pLysS was transformed with pT7CTA1h6, an inducible CTA1-His6 expression plasmid (31). 13C-labeled toxin was generated by culturing the strain at 37 °C in M9 minimal media supplemented with uniformly 13C-labeled 13C6-d-glucose (Cambridge Isotope, Andover, MA) as the sole metabolic carbon source. The culture was shifted to 22 °C when it reached an A600 of 0.6, and protein expression was induced overnight by the addition of 1 mm IPTG. The expressed protein was then purified in 8 m urea buffer as described previously (26). The purity of the protein sample was assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie staining. In order to facilitate protein refolding, slide-a-lyzer 3500 MWCO cassettes (Pierce) were used for dialysis as previously described (27). After dialysis, CTA1-His6 was lyophilized for FTIR experiments. Control experiments using circular dichroism (CD) confirmed that lyophilization did not alter the structure of CTA1.
Determination of Protein Secondary Structure by Fourier Transform Infrared (FTIR) Spectroscopy
Protein samples for FTIR measurements were prepared in a D2O-based buffer containing 10 mm sodium borate pH* 6.6 that corresponds to pH 7.0. The samples contained either unlabeled PDI, uniformly 13C-labeled CTA1, or both PDI and 13C-labeled CTA1 at a 1:1 molar ratio. FTIR spectra were measured on a Jasco 4200 FTIR spectrometer at 0.964 cm−1 spectral resolution and a set resolution of 1 cm−1. The absorbance spectra were calculated using the respective buffers as reference and corrected by subtraction of H2O vapor contribution, smoothing, and baseline correction in the amide I region. Curve fitting was conducted using the component frequencies from the second derivative spectra, as described in (32, 33) and elaborated upon in the supplemental data. No appreciable signals from the second derivative spectra were detected in the region above 1700 cm−1.
Toxicity Assay
Cells were seeded into 24-well plates and grown overnight to 80% confluency. The medium was removed and replaced with serum-free medium containing various concentrations of CT. After 2 h, the cells were processed for determination of cAMP levels as described in Ref. 34. The basal levels of cAMP determined from unintoxicated cells were background-subtracted from the values obtained for toxin-treated cells; the maximal response from the parental control cells was arbitrarily set to 100%; and all other results were expressed as ratios of that 100% value. All conditions were performed in triplicate.
CTA1 Transfection Assays
Cells seeded to 80% confluency in 6-well plates were transfected with pcDNA3.1/ssCTA1 (35) using Lipofectamine (Invitrogen) according as per manufacturer's instructions. For the translocation assay, cells were incubated at 24 h post-transfection in methionine-free medium for 1 h before [35S]methionine was added for another hour. Digitonin was then used to generate separate membrane and cytosolic fractions as previously described (34). Both cellular fractions were immunoprecipitated with an anti-CTA antibody. SDS-PAGE with PhosphorImager analysis was used to visualize and quantify the immunoisolated material. The extent of CTA1 translocation for each cell line was calculated with the following equation: % cytosolic CTA1 = CTA1 supernatant signal/[CTA1 supernatant signal + CTA1 pellet signal].
For the toxicity assay, cells were exposed to ice-cold acidic ethanol at 3 h post-transfection and were processed for determination of cAMP levels as described in (34). An additional set of transfected cells were chased overnight to determine the relative levels of CTA1 expression via metabolic labeling and immunoprecipitation. Values obtained from the cAMP assay were standardized to CTA1 expression levels before expressing the data as percentages of the signal obtained from the parental control cells.
RESULTS
PDI-mediated Disassembly of the CT Holotoxin
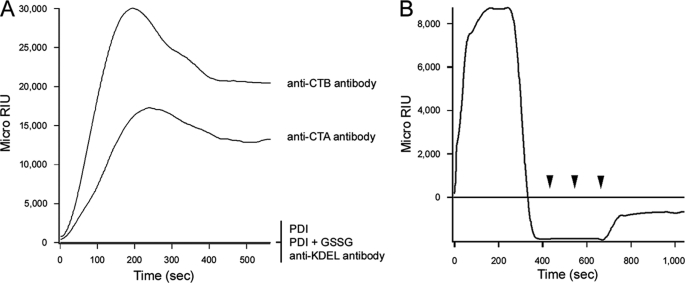
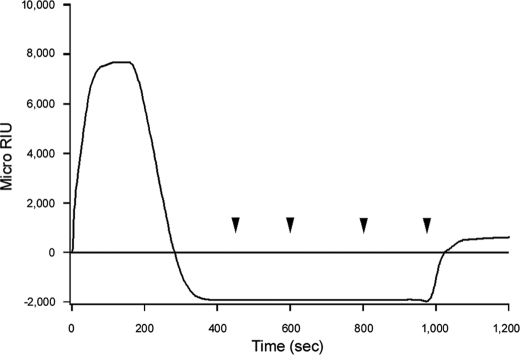
SPR was used to examine the interaction between CT and PDI (Fig. 1). CT was appended to a GM1-coated sensor slide, and anti-CTA or anti-CTB antibodies were perfused over the slide for control measurements (Fig. 1A). Both antibodies generated an increase in the refractive index of the sensor slide, thus indicating direct antibody binding to the CT holotoxin. The anti-CTB antibody produced a stronger signal than the anti-CTA antibody because, given the AB5 structure of the holotoxin, there was 5-fold more CTB than CTA on the sensor slide. Differences in antibody affinity could also account for some of the differences in anti-CTB versus anti-CTA signal strength. The anti-KDEL antibody did not produce a positive signal (Fig. 1A), possibly because the presence of the CTA1 subunit blocked antibody access to the KDEL tag at the C terminus of CTA2 (supplemental Fig. S1). PDI and oxidized PDI did not bind to the CT holotoxin (Fig. 1A), but an interaction between reduced PDI and CT was detected (Fig. 1B). These observations were consistent with a published report on the redox-dependent interplay between PDI and CT (9).
FIGURE 1.
Dissociation of the CT holotoxin by reduced PDI. A, following ligands were perfused over a CT-coated SPR sensor slide at 37 °C: PDI, oxidized PDI (PDI + 1 mm GSSG), an anti-CTA antibody, an anti-CTB antibody, and an anti-KDEL antibody. B, after taking a baseline measurement corresponding to the mass of the sensor-bound holotoxin, reduced PDI (PDI + 1 mm GSH) was perfused over the CT-coated sensor slide at 37 °C. PDI was removed 400 s into the experiment and replaced with sequential additions of anti-PDI, anti-CTA, and anti-KDEL antibodies as indicated by the arrowheads.
Previous work has shown that reduced PDI can remove CTA1 from the CT holotoxin (9). This event was documented by our SPR system as well (Fig. 1B). After PDI bound to the CT holotoxin, a time-dependent loss of signal was observed. This indicated that PDI was released from the CT sensor slide after its initial binding. If only PDI was lost from the sensor, then the signal strength would revert to the baseline value which was derived from the mass of the bound holotoxin. However, the signal actually dropped to a negative value. This indicated that a portion of the holotoxin had also been released from the sensor slide. To confirm that reduced PDI had displaced CTA1 from the holotoxin, anti-CTA antibodies were perfused over the CT sensor slide. In contrast to our control experiment with the intact CT holotoxin (Fig. 1A), the anti-CTA antibody did not generate an increase in the refractive index of the PDI-treated holotoxin (Fig. 1B). CTA1 was therefore absent from the PDI-treated CT sensor slide. Perfusion of an anti-PDI antibody over the sensor slide also demonstrated that PDI was absent from the plate (Fig. 1B). Previous reports have demonstrated that reduction of the CTA1/CTA2 disulfide bond is not sufficient to remove CTA1 from the holotoxin (9, 36), which was also documented in our SPR system: the CT holotoxin remained intact when exposed to a reducing perfusion buffer containing 1 mm GSH (data not shown). Thus, reduced PDI displaced CTA1 from the CT holotoxin. This effect was documented with a range of PDI concentrations, and the amount of reduced PDI in the perfusion buffer was directly related to the kinetics of holotoxin disassembly (supplemental Fig. S2).
To determine the fate of the CTA2 polypeptide after holotoxin disassembly, we perfused an anti-KDEL antibody over the PDI-treated holotoxin (Fig. 1B). The anti-KDEL antibody did not generate an increase in the refractive index when applied to the intact CT holotoxin (Fig. 1A), but a positive signal was obtained when the antibody was applied to the PDI-treated holotoxin (Fig. 1B). Removal of the CTA1 subunit thus allowed the anti-KDEL antibody to reach the KDEL tag on the CTA2 subunit that remained associated with the sensor-bound CTB pentamer. Reduced PDI thus displaced CTA1 from the CT holotoxin but did not separate CTA2 from CTB5.
Structure-dependent Interactions between PDI and CTA1
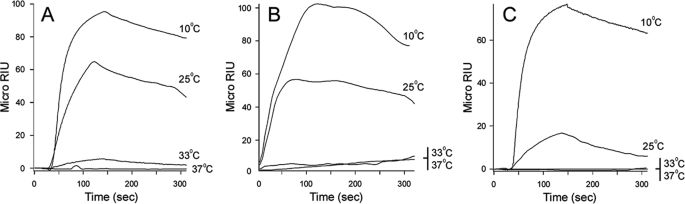
Reduced PDI did not bind to CTA2/CTB5 (supplemental Fig. S3), but it did interact with the isolated CTA1 subunit (Fig. 2). As seen with the CT holotoxin, only reduced PDI could interact with CTA1: neither PDI nor oxidized PDI increased the refractive index when perfused over a CTA1 sensor slide (data not shown). Binding of reduced PDI to CTA1 was temperature-dependent. A strong interaction was detected at 10 °C, a weaker interaction was detected at 25 °C, and little to no interactions were detected at 33 °C or the physiological temperature of 37 °C (Fig. 2A). The isolated CTA1 subunit is a thermally unstable protein with a partially perturbed secondary structure and a disordered tertiary structure at 37 °C (17). It thus appeared that PDI can interact with the folded conformations of CTA1 present at low temperatures but not with the unfolded CTA1 conformations present at physiological or near-physiological temperatures. Thermal instability in the CTA1 polypeptide is only apparent after dissociation from the holotoxin (17, 37, 38), which explains why reduced PDI could interact with CTA1 when it was present in the CT holotoxin at 37 °C (Fig. 1B).
FIGURE 2.
Conformation-dependent interactions between PDI and CTA1. Reduced PDI (PDI + 1 mm GSH) was perfused at the indicated temperatures over SPR sensor slides coated with (A) CTA1, (B) CTA11–168, or (C) CTA11–133. PDI was removed from the perfusion buffer 150 s into the experiment.
The C-terminal A13 subdomain of CTA1 has been proposed to act as the trigger for ERAD-mediated translocation and to serve as the site for PDI binding (12, 39).However, a CTA1 variant lacking most of the A13 subdomain (CTA11–169) can still move from the ER to the cytosol (31). We found that a CTA11–168 variant could also interact with reduced PDI (Fig. 2B). As with the full-length CTA1, PDI binding to CTA11–168 was temperature-dependent: a strong interaction was detected at 10 °C, a weaker interaction was detected at 25 °C, and no interactions were detected at 33 °C or 37 °C (Fig. 2B). A similar pattern of temperature-dependent binding was recorded for the interaction between PDI and CTA11–133, a CTA1 variant lacking both the A12 and A13 subdomains (Fig. 2C). Increasing the temperature from 10 to 37 °C results in a progressive loss of structure for CTA1, CTA11–168, and CTA11–133 (27). PDI thus appeared to recognize a folded region in the A11 subdomain of CTA1. The CTA11–133 construct is less stable than either CTA1 or CTA11–168 (27), which explains the weaker binding of PDI to CTA11–133 at 25 °C in comparison to CTA1 and CTA11–168. These experiments highlighted a unique aspect of PDI-CTA1 interactions, as PDI usually binds to unfolded but not folded protein conformations (19, 20).
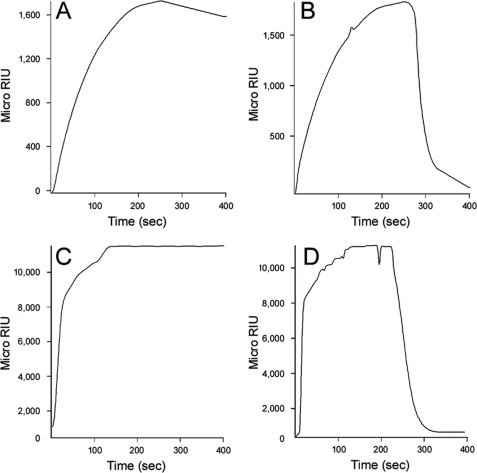
When exposed to mildly acidic pH, the isolated CTA1 subunit retains a substantial amount of its native structure at physiological temperature (27). Consistent with this observation, we found that an acidic perfusion buffer allowed reduced PDI to effectively bind free CTA1 at 37 °C (Fig. 3A). This again suggested that PDI recognizes a structure in the folded conformation of CTA1. It also demonstrated, along with the holotoxin study of Fig. 1B, that reduced PDI was functional in our SPR system at 37 °C. Interestingly, PDI was rapidly displaced from CTA1 when the acidic buffer was replaced with neutral buffer (Fig. 3B). The high affinity interaction between CTA1 and PDI that occurred at pH 6.5 was completely lost, as reflected by the return of the signal to baseline value, when the 37 °C buffer was switched to neutral pH. The disruption of PDI binding occurred despite the continued presence of PDI in the pH 7.0 perfusion buffer. Furthermore, the loss of PDI interaction was not simply time-dependent: when the perfusion buffer was maintained at pH 6.5, PDI remained stably associated with CTA1 (Fig. 3A). CTA1 would shift from a folded to an unfolded conformation when the buffer was switched from pH 6.5 to pH 7.0. It therefore appeared that the thermal unfolding of CTA1 results in displacement of bound PDI.
FIGURE 3.
CTA1 unfolding displaces toxin-bound PDI. A and B, reduced PDI (PDI + 1 mm GSH) was perfused over a CTA1-coated sensor slide at 37 °C in pH 6.5 buffer. After 250 s, the pH of the perfusion buffer was either (A) maintained at pH 6.5 or (B) shifted to pH 7.0. PDI was present in the perfusion buffer throughout the experiment. C and D, reduced PDI (PDI + 1 mm GSH) was perfused over a CTA1-coated sensor slide at 37 °C in buffer containing 10% glycerol. After 250 s, glycerol was either (C) maintained or (D) removed from the perfusion buffer. PDI was present in the perfusion buffer throughout the experiment.
To ensure that our pH data resulted from structural alterations to CTA1, we repeated the PDI/CTA1 binding assay with another condition that stabilizes the structure of CTA1: 10% glycerol. Like acidic pH, treatment with 10% glycerol increases the thermal stability of CTA1 and allows the toxin to maintain a substantial amount of its native structure at 37 °C (26). Glycerol treatment also allowed reduced PDI to effectively bind CTA1 at 37 °C (Fig. 3C). This interaction was lost when glycerol, but not PDI, was removed from the perfusion buffer (Fig. 3D). In contrast, PDI remained stably associated with CTA1 when 10% glycerol was kept in the perfusion buffer for the duration of the experiment (Fig. 3C). CTA1 would shift from a folded to an unfolded conformation when glycerol was removed from the 37 °C perfusion buffer. Thus, two independent experimental conditions both indicated that toxin unfolding will displace PDI from its CTA1 binding partner.
PDI-mediated Disassembly of the CT Holotoxin Does Not Require CTA1 Unfolding
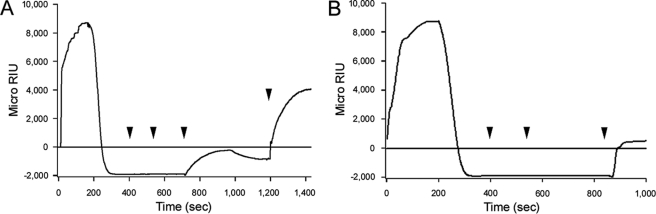
PDI is thought to remove reduced CTA1 from CTA2/CTB5 by unfolding the holotoxin-associated A1 subunit (9). According to this model, the resulting conformational change in CTA1 would disrupt its non-covalent interactions with CTA2/CTB5 and thereby promote its dissociation from the holotoxin. To determine if PDI could remove CTA1 from CTA2/CTB5 without unfolding CTA1, we added reduced PDI to a CT-coated sensor slide at pH 6.5 (Fig. 4A). The stable interaction between PDI and CTA1 at 37 °C and pH 6.5 (Fig. 3A) indicated that PDI does not unfold CTA1 under these conditions - otherwise, as shown in Fig. 4B, CTA1 unfolding would displace the bound PDI. At both pH 7.0 (Fig. 1B) and pH 6.5 (Fig. 4A), reduced PDI displaced CTA1 from the CT holotoxin. Likewise, reduced PDI could displace CTA1 from the CT holotoxin in the presence of 10% glycerol (26). It therefore appeared that PDI can dislodge CTA1 from CTA2/CTB5 without unfolding the A1 subunit. To further examine this possibility, we added PDI to the CT holotoxin at 10 °C or 4 °C. PDI does not induce CTA1 to assume a protease-sensitive conformation at 4 °C, and this has been interpreted to mean that PDI cannot unfold CTA1 at low temperatures (29). However, PDI could still promote the dissociation of CTA1 from CTA2/CTB5 at either 10 °C (Fig. 4B) or 4 °C (supplemental Fig. S4). This again demonstrated that CTA1 unfolding was not required for its dissociation from the CT holotoxin.
FIGURE 4.
CT disassembly under conditions that prevent the unfolding of CTA1. After appending CT to a GM1-coated sensor slide, a baseline measurement corresponding to the mass of the bound holotoxin was recorded. Reduced PDI (PDI + 1 mm GSH) was then perfused over the CT-coated sensor at either (A) 37 °C in pH 6.5 buffer or (B) 10 °C in pH 7.0 buffer. PDI was removed 400 s into the experiment and replaced with sequential additions of anti-PDI, anti-CTA, and anti-KDEL antibodies as indicated by the arrowheads. In panel A, an anti-CTB antibody was also perfused over the slide as indicated by the final arrowhead.
Heating the reduced CT holotoxin to 65 °C, a temperature that results in irreversible denaturation of the holotoxin-associated CTA1 subunit (37), did not result in the displacement of CTA1 from CTA2/CTB5 (supplemental Fig. S5). It therefore appeared that unfolding alone is insufficient to displace CTA1 from CTA2/CTB5. Collectively, these results suggested that PDI removes CTA1 from CTA2/CTB5 by a mechanism that does not involve unfolding of the holotoxin-associated CTA1 subunit.
To determine the fate of PDI after it facilitates the separation of CTA1 from CTA2/CTB5, we repeated the disassembly assay with a holotoxin that was appended to the SPR sensor slide with a rabbit anti-CTA antibody. CTA2, CTB5, and PDI were absent from the sensor slide after PDI-mediated disassembly of the holotoxin at 37 °C (Fig. 5). Use of an anti-CTA monoclonal antibody confirmed that CTA1 was still on the plate. This experiment demonstrated that PDI does not remain associated with CTA1 after holotoxin disassembly.
FIGURE 5.
Association of PDI with CTA1 after holotoxin disassembly. CT was appended to a SPR sensor slide coated with a polyclonal anti-CTA antibody, and a baseline measurement corresponding to the mass of the bound holotoxin was recorded. Reduced PDI (PDI + 1 mm GSH) was then perfused over the CT-coated sensor at 37 °C. PDI was removed from the perfusion buffer 400 s into the experiment and replaced with sequential additions of an anti-PDI antibody, an anti-CTB antibody, an anti-KDEL antibody, and an anti-CTA monoclonal antibody as indicated by the arrowheads. An identical result was obtained when CT was initially appended to a sensor slide coated with the monoclonal anti-CTA antibody and later detected with the anti-CTA polyclonal antibody.
PDI Does Not Unfold the CTA1 Polypeptide
To directly examine the effect of PDI binding on the conformational state of CTA1, we performed structural studies using isotope-edited FTIR spectroscopy. This method allows the conformation of a 13C-labeled protein to be monitored in the presence of a second unlabeled protein. 13C labeling does not alter the conformation of a protein. However, the heavier nuclear mass of the stable 13C isotope generates a spectral downshift which allows the FTIR spectrum of a 13C-labeled protein to be resolved from the spectrum of an unlabeled protein (32, 40). The FTIR spectra of 13C-labeled CTA1 accordingly exhibited a 50 cm−1 shift in comparison to the spectra of unlabeled CTA1 (supplemental Fig. S6A). As expected, no structural differences between labeled and unlabeled CTA1 were detected by CD over a range of temperatures (supplemental Fig. S6, B and C).
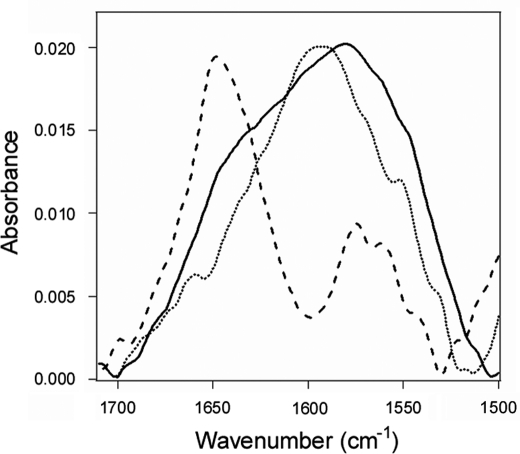
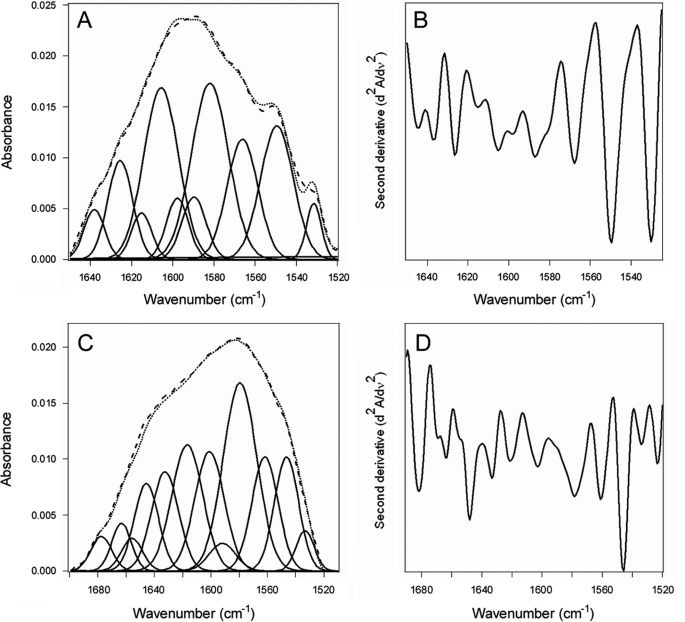
The down-shifted FTIR spectra of 13C-labeled CTA1 could be readily distinguished from the spectra of PDI (Fig. 6), allowing us to directly examine the impact of PDI on CTA1 structure. Analysis of the 10 °C FTIR spectrum from CTA1 alone indicated that the folded toxin contains 37% α-helical and 45% β-sheet content (Fig. 7, A and B, Table 1). This result was consistent with the 36% α-helix/51% β-sheet content predicted from the crystal structure of the folded, holotoxin-associated CTA1 subunit (41). The α-helical, β-sheet, and irregular (often referred to as “random”) structures of CTA1 were unaltered in the presence of PDI (Fig. 7, C and D, Table 1). Thus, the interaction between PDI and CTA1 at 10 °C which displaces CTA1 from the CT holotoxin (Fig. 4B) did not result in any significant disordering of the CTA1 polypeptide. In contrast, because of its intrinsic thermal instability, heating CTA1 alone at neutral pH (supplemental Fig. S7) shifted the percentage of its irregular structure from 8% at 10 °C to 53% at 37 °C (Table 1). The gain in irregular structure was accompanied by a loss of both α-helical and β-sheet structure. These observations were consistent with previous CD measurements that documented the partially disordered conformation of CTA1 at 37 °C (17, 26, 27). Our FTIR data thus demonstrated that physiological temperature alone has a dramatic disordering effect on CTA1 structure, while the interaction with PDI exerts little effect on the conformation of CTA1.
FIGURE 6.
Effect of PDI binding on CTA1 structure. FTIR spectra were recorded for PDI (dashed line), 13C-labeled CTA1 (dotted line), or a 1:1 molar ratio of PDI + 13C-labeled CTA1 (solid line). All measurements were taken at 10 °C with 1 mm GSH in the pH 7.0 buffer.
FIGURE 7.
Structure of CTA1 in the absence or presence of PDI as evaluated by FTIR at 10 °C. Curve-fitting (A and C) and second derivatives (B and D) for the FTIR spectrum of 13C-labeled CTA1 recorded in the absence (A and B) or presence (C and D) of PDI are shown. In panels A and C, the dotted line represents the sum of all deconvoluted components (solid lines) from the measured spectrum (dashed line). Note that different wavenumber scales are used in panels A and B than in panels and C and D.
TABLE 1.
PDI does not induce unfolding of the CTA1 polypeptide
Deconvolution of the conformation-sensitive amide I bands from FTIR data presented in Figs. 7, S7, S8, and S9 was used to calculate, as detailed in supplemental data, the percentages of CTA1 structure under various conditions. The means ± standard deviations from four separate curve-fitting iterations are shown.
| Condition | % of CTA1 structure |
|||
|---|---|---|---|---|
| α-Helix | β-Sheet | Irregular | Other | |
| 10 °C | 37 ± 1 | 45 ± 3 | 8 ± 2 | 9 ± 2 |
| 10 °C + PDI | 36 ± 1 | 44 ± 2 | 8 ± 1 | 10 ± 2 |
| 30 °C | 34 ± 4 | 35 ± 2 | 29 ± 4 | 12 ± 2 |
| 30 °C + PDI | 27 ± 2 | 38 ± 3 | 25 ± 2 | 10 ± 1 |
| 37 °C pH 6.5 | 29 ± 2 | 37 ± 4 | 25 ± 3 | 9 ± 2 |
| 37 °C pH 6.5 + PDI | 24 ± 2 | 35 ± 3 | 30 ± 1 | 11 ± 4 |
| 37 °C | 21 ± 2 | 15 ± 4 | 53 ± 2 | 11 ± 4 |
To determine if the putative unfoldase activity of PDI could be detected at temperatures greater than 10 °C, we performed additional experiments in which reduced PDI was added to CTA1 at 30 °C (supplemental Fig. S8) or to an acid-stabilized CTA1 subunit at 37 °C (supplemental Fig. S9). Although a temperature of 30 °C does not promote an optimal interaction between PDI and CTA1 (Fig. 2), we chose this temperature because most of the CTA1 protease sensitivity assays have been performed at 30 °C (9, 13, 23, 29). PDI and CTA1 do not normally interact at 37 °C (Fig. 2), so we used acidified medium as a means to facilitate PDI-CTA1 interactions at physiological temperature (Fig. 3A). Previous CD analysis has shown that CTA1 undergoes a progressive thermal disordering which is inhibited by mildly acidic pH (27). The FTIR data were consistent with this observation: the conformations of CTA1 at 30 °C and 37 °C at pH 6.5 contained less secondary structure than the 10 °C toxin but more secondary structure than the toxin incubated at 37 °C pH 7.0 (Table 1). At both 30 °C in neutral buffer and 37 °C in acidified buffer, the FTIR data demonstrated that reduced PDI does not substantially increase the percentage of irregular structure in CTA1 (Table 1). Thus, the earlier data that led to the unfoldase hypothesis may be interpreted in terms of a PDI-induced effect on CTA1 at 30 °C that yields an ordered but trypsin-sensitive toxin conformation. The ability of PDI to displace CTA1 from the CT holotoxin at 30 °C in neutral buffer (9) and at 37 °C in pH 6.5 buffer (Fig. 4A) again indicated that an unfoldase activity is not required for PDI to remove CTA1 from CTA2/CTB5.
PDI Is Required for CT Intoxication But Not for CTA1 Translocation
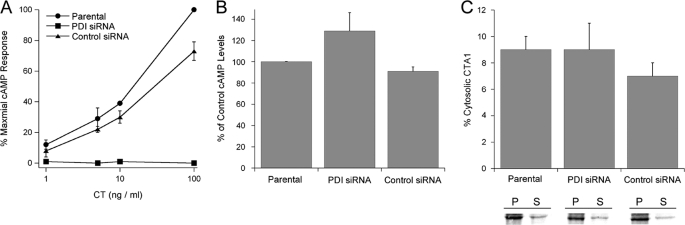
To examine the in vivo interaction between PDI and CT, we performed a CT toxicity assay in the TZM 1–2 cell line that is stably transfected with a PDI siRNA (42, 43). Experiments were also conducted with the parental TZM cells and with a TZM 5-1 cell line stably transfected with a nonspecific control siRNA (Fig. 8A). As expected, TZM 1-2 cells were completely resistant to CT. TZM 5-1 cells transfected with the control siRNA only exhibited a minor degree of toxin resistance at the highest CT concentration, thus confirming that CT resistance in the TZM 1-2 cell line was specifically due to the loss of PDI expression. PDI was therefore essential for holotoxin disassembly and productive CT intoxication.
FIGURE 8.
In vivo role of PDI in CT intoxication and CTA1 translocation. A, parental TZM cells, TZM 1-2 cells stably transfected with PDI siRNA, and TZM 5-1 cells stably transfected with a nonspecific control siRNA were exposed to the stated concentrations of CT for 2 h. Toxicity was then assessed from the rise in intracellular cAMP. The averages ± standard deviations of three independent experiments with triplicate samples are shown. B, TZM, TZM 1-2, and TZM 5-1 cells were transfected with a plasmid encoding a CTA1 subunit appended with an ER-targeting sequence. cAMP levels recorded at 3 h post-transfection were standardized to CTA1 expression levels, and the resulting data were plotted as percentages of the control response from parental TZM cells. The averages ± ranges of two independent experiments are shown. C, TZM, TZM 1-2, and TZM 5-1 cells were transfected with a plasmid encoding a CTA1 subunit appended with an ER-targeting sequence. CTA1 immunoprecipitated from the membrane (P, pellet) and cytosolic (S, supernatant) fractions of metabolically labeled cells were resolved by SDS-PAGE. The averages ± ranges of cytosolic CTA1 calculated from two independent experiments are shown in the graph.
Our collective data suggested that PDI is only needed to displace CTA1 from CTA2/CTB5; the dissociated CTA1 subunit can then unfold spontaneously to trigger its ERAD-mediated dislocation. To test this model, we performed a transfection-based CTA1 toxicity assay in the TZM, TZM 1–2, and TZM 5–1 cell lines (Fig. 8B). A plasmid-based system was used to express CTA1 directly in the ER of transfected cells (35). cAMP levels were then quantified at 3 h post-transfection. To control for possible differences in transfection efficiencies between the cell lines, CTA1 was radiolabeled and immunoprecipitated from parallel sets of transfected cells. Results from the cAMP assay were subsequently standardized to CTA1 expression levels. Although TZM 1–2 cells were completely resistant to exogenously applied CT holotoxin, they exhibited no resistance to the ER-localized CTA1 construct. In fact, the cAMP levels in TZM 1-2 cells expressing ER-localized CTA1 were slightly elevated in comparison to the cAMP levels generated from CTA1 expression in either the parental TZM cells or TZM 5-1 control cells. Collectively, this data indicated that PDI function is not required for ER-localized CTA1 to enter the cytosol.
Previous work with metabolically labeled cells has demonstrated that the entire detectable pool of plasmid-expressed CTA1 is inserted into the ER (44). Yet, if the co-translational targeting of our ER-localized CTA1 construct was not completely efficient, then the CTA1 toxicity assay would be compromised by the small fraction of CTA1 that was synthesized in the cytosol rather than in the ER. To control for this possibility, we directly monitored the ER-to-cytosol translocation of transfected CTA1 in the parental TZM, TZM 1-2, and TZM 5-1 cell lines. Transfected, radiolabeled cells were partitioned into separate membrane and cytosolic fractions according to established protocols (26, 29, 34, 45). Material immunoprecipitated from each fraction with an anti-CTA antibody was then resolved by SDS-PAGE and quantified using PhosphorImager analysis. As shown in Fig. 8C, approximately equivalent amounts of CTA1 were recovered from the cytosolic fractions of all three cell lines. Thus, the lack of PDI in TZM 1-2 cells did not inhibit the ER-to-cytosol export of transfected CTA1. The results of Fig. 8 demonstrated that PDI is required for productive intoxication with exogenously applied CT but is not needed for translocation of the isolated CTA1 subunit. As such, it appears that the only role of PDI in cholera intoxication is to displace CTA1 from the CT holotoxin.
DISCUSSION
For CTA1 translocation, PDI has been proposed to act as a redox-dependent chaperone that actively unfolds the holotoxin-associated CTA1 subunit to separate CTA1 from the rest of the toxin (9). The unfoldase activity of PDI in this process is, to the best of our knowledge, a unique property that has not been reported for PDI interactions with any other substrate. Furthermore, the proposed unfoldase activity of PDI is based upon a protease sensitivity assay that only serves as an indirect measure of protein folding (9, 13, 23, 28, 29). In this work, we demonstrated that PDI is required for disassembly of the CT holotoxin but does not unfold the CTA1 subunit. CTA1 unfolds spontaneously after its dissociation from CTA2/CTB5 at physiological temperature (17, 26). PDI-mediated disassembly of the CT holotoxin thus leads to the spontaneous unfolding of dissociated CTA1, but PDI itself does not function as an unfoldase to actively unfold CTA1.
Using SPR, we were able to monitor the PDI-mediated disassembly of the CT holotoxin in real time. The kinetics of holotoxin disassembly were linked to the available concentration of PDI (supplemental Fig. S2). Holotoxin disassembly required the reduced form of PDI and did not occur in the presence of reducing agent alone (Fig. 1). These results were consistent with previous reports (9, 36), as was the observed retention of CTA2 with CTB5 after release of CTA1 from the holotoxin (Fig. 1B) (46). We could thus faithfully reconstitute the process of CT disassembly on an SPR sensor slide.
Conditions that block CTA1 unfolding in vitro also block the ER-to-cytosol export of CTA1 in vivo (26, 27). However, conditions that block CTA1 unfolding do not block CTA1 dissociation from the CT holotoxin: reduced PDI could separate CTA1 from CTA2/CTB5 in the presence of acidic pH (Fig. 4A) or 10% glycerol (26). Secretion of the free A1 subunit from glycerol-treated cells (26) further suggested that in vivo unfolding of CTA1 is unnecessary for toxin disassembly: only folded proteins exit the ER for secretory transport, so the secreted pool of CTA1 must have been in a folded conformation. CT disassembly could also occur at 4 °C (supplemental Fig. S4), a temperature that maintains CTA1 in a folded conformation and prevents the PDI-induced shift of CTA1 to a protease-resistant conformation (29). Collectively, these results strongly suggested that CTA1 unfolding is not required for CTA1 dissociation from the rest of the toxin. In further support of this interpretation, we demonstrated that the thermal denaturation of reduced but holotoxin-associated CTA1 did not displace CTA1 from its non-covalent assembly in the CT holotoxin (supplemental Fig. S5). A similar observation was previously made by Goins and Friere with the technique of differential scanning calorimetry (37). Thus, PDI apparently removes CTA1 from CTA2/CTB5 by a mechanism that does not involve unfolding of the holotoxin-associated CTA1 subunit.
To directly examine the putative unfoldase activity of PDI, we employed the structural technique of isotope-edited FTIR spectroscopy. Experiments were performed under three conditions that facilitate PDI-CTA1 interactions and support PDI-mediated disassembly of the CT holotoxin: 10 °C, 30 °C, and 37 °C at pH 6.5. For each condition, deconvolution of the conformation-sensitive amide I bands demonstrated that PDI did not substantially alter the percentage of irregular structure in CTA1 and, thus, did not unfold the CTA1 polypeptide (Table 1). In contrast, heating CTA1 alone at neutral pH shifted the percentage of its irregular structure from 8% at 10 °C to 53% at 37 °C at the expense of both α-helix and β-sheet structures (Table 1). CT disassembly therefore occurs under conditions that do not involve substantial PDI-induced disordering of the CTA1 polypeptide. These results, which represent the first direct examination of the impact of PDI binding on CTA1 structure, demonstrated that CTA1 is unfolded by physiological temperature (due to its intrinsic thermal instability) but not by PDI.
The unfoldase activity of PDI has only been described with a biochemical protease sensitivity assay. Using this method, the folding state of a protein is inferred from its susceptibility to proteolysis: a folded protein is generally more resistant to proteolysis than an unfolded variant of the same protein. The resistance of CTA1 to trypsin-mediated proteolysis, combined with the sensitivity of PDI-treated CTA1 to proteolysis, thus yielded the conclusion that PDI actively unfolds the CTA1 subunit. Unfortunately, interpretation of these experiments is complicated by the conformational shift in CTA1 that occurs upon its separation from the holotoxin and by the unstable, heat-labile nature of the free A1 subunit (17). Protease concentration can also affect the outcome of the experiment, as can the temperature of the experiment (17, 26, 28). It should be noted that most experiments to monitor the “unfoldase” activity of PDI have been performed at 30 °C (9, 13, 23, 29), but incubation of CTA1 alone at 37 °C will induce the toxin to assume a protease-sensitive conformation (17, 26) (supplemental Fig. S10). Furthermore, the sensitivity of PDI-treated CTA1 to proteolysis depends upon the protease used: the toxin is more sensitive to trypsin (9) than to the thermolysin protease which cleaves bulky and aromatic residues (supplemental Fig. S10). These issues suggest that protease sensitivity is a poor, indirect measure of the purported conformational change required for CTA1 dissociation from the CT holotoxin. More important, however, is the disconnect between the PDI-induced shift of CTA1 to a trypsin-sensitive conformation and the PDI-mediated disassembly of the CT holotoxin. PDI does not induce CTA1 to assume a protease-sensitive conformation at 4 °C (29), yet PDI can still displace CTA1 from CTA2/CTB5 at 4 °C (supplemental Fig. S4). These results demonstrate that the PDI-induced shift of CTA1 to a protease-sensitive conformation, which has been interpreted as an unfolding event (9, 13, 23, 28, 29), does not correlate to the PDI-induced separation of CTA1 from CTA2/CTB5.
PDI binds to folded but not unfolded conformations of CTA1 (Figs. 1–3). The spontaneous unfolding of CTA1 that occurs upon holotoxin disassembly at 37 °C thus displaces the toxin-bound PDI (Figs. 3 and 5). This observation is consistent with a previous report that concluded PDI-substrate interactions can be disrupted by changes to the structure of the substrate (24). However, our results conflict with the model of CTA1-PDI interactions that proposes PDI oxidation by Ero1p is responsible for displacing toxin-bound PDI (23). That model is based upon studies performed at temperatures below 37 °C. While Ero1p clearly regulates the redox status of PDI (22, 23, 47, 48), our results strongly suggest that Ero1p function is not required to release PDI from CTA1 at physiological temperature.
As demonstrated with cell-based assays, CTA1 translocation does not require PDI function after disassembly of the holotoxin. PDI-deficient cell lines were completely resistant to exogenously applied CT, as expected from its essential role in separating CTA1 from CTA2/CTB5. However, the same PDI-deficient cells displayed wild-type sensitivity to a CTA1 construct that was directly expressed in the ER of transfected cells. This experimental condition, which mimicked the status of ER-localized CTA1 after its dissociation from the holotoxin, demonstrated that PDI is not required for the ER-to-cytosol export of free CTA1. Direct monitoring of the CTA1 translocation event provided further support for this conclusion.
Based on the available biophysical, biochemical, and cell biological data, we propose the following revised model of CTA1 translocation. CT travels from the cell surface to the ER as an intact holotoxin (4). The disulfide bond linking CTA1 to CTA2/CTB5 is reduced at the resident redox state of the ER (8). However, at this point CTA1 remains anchored to the holotoxin through non-covalent interactions (9, 36). Reduced PDI then binds to a region in the A11 subdomain of CTA1 and physically displaces CTA1 from the holotoxin. Released from the structural constraints of its non-covalent interactions with CTA2/CTB5, the dissociated CTA1 subunit undergoes a thermal transition to a partially unfolded conformation (17). This unfolding event, which occurs spontaneously at 37 °C, displaces the toxin-bound PDI and identifies free CTA1 as a misfolded protein for processing by the ERAD system. CTA1 is consequently delivered to the Sec61p and/or Derlin-1 pores for Hsp90-mediated extraction to the cytosol (21, 25, 34, 49, 50). An interaction with ARF and possibly other host factors then allows the cytosolic pool of CTA1 to regain an active conformation for the ADP-ribosylation of its Gsα target (16–18).
Toxin-ERAD interactions involving pertussis toxin (PT) and ricin may follow a similar pattern to CT. Like CTA1, the catalytic PTS1 subunit shifts to a disordered conformation after separation from its B oligomer at physiological temperature (51). This conformational shift would identify PTS1 as an ERAD substrate and thereby facilitate PTS1 passage into the cytosol. Ricin A chain is more stable than either CTA1 or PTS1 (52), but an interaction with anionic phospholipids of the ER membrane at 37 °C induces ricin A chain to assume a disordered conformation that would be treated as an ERAD substrate (53, 54). Furthermore, the interaction between PDI and ricin mirrors the interaction between PDI and CT: PDI is responsible for disassembly of the ricin holotoxin but does not unfold ricin A chain (55, 56). Our results are thus consistent with a general model of toxin translocation in which PDI-mediated toxin disassembly and temperature-induced A chain unfolding are used in similar fashion by multiple ERAD-exploiting toxins.
Supplementary Material
Acknowledgment
We thank Dr. Jonathan Silver (Laboratory of Molecular Microbiology, NIAID, NIH, Bethesda, Maryland) for the kind gift of TZM, TZM 1-2, and TZM 5-1 cells.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI073783 (to K. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10.
- CT
- cholera toxin
- CD
- circular dichroism
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- FTIR
- Fourier transform infrared
- PT
- pertussis toxin
- PDI
- protein-disulfide isomerase
- SPR
- surface plasmon resonance.
REFERENCES
- 1. De Haan L., Hirst T. R. (2004) Mol. Membr. Biol. 21, 77–92 [DOI] [PubMed] [Google Scholar]
- 2. Sánchez J., Holmgren J. (2008) Cell Mol. Life Sci. 65, 1347–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang R. G., Scott D. L., Westbrook M. L., Nance S., Spangler B. D., Shipley G. G., Westbrook E. M. (1995) J. Mol. Biol. 251, 563–573 [DOI] [PubMed] [Google Scholar]
- 4. Wernick N. L., Chinnapen D. J., Cho J. A., Lencer W. I. (2010) Toxins 2, 310–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lencer W. I., Constable C., Moe S., Jobling M. G., Webb H. M., Ruston S., Madara J. L., Hirst T. R., Holmes R. K. (1995) J. Cell Biol. 131, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lencer W. I., de Almeida J. B., Moe S., Stow J. L., Ausiello D. A., Madara J. L. (1993) J. Clin. Invest. 92, 2941–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Orlandi P. A. (1997) J. Biol. Chem. 272, 4591–4599 [PubMed] [Google Scholar]
- 8. Majoul I., Ferrari D., Söling H. D. (1997) FEBS Lett. 401, 104–108 [DOI] [PubMed] [Google Scholar]
- 9. Tsai B., Rodighiero C., Lencer W. I., Rapoport T. A. (2001) Cell 104, 937–948 [DOI] [PubMed] [Google Scholar]
- 10. Teter K., Holmes R. K. (2002) Infect. Immun. 70, 6172–6179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Teter K., Jobling M. G., Holmes R. K. (2003) Traffic 4, 232–242 [DOI] [PubMed] [Google Scholar]
- 12. Hazes B., Read R. J. (1997) Biochemistry 36, 11051–11054 [DOI] [PubMed] [Google Scholar]
- 13. Rodighiero C., Tsai B., Rapoport T. A., Lencer W. I. (2002) EMBO Rep. 3, 1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deeks E. D., Cook J. P., Day P. J., Smith D. C., Roberts L. M., Lord J. M. (2002) Biochemistry 41, 3405–3413 [DOI] [PubMed] [Google Scholar]
- 15. Worthington Z. E., Carbonetti N. H. (2007) Infect Immun. 75, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murayama T., Tsai S. C., Adamik R., Moss J., Vaughan M. (1993) Biochemistry 32, 561–566 [DOI] [PubMed] [Google Scholar]
- 17. Pande A. H., Scaglione P., Taylor M., Nemec K. N., Tuthill S., Moe D., Holmes R. K., Tatulian S. A., Teter K. (2007) J. Mol. Biol. 374, 1114–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ampapathi R. S., Creath A. L., Lou D. I., Craft J. W., Jr., Blanke S. R., Legge G. B. (2008) J. Mol. Biol. 377, 748–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferrari D. M., Söling H. D. (1999) Biochem. J. 339, 1–10 [PMC free article] [PubMed] [Google Scholar]
- 20. Hatahet F., Ruddock L. W. (2007) FEBS J 274, 5223–5234 [DOI] [PubMed] [Google Scholar]
- 21. Bernardi K. M., Forster M. L., Lencer W. I., Tsai B. (2008) Mol. Biol. Cell 19, 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moore P., Bernardi K. M., Tsai B. (2010) Mol. Biol. Cell 21, 1305–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsai B., Rapoport T. A. (2002) J. Cell Biol. 159, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lumb R. A., Bulleid N. J. (2002) EMBO J. 21, 6763–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saslowsky D. E., Cho J. A., Chinnapen H., Massol R. H., Chinnapen D. J., Wagner J. S., De Luca H. E., Kam W., Paw B. H., Lencer W. I. (2010) J. Clin. Invest. 120, 4399–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Massey S., Banerjee T., Pande A. H., Taylor M., Tatulian S. A., Teter K. (2009) J. Mol. Biol. 393, 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Banerjee T., Pande A., Jobling M. G., Taylor M., Massey S., Holmes R. K., Tatulian S. A., Teter K. (2010) Biochemistry 49, 8839–8846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Forster M. L., Sivick K., Park Y. N., Arvan P., Lencer W. I., Tsai B. (2006) J. Cell Biol. 173, 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Forster M. L., Mahn J. J., Tsai B. (2009) J. Biol. Chem. 284, 13045–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holmes R. K., Twiddy E. M. (1983) Infect Immun. 42, 914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Teter K., Jobling M. G., Sentz D., Holmes R. K. (2006) Infect Immun. 74, 2259–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tatulian S. A. (2003) Biochemistry 42, 11898–11907 [DOI] [PubMed] [Google Scholar]
- 33. Tatulian S. A., Cortes D. M., Perozo E. (1998) FEBS Lett. 423, 205–212 [DOI] [PubMed] [Google Scholar]
- 34. Taylor M., Navarro-Garcia F., Huerta J., Burress H., Massey S., Ireton K., Teter K. (2010) J. Biol. Chem. 285, 31261–31267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Teter K., Jobling M. G., Holmes R. K. (2004) Infect Immun. 72, 6826–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mekalanos J. J., Collier R. J., Romig W. R. (1979) J. Biol. Chem. 254, 5855–5861 [PubMed] [Google Scholar]
- 37. Goins B., Freire E. (1988) Biochemistry 27, 2046–2052 [DOI] [PubMed] [Google Scholar]
- 38. Surewicz W. K., Leddy J. J., Mantsch H. H. (1990) Biochemistry 29, 8106–8111 [DOI] [PubMed] [Google Scholar]
- 39. Lencer W. I., Tsai B. (2003) Trends Biochem. Sci. 28, 639–645 [DOI] [PubMed] [Google Scholar]
- 40. Tatulian S. A. (2010) Spectroscopy Int. J. 24, 37–43 [Google Scholar]
- 41. O'Neal C. J., Jobling M. G., Holmes R. K., Hol W. G. (2005) Science 309, 1093–1096 [DOI] [PubMed] [Google Scholar]
- 42. Gilbert J., Ou W., Silver J., Benjamin T. (2006) J. Virol. 80, 10868–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ou W., Silver J. (2006) Virology 350, 406–417 [DOI] [PubMed] [Google Scholar]
- 44. Teter K., Allyn R. L., Jobling M. G., Holmes R. K. (2002) Infect Immun. 70, 6166–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kaiser E., Pust S., Kroll C., Barth H. (2009) Cell Microbiol. 11, 780–795 [DOI] [PubMed] [Google Scholar]
- 46. Wernick N. L., De Luca H., Kam W. R., Lencer W. I. (2010) J. Biol. Chem. 285, 6145–6152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frand A. R., Kaiser C. A. (1999) Mol. Cell 4, 469–477 [DOI] [PubMed] [Google Scholar]
- 48. Inaba K., Masui S., Iida H., Vavassori S., Sitia R., Suzuki M. (2010) EMBO J. 29, 3330–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schmitz A., Herrgen H., Winkeler A., Herzog V. (2000) J. Cell Biol. 148, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dixit G., Mikoryak C., Hayslett T., Bhat A., Draper R. K. (2008) Exp. Biol. Med. 233, 163–175 [DOI] [PubMed] [Google Scholar]
- 51. Pande A. H., Moe D., Jamnadas M., Tatulian S. A., Teter K. (2006) Biochemistry 45, 13734–13740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Argent R. H., Parrott A. M., Day P. J., Roberts L. M., Stockley P. G., Lord J. M., Radford S. E. (2000) J. Biol. Chem. 275, 9263–9269 [DOI] [PubMed] [Google Scholar]
- 53. Day P. J., Pinheiro T. J., Roberts L. M., Lord J. M. (2002) Biochemistry 41, 2836–2843 [DOI] [PubMed] [Google Scholar]
- 54. Mayerhofer P. U., Cook J. P., Wahlman J., Pinheiro T. T., Moore K. A., Lord J. M., Johnson A. E., Roberts L. M. (2009) J. Biol. Chem. 284, 10232–10242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bellisola G., Fracasso G., Ippoliti R., Menestrina G., Rosén A., Soldà S., Udali S., Tomazzolli R., Tridente G., Colombatti M. (2004) Biochem. Pharmacol. 67, 1721–1731 [DOI] [PubMed] [Google Scholar]
- 56. Spooner R. A., Watson P. D., Marsden C. J., Smith D. C., Moore K. A., Cook J. P., Lord J. M., Roberts L. M. (2004) Biochem. J. 383, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.