Abstract
CD38 catalyzes the synthesis of cyclic ADP-ribose (cADPR), a Ca2+ messenger responsible for regulating a wide range of physiological functions. It is generally regarded as an ectoenzyme, but its intracellular localization has also been well documented. It is not known if internal CD38 is enzymatically active and contributes to the Ca2+ signaling function. In this study, we engineered a novel soluble form of CD38 that can be efficiently expressed in the cytosol and use cytosolic NAD as a substrate to produce cADPR intracellularly. The activity of the engineered CD38 could be decreased by mutating the catalytic residue Glu-226 and increased by the double mutation E146A/T221F, which increased its cADPR synthesis activity by >11-fold. Remarkably, the engineered CD38 exhibited the ability to form the critical disulfide linkages required for its enzymatic activity. This was verified by using a monoclonal antibody generated against a critical disulfide, Cys-254–Cys-275. The specificity of the antibody was established by x-ray crystallography and site-directed mutagenesis. The engineered CD38 is thus a novel example challenging the general belief that cytosolic proteins do not possess disulfides. As a further refinement of this approach, the engineered CD38 was placed under the control of tetracycline using an autoregulated construct. This study has set the stage for in vivo manipulation of cADPR metabolism.
Keywords: Calcium Intracellular Release, Cellular Regulation, Crystallography, Cyclic Nucleotides, Enzyme Mechanisms, Enzyme Structure, NAD, ADP-ribosyl Cyclase, CD38, Cyclic ADP-ribose
Introduction
Mobilization of Ca2+ from intracellular stores is of fundamental importance in virtually all aspects of cellular activity. The major intracellular Ca2+ stores are in the endoplasmic reticulum (ER)2 and are mobilized by specific messenger molecules, inositol trisphosphate and cyclic ADP-ribose (cADPR). The latter is a novel cyclic nucleotide derived from NAD. It was first described in sea urchin eggs (1, 2) but has since been established as a second messenger molecule responsible for regulating a wide range of physiological functions as diverse as abscisic acid signaling in plants (3) and sponges (4) and social behavior in mice (Ref. 5; reviewed in Refs. 6 and 7). It targets the ryanodine receptor of the endoplasmic Ca2+ stores.
The synthesis and hydrolysis of cADPR in mammalian cells are catalyzed by CD38 (8), a transmembrane protein ubiquitously expressed in virtually all tissues (reviewed in Ref. 9). Gene knock-out studies have established that CD38 plays a critical role in a wide range of physiological functions, including insulin secretion (10), susceptibility to bacterial infection (11), and social behavior of mice through modulating neuronal oxytocin secretion (5).
CD38 is a membrane protein with a short N-terminal tail, a single transmembrane segment, and a large C-terminal domain containing all the enzymatic activities (8, 12, 13). The crystal structure of the catalytic domain of CD38 has been solved, and the mechanism of how it catalyzes the multiple reactions responsible for metabolizing cADPR has also been elucidated to atomic resolution by x-ray crystallography (14–16). It is established that the substrate NAD enters the active site pocket located near the middle of CD38 with the nicotinamide end first. The interaction with the catalytic residue Glu-226 results in the release of the nicotinamide group, forming an intermediate. Subsequent linkage between N1 of the adenine and the anomeric carbon of the terminal ribose results in cyclization and the formation of cADPR.
It is generally believed that CD38 is expressed on the cell surface as a type II membrane protein with its catalytic domain located outside the cell. Paradoxically, it is also well established that not only is its substrate (NAD) cytosolic but that its product (cADPR) targets the endoplasmic Ca2+ stores (17, 18). This represents a topological paradox for the signaling functions of CD38 that has not been fully resolved (19). Elaborate mechanisms have been advanced to show that NAD can be released from cells via the connexin hemichannels, whereas nucleoside transporters present in the cytoplasmic membrane can transport its product, cADPR, back into the cell to effect Ca2+ release from the endoplasmic stores (20–22).
On the other hand, it is equally well documented that CD38 is also expressed intracellularly (23–27). The orientation of intracellular CD38 has not been definitively determined, but evidence suggests that its catalytic domain may face the cytosol or its topological equivalents (23, 24). That one protein can be expressed in two alternative membrane orientations has precedence: the prion proteins (28–30) and the multidrug resistance transporter EmrE (31, 32). The topological problem can be easily resolved if internal CD38 is enzymatically active in synthesizing cADPR using cytosolic NAD as a substrate. However, it is generally believed that cytosolic proteins, or cytosol-facing membrane proteins, do not contain disulfide. On the other hand, CD38 has six disulfides that are important for its catalytic activities (16, 33). It is thus important to determine whether internal CD38 can form the critical disulfides and can catalyze the synthesis of cADPR intracellularly.
In this study, we show that native CD38, a transmembrane protein, can be engineered for expression in the cytosol in a soluble form and is fully active in elevating cellular cADPR levels. This finding not only provides an important clue for resolving the topological paradox of the signaling function of CD38 but also sets the stage for using the engineered CD38 to manipulate the cADPR levels in cells and tissues.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293T and NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 mg/ml streptomycin in a humidified 5% CO2 atmosphere at 37 °C. For cells infected with lentivirus containing tetracycline-inducible vectors, tetracycline-free serum (catalogue no. 10437028, Invitrogen) was used. LipofectamineTM 2000 (Invitrogen) was used to transfect HEK293T cells.
Constructs and Lentivirus Production and Infection
To make the construct pEGFPN1-CD38, the cDNA of human CD38 was amplified from Jurkat cell cDNA and cloned into pEGFP-N1 (Clontech) with KpnI and HindIII. To construct pEGFP-sCD38, the DNA encoding amino acids 46–300 of human CD38 was subcloned into pEGFP-C1 (Clontech) with KpnI and SacI. To construct pEGFP-sCD38(DM), the double mutant CD38(E146A/T221F) (14) was used as a template for amplification and subcloned into pEGFP-C1. To construct pEGFP-sCD38 with various cysteine-to-serine mutations (C67S, C99S, C160S, C201S, C254S, and C296S), mutagenesis was performed on the pEGFP-sCD38 parent plasmid using a QuikChange II XL site-directed mutagenesis kit (Stratagene). To make the pCHMWS series lentivector constructs, wtCD38-EGFP, EGFP-sCD38, and EGFP-sCD38(DM) were subcloned from the corresponding pEGFP vector into pCHMWS-EGFP, which was a gift from the Pasteur Research Centre of The University of Hong Kong. For the tetracycline-inducible expression lentivector, EGFP-sCD38 and EGFP-sCD38(DM) were amplified by PCR and ligated with AgeI/PstI into TREAuto-V14, which was a gift from N. Legrand (University of Amsterdam) (34). pDsRed was purchased from Clontech. pDsRed-ER was a gift from J. Marchant (University of Minnesota).
The second generation of the packaging system from Dr. Didier Trono's laboratory (University of Geneva) was used to generate lentivirus. HEK293T cells were cotransfected with the pCHMWS or TREAuto-V14 vector and the two packaging plasmids (pPAX2 and pMD2.G). The medium was replaced with fresh medium 8 h post-transfection. Approximately 72 h after infection, the supernatants were harvested, cleared by brief centrifugation, and filtrated with 0.45-μm filters (catalogue no. PN 4614, Pall Corp.). Aliquots of the virus were then stored at −80 °C.
HEK293T or NIH 3T3 cells (2 × 104) were incubated for 18 h with 0.5 ml of viral supernatants (containing ∼2 × 104 functional viral particles) in the presence of Polybrene (5 μg/ml). The infected cells were then cultured in the regular complete medium for 48 h before they were analyzed for protein expression, cADPR production, and cell growth.
Antibodies and Immunofluorescence Microscopy
The polyclonal antibody against human CD38 (Ab77) was a gift from Richard M. Graeff (University of Minnesota). Monoclonal antibody HB7 was purified from the ascites fluid produced in nude mice that were injected with the HB7 hybridoma cells (catalogue no. HB-136, American Type Culture Collection). Anti-GRP94 antibody was a product of Thermo Scientific (catalogue no. MA3-016). For confocal imaging, HEK293T cells were transfected with different constructs in FluoroDish with a cover glass bottom (World Precision Instruments, Inc.). 48 h post-transfection, images were acquired by Zeiss LSM 510 META confocal laser scanning.
Reducing and Nonreducing SDS-PAGE
After cells were lysed in lysis buffer (PBS, 0.5% Triton X-100, 100 μm PMSF, 10 μg/ml leupeptin, 10 μg/ml pepstatin, and 10 μg/ml aprotinin), the supernatant was collected, and the protein concentration was determined by the Bradford method (35) using BSA as a standard. Under reducing conditions, samples were mixed with Laemmli sample buffer with 100 mm DTT replacing the regular 2-mercaptoethanol. Under nonreducing conditions, Laemmli buffer without 2-mercaptoethanol was used. After incubation at 65 °C for 15 min, protein samples were analyzed by 10% SDS-PAGE and electrotransferred onto an Immobilon-P blotting membrane (Millipore). After blocking with 10% (w/v) nonfat milk in saline buffered with 30 mm Tris-HCl (pH 8.0) and containing 0.05% Tween 20 (TBST), the membranes were incubated with the respective antibodies (1:1000 dilution for antibody HB7 in 3% milk/TBST, 1:2000 dilution for antibody Ab77, and 1:5000 dilution for anti-GRP94 antibody) at 4 °C overnight, followed by incubation with horseradish peroxidase-conjugated anti-rabbit (1:10,000 dilution for antibody Ab77), anti-mouse (1:5000 dilution for antibody HB7), or anti-rat (1:10,000 dilution for anti-GRP94 antibody) antibodies for 2 h. Chemiluminescence detection of horseradish peroxidase was performed by incubation of the membranes with ECL (Roche Applied Science) or ImmobilonTM Western (Millipore) reagent for 1–2 min, followed by detection of the chemiluminescence signals using x-ray films (Fuji). The digitized band intensities were analyzed using ImageJ software.
Intracellular cADPR Content Measurements
The cADPR contents were measured by the cycling assay described previously (36). Briefly, cells were washed twice with PBS, pelleted at 5000 × g for 15 s, and lysed in 0.3 ml of ice-cold perchloric acid (0.6 m). The lysates were centrifuged at 14,000 × g for 5 min to remove precipitates. Perchloric acid was removed by mixing the aqueous supernatants with chloroform/tri-n-octylamine (3:1), and the aqueous phase was collected and neutralized with 2 mm Tris base. To remove contaminating nucleotides, the samples were incubated overnight at 37 °C with 0.44 units/ml nucleotide pyrophosphatase in 100 mm sodium phosphate buffer (pH 7.0) containing 2 mm MgCl2. The enzyme were removed by filtration using MultiScreenHTSTM (catalogue no. MSIPN4510, Millipore). To measure the cADPR contents, samples (20 μl/well) were incubated in 96-well flat white plates (catalogue no. 3912, Corning Costar) with 100 μl the cycling reagent, which contained 2% ethanol, 0.1 mg/ml alcohol dehydrogenase, 20 μm resazurin, 11 μg/ml diaphorase, 10 μm riboflavin 5′-phosphate, and 10 mm nicotinamide with or without 0.2 μg/ml Aplysia ADP-ribosyl cyclase in 100 mm sodium phosphate (pH 7.0). An increase in the resorufin fluorescence was measured at 544 nm excitation and 590 nm emission using a fluorescence plate reader (Infinite M200, Tecan). Various cADPR standards with known concentrations were assayed in parallel to generate a standard curve. The results are presented as picomoles of cADPR/mg of protein or picomoles of cADPR/unit of recombinant protein. The unit of recombinant protein was calculated by the band intensity measured in Western blots of the cell extracts using anti-CD38 antibody Ab77.
Cell Fractionation
To generate stable cell lines expressing EGFP-sCD38, EGFP-sCD38(DM), or wtCD38-EGFP, HEK293T cells were infected with pCHMWS virus carrying the corresponding genes. These cells were then used to determine the subcellular localization of the expressed proteins. For differential fractionation, cells were treated with 0.02% digitonin for 5 min on ice and centrifuged at 1500 × g for 10 min at 4 °C. The pellets (P1.5) were saved, and the supernatants were further centrifuged at 10,000 × g and then at 100,000 × g for 30 min to 1 h at 4 °C. The pellets (P1.5, P10, and P100) and the final supernatant (S100) were used for Western analyses. The P1.5 samples were dissolved by sonication in PBS containing 0.5% Triton X-100 and protease inhibitors; the P10 and P100 samples were directly dissolved in Laemmli sample buffer.
Cell Permeabilization with Digitonin
HEK293T cells cultured in 10-cm plates were cotransfected with pEGFP-sCD38 and pDsRed/pDsRed-ER. 48 h post-transfection, cells were trypsinized, washed with PBS, and treated with different concentrations of digitonin on ice for 5 min. After centrifugation at 5000 × g for 5 min at 4 °C, the green and red fluorescence in the supernatant was measured using the fluorescence plate reader. The results were normalized to the maximal fluorescence intensity measured at the highest detergent concentrations.
Tetracycline-inducible Expression System
NIH 3T3 cells were infected with lentivirus carrying the genes for EGFP-sCD38(DM) in the TREAuto-V14 backbone. The cells were maintained in tetracycline-free medium for 3 days, seeded in 6-well plates, and used to determine the induction kinetics. Cells were incubated with 1 μg/ml tetracycline for different time periods and harvested to determine protein expression by EGFP fluorescence and intracellular cADPR contents as described above. To determine the time course for turning off the tetracycline induction, cells were first induced with 1 μg/ml tetracycline for 24 h. The medium was then changed to one without tetracycline. Samples were collected periodically afterward and similarly assayed for EGFP fusion protein expression and cADPR contents.
Recombinant CD38 Production
A yeast expression system including the pPICZαA expression vector and Pichia pastoris yeast (Invitrogen) was used to prepare wild-type CD38 as described previously (13, 17).
Monoclonal Antibody Production
Monoclonal antibody HB7 was produced from hybridoma cells. The cells were cultured in Hybri-CareTM medium (catalogue no. 46-X, American Type Culture Collection) and inoculated into BALB/c nude mice. The antibody was purified from the ascites fluids using a protein G column. The Fab fragment of HB7 was obtained by papain digestion in the presence of recombinant CD38 at a molar ration of 1:1. The HB7-CD38 complex was further purified on a Superdex 200 size exclusion column and concentrated up to 18 mg/ml in 20 mm HEPES (pH 7.0) and 50 mm sodium chloride.
Crystallization, Data Collection, and Structure Refinement
The purified HB7-CD38 complex was used in the crystallization screening. Crystals were obtained by the hanging drop vapor diffusion method with reservoir buffer in 3.6 m sodium formate (pH 7.0). They were harvested and flash-frozen in liquid nitrogen. The diffraction data were collected at 100 K at beamline BL17U at the Shanghai Synchrotron Radiation Facility and processed with HKL2000 (37). Molecular replacement was performed using the program Phaser (38) from CCP4 (39) with human wild-type CD38 (Protein Data Bank code 1YH3) as the first searching model. A protein sequence search using BLAST showed that the heavy chain of the antibody in Protein Data Bank 1T2Q and the light chain of the antibody in Protein Data Bank 3CVI have higher sequence identity to the heavy and light chains of HB7, respectively. A chimeric Fab V domain and a Fab C domain were generated by superimposing the two coordinates and were used as the second and third searching models in molecular replacement, respectively. The model was refined with Phenix (40) and then cycled with rebuilding in Coot (41). The final model was analyzed using MolProbity (42). Data collection and model refinement statistics are summarized in Table 1.
TABLE 1.
Crystallographic data and refinement statistics
Values in parentheses are from the highest resolution shell. NA, not applicable; r.m.s.d., root mean square deviation.
| Data collection | |
| Cell dimensions | a = 59.66, b = 271.52, c = 136.66 Å; α = β = γ = 90° |
| Space group | C222(1) |
| Resolution (Å) | 3.05 |
| Unique reflections | 19,878 |
| Multiplicity | 5.5 (4.5) |
| I/σ | 24.5 (2.6) |
| Rmerge (%)a | 0.085 (0.445) |
| Completeness (%) | 92.4 (72.5) |
| Refinement | |
| Resolution (Å) | 50–3.0 |
| R factor (%)b | 20.49 |
| Rfree factor (%)c | 27.5 |
| Protein atoms | 5034 |
| Ligands | 0 |
| Mean B-factor for protein atoms (Å2) | 98.4 |
| B-factor for ligands (Å2) | NA |
| r.m.s.d. | |
| Bond lengths (Å) | 0.01 |
| Bond angles | 1.36° |
a Rmerge = Σ|I − 〈I〉|/ΣI, where I is the integrated intensity of a given reflection.
b R = Σ‖Fo| − |Fc‖/Σ|Fo|.
c Rfree was calculated using 5% of data excluded from refinement.
RESULTS
Engineering Cytosolic CD38
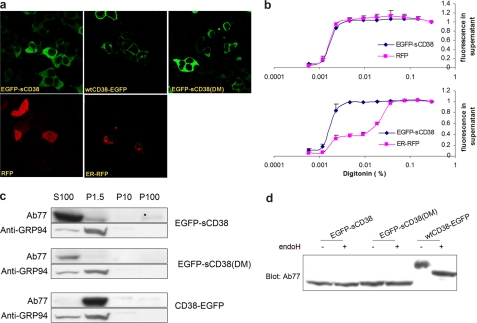
We first deleted the tail and transmembrane segment of native CD38. Unexpectedly, the resulting protein was expressed in organelles instead of the cytosol (data not shown). We next attached an EGFP tag to the N terminus of truncated CD38 to produce an EGFP-sCD38 fusion protein. EGFP is a soluble protein, and its attachment to the N terminus should ensure that the fusion protein is soluble and expressed in the cytosol. This was found to be the case as shown in Fig. 1a. The cytosolic expression of EGFP-sCD38 in HEK293 cells was similar to that in control cells expressing a cytosolic red fluorescent protein (RFP) (Fig. 1a) but was distinct from the expression of an ER luminal marker (ER-RFP). The ER-RFP marker used was RFP with the signal sequence of calreticulin, an ER luminal protein (43). The fluorescence micrographs also show that RFP was small enough to enter the nuclei, whereas the larger EGFP-sCD38 fusion protein was excluded. For comparison, we also expressed wild-type CD38 with both the transmembrane segment and N-terminal tail intact but with an EGFP tag at the C terminus (wtCD38-EGFP). The plasma membrane expression of wtCD38-EGFP was clearly evident, as all cells exhibited a ring-like fluorescence pattern distinct from that of cytosolic EGFP-sCD38.
FIGURE 1.
Cytosolic expression of the engineered CD38. a, fluorescence microscopy of the engineered CD38. CD38 constructs were expressed in HEK293 cells and visualized by fluorescence microscopy. sCD38 or its double mutant (sCD38(E146A/T221F)) tagged with EGFP at the N terminus (EGFP-sCD38(DM)) was expressed in the cytosol. Their locations were similar to those in control cells expressing a cytosolic marker (RFP) and distinct from those in cells expressing an ER luminal marker (ER-RFP). Wild-type CD38 with its transmembrane segment and N-terminal tail intact and tagged with EGFP at the C terminus (wtCD38-EGFP) was expressed on the plasma membrane. b, release of the engineered CD38 by membrane permeabilization. Upper panel, HEK293 cells were cotransfected with the soluble CD38 construct and RFP and permeabilized by various concentrations of the detergent digitonin. The amounts of EGFP-sCD38 and RFP released were determined by measuring the fluorescence of the released EGFP tag of sCD38 and RFP in the supernatants and normalized to values obtained with the highest digitonin concentration. Lower panel, same as described for the upper panel, except that the cells were cotransfected with an ER luminal marker (ER-RFP) instead of the cytosolic marker. c, fractionation of the engineered CD38 by centrifugation. HEK293 cells expressing EGFP-sCD38, EGFP-sCD38(DM), or wtCD38-EGFP were permeabilized by 0.005% digitonin and centrifuged at 15000 × g for 10 min. The pellet (P1.5) was collected, and the supernatant was centrifuged at 10,000 × g for 30 min to produce the P10 pellet. The supernatant was further centrifuged at 100,000 × g for 30 minutes to produce the P100 pellet and the S100 supernatant. The fractions were analyzed by immunoblotting using anti-CD38 antibody Ab77 or an antibody against the ER marker GRP94. d, insensitivity of the engineered CD38 to deglycosylation. Extracts from HEK293 cells expressing EGFP-sCD38, EGFP-sCD38(DM), or wtCD38-EGFP were treated with (+) or without (−) the deglycosylating enzyme endoglycosidase H (endoH) and analyzed by immunoblotting using antibody Ab77 against CD38.
Three additional methods were used to verify the cytosolic expression of the construct because cytosolic localization is the main feature of the approach. First, EGFP-sCD38 could be released by permeabilizing the plasma membrane. Fig. 1b (upper panel) shows that EGFP-sCD38 could be released from the cells at the same concentrations of digitonin that also released the cytosolic protein RFP coexpressed in the same cells. In contrast, the ER luminal marker (ER-RFP) required much higher detergent concentrations for release (Fig. 1b, lower panel).
Second, fractionation by centrifugation was used to show that EGFP-sCD38 released by membrane permeabilization remained in the supernatant even after centrifugation at an ultra-high speed of 100,000 × g for 1 h (S100) (Fig. 1c). For comparison, the ER luminal marker GRP94 (glucose-regulated protein 94) was spun down with the ER into the pellets at even a lower speed of 1500 × g for 10 min, which was also sufficient to spin down wild-type CD38 (wtCD38-EGFP) expressed in the plasma membrane (Fig. 1c, P1.5 lane).
The third test for cytosolic expression is to determine whether the protein is glycosylated. Wild-type native CD38 is glycosylated at four sites distributed in the C-terminal domain (12). Glycosylation generally occurs inside the ER. If EGFP-sCD38 is expressed in the cytosol, it should not be glycosylated. Fig. 1d shows the positive control using wtCD38-EGFP. Its size was sensitive to treatment with a deglycosylating enzyme (endoglycosidase H), which reduced its size. On the other hand, EGFP-sCD38 was smaller than wtCD38-EGFP, and its size was not sensitive to endoglycosidase H, indicating that it was not glycosylated. On the Western blot shown, it can be seen that EGFP-sCD38 was smaller than the deglycosylated wtCD38-EGFP fusion protein. This is because EGFP-sCD38 lacks the N-terminal tail and transmembrane segment, totaling ∼4.7 kDa. These results confirm that EGFP-sCD38 was indeed expressed in the cytosol and is a soluble protein.
The Engineered Cytosolic CD38 Contains Disulfides
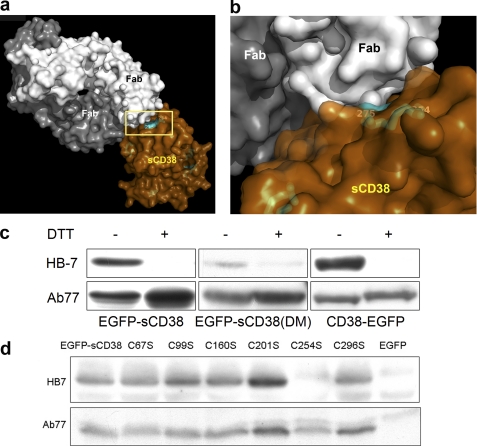
The disulfide issue was addressed next by generating a monoclonal antibody (HB7) that can recognize a specific disulfide of CD38. This approach for detecting protein disulfide is novel and is preferred because of the minimal sample manipulation required compared with the conventional method using mass spectrometry. Fig. 2a shows the crystal structure of the Fab fragment of HB7 (white and dark gray) complexed with the C-terminal domain of CD38 produced by a yeast expression system (brown). We have previously shown that the C-terminal domain has all 12 cysteine residues paired as disulfides (16). The close-up in Fig. 2b shows that HB7 directly bound to and specifically recognized the disulfide Cys-254–Cys-275 (cyan).
FIGURE 2.
Engineered CD38 possesses intact disulfides. a, crystal structure of CD38 complexed with antibody HB7. The catalytic domain of CD38 (brown) was produced in yeast and complexed with the Fab fragments of HB7 (white and gray). b, close-up view of HB7 binding specifically to the surface region of CD38 defined by the disulfide linkage Cys-254–Cys-275 (cyan). c, specific recognition of the nonreduced form of the engineered CD38 by HB7. Extracts from HEK293 cells expressing EGFP-sCD38, EGFP-sCD38(DM), or wtCD38-EGFP were analyzed by immunoblotting using either monoclonal antibody HB7 or polyclonal antibody Ab77 against CD38. SDS-PAGE was performed either under reducing (+DTT) or nonreducing (−DTT) conditions. HB7 recognizes wild-type or engineered CD38 only under nonreducing conditions. d, extracts from HEK293 cells expressing EGFP-sCD38, its cysteine mutants (C67S, C99S, C160S, C201S, C254S, or C296S), or only EGFP as a negative control were analyzed by immunoblotting using either monoclonal antibody HB7 or polyclonal antibody Ab77 against CD38 under nonreducing conditions (without DTT). HB7 recognizes EGFP-sCD38 and all its cysteine mutants except C254S.
Fig. 2c shows that HB7 recognized cytosolic EGFP-sCD38 as well as wtCD38-EGFP but only when the Western analyses were done without DTT such that all the protein disulfides were intact. Reducing the disulfides with DTT destroyed HB7 recognition. Recognition by polyclonal antibody Ab77 was not affected by the presence of DTT. The specificity of HB7 was further verified by mutating individually all six cysteines that are involved in forming the six disulfides in cytosolic EGFP-sCD38. Under nonreducing conditions, HB7 recognized EGFP-sCD38 and all mutants except C254S (Fig. 2d); the C254S mutation should destroy the Cys-254–Cys-275 disulfide targeted by HB7. The results indicate that Cys-254–Cys-275 is intact in cytosolic EGFP-sCD38. As shown in Fig. 2 (a and b), the linkage is on the surface of the protein, which is exposed to the reducing environment of the cytosol. The fact that this disulfide remains intact in EGFP-sCD38 suggests that all other more sheltered disulfides are likely to be intact as well. This was tested further as described below.
The Engineered CD38 Effectively Elevates Cytosolic cADPR
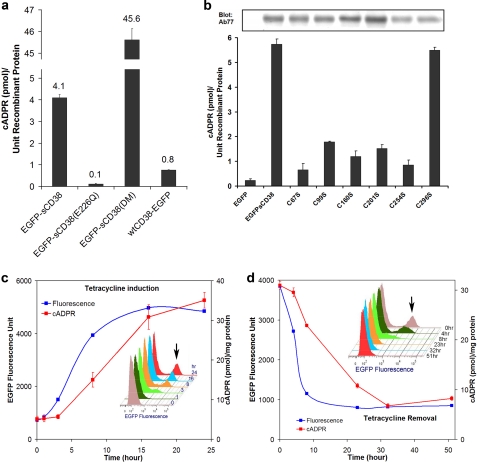
We next tested whether the constructs were active. EGFP-sCD38 was found to be enzymatically active, and its expression in the cells raised the cellular cADPR levels (Fig. 3a). As a negative control, an inactive mutant, EGFP-sCD38(E226Q) was also expressed for comparison (Fig. 3a). We have previously shown that Glu-226 is the catalytic residue and that its mutation to Gln inactivates all enzymatic activities of CD38 (13). The increased cADPR levels in cells expressing EGFP-sCD38 were, in fact, >5-fold higher than in cells expressing wild-type CD38 (wtCD38-EGFP). The cADPR-synthesizing activity was further enhanced by mutating two residues in EGFP-sCD38. We have previously shown that mutating Glu-146 to Ala and Thr-221 to Phe greatly stimulates the cADPR-synthesizing activity of CD38 (14). Consistently, the EGFP-sCD38(DM) double mutant greatly increased the endogenous cADPR concentration by >11-fold compared with cells expressing EGFP-sCD38 (Fig. 3a). The results shown in Fig. 1 (a, c and d) verified that EGFP-sCD38(DM) was also expressed in the cytosol and was soluble based on tests as described for EGFP-sCD38. Similar to EGFP-sCD38, it was recognized by HB7 only under nonreducing conditions (Fig. 2c), indicating that its disulfides were also intact. The cADPR levels shown in Fig. 3a were normalized to protein expression as determined by Western analyses. The values thus reflect the endogenous efficiency of the various forms of CD38 in synthesizing cADPR inside the cells.
FIGURE 3.
Engineered CD38 is biologically active. a, elevation of intracellular cADPR levels in cells expressing the engineered CD38. The intracellular cADPR levels in HEK293 cells expressing various forms of the engineered CD38 were measured. The enzymatically inactive mutation EGFP-sCD38(E226Q) served as a negative control. The cADPR levels shown were normalized to the protein expression levels of the engineered CD38 determined by immunoblotting (cf. b). b, intact disulfides are required for the activity of the engineered CD38. The intracellular cADPR levels in HEK293 cells expressing either EGFP-sCD38 or a cysteine mutant (C67S, C99S, C160S, C201S, C254S, or C296S) were measured and normalized to the protein expression levels of the engineered CD38 determined by immunoblotting (gel). All disulfides are critical for the activity of the engineered CD38 except the last one, Cys-287–Cys-296. c, tetracycline induction of EGFP-sCD38(DM) expression. NIH 3T3 cells were infected with lentivirus carrying the tetracycline-inducible vector containing EGFP-sCD38(DM). The inset shows flow cytometry analyses at various time points during the induction process and depicts the increasing number of cells expressing EGFP fluorescence of the fusion protein. d, time course of turning off the tetracycline induction. NIH 3T3 cells were induced with 1 μg/ml tetracycline for 24 h and washed with a culture medium free of tetracycline. The turn-off time course is shown. The inset shows flow cytometry analyses at the various times after tetracycline removal.
Fig. 3b shows that the disulfides are crucial for the cADPR-synthesizing activity of EGFP-sCD38. The cADPR levels in cells expressing each of the cysteine mutations of EGFP-sCD38 were measured and normalized to the expression level of each individual mutant protein as determined by Western analyses using polyclonal antibody Ab77. Destroying even a single disulfide by mutating the corresponding cysteine residue greatly depressed the activity of the protein, including Cys-254–Cys-275, which was detected by HB7 (cf. Fig. 2a). The least important disulfide is Cys-287–Cys-296, which is close to the C terminus (residue 300) of the protein. As determined by x-ray crystallography, the C-terminal region after Tyr-279 is highly disordered and lacks definite structure, consistent with the last disulfide (Cys-287–Cys-296) being least important for the enzymatic activity. The fact that EGFP-sCD38 has much higher cADPR-synthesizing activity compared with each of the mutants provides strong support that it is capable of forming all the disulfides while expressed in the cytosol. Although cytosolic proteins containing disulfides have been described (44–46), that the engineered CD38 most likely possesses all six of its disulfides is perhaps a unique example challenging the popular belief that cytosolic proteins should be devoid of disulfide.
To provide better control for the expression of the engineered CD38, EGFP-sCD38(DM) was cloned into the TREAuto-V14 backbone. This tetracycline-inducible vector is an improvement over the widely used commercial Tet-On system. It contains an optimized reverse tetracycline-responsive transactivator and combines all the elements required for Tet-On regulation into a single cassette with an autoregulatory loop for the expression of reverse tetracycline-responsive transactivator (34, 47). The autoregulatory expression of the reverse tetracycline-responsive transactivator was shown to be superior to the commercial Tet-On system, resulting in higher viral titers, low background, improved induction kinetics, and increased induction levels (47). Fig. 3c shows the time course of induction. NIH 3T3 cells were infected with the viral vector and treated with 1 μg/ml tetracycline. A progressive increase in EGFP fluorescence, reflecting expression of the fusion protein EGFP-sCD38(DM), was detectable within 3 h and reached a maximum in ∼15 h. The intracellular cADPR contents of the cells concomitantly increased but lagged slightly behind expression of the fusion protein. Flow cytometry of the cells at various time points showed the increases in the number of cells expressing EGFP fluorescence (indicated by the arrow in the inset of Fig. 3c). Fig. 3d shows the turn-off kinetics. The removal of tetracycline effected a progressive decrease in the expression of EGFP-sCD38(DM) (EGFP fluorescence). The flow cytometry results in the inset show that the number of cells expressing the protein likewise decreased. The cADPR contents corresponding decreased but with a delayed time course.
DISCUSSION
In this study, we have shown that native CD38, a transmembrane protein, can be engineered for expression in the cytosol in a soluble form and is fully active in elevating cellular cADPR levels. This is a remarkable feat considering that the engineered CD38 needs to form six disulfide linkages in the cytosol to retain its enzymatic activity. This finding indicates that the endogenous CD38 that is expressed intracellularly should also be fully active, even if its catalytic domain is facing the cytosol. CD38 is thus a truly remarkable signaling enzyme that can function both in the reductive cytosol and in the oxidative ectoenvironment. Furthermore, intracellular CD38 is likely to be more functionally relevant because it is also amenable to regulation by a variety of intracellular mechanisms, such as protein phosphorylation. The results of this study thus provide important information for addressing the topological paradox of CD38 signaling.
The main evidence that we provided for the presence of intact disulfides in cytosolic CD38 was obtained using a remarkable antibody, HB7, which can recognize a specific disulfide, Cys-254–Cys-275. From the crystal structure, it is easy to understand the specificity. HB7 binds directly to the region of the disulfide, which is clearly important for determining the structure of the region. Breaking the linkage is likely to result in unraveling of the polypeptide, at least locally, destroying the epitope and the recognition.
Further support for the presence of disulfides in EGFP-sCD38 was provided by mutagenesis studies. The results from the six cysteine mutants indicate that EGFP-sCD38 most likely forms all six disulfides critical for its enzymatic activity. It is generally believed that disulfide formation occurs only inside the ER and that cytosolic proteins do not have disulfide linkages. CD38 thus belongs to a group of unusual cytosolic proteins that do possess disulfides. Although rare, this group does contain a wide range of examples from bacteria to mammalian cells and includes proteins involved in chaperone function, signal transduction, and cell growth (44–46). One possible reason for this unusual property of CD38 in forming the disulfides in the cytosol is that the information for its disulfide formation may be encoded in its primary sequence. For example, during translation and folding of CD38, the appropriate cysteines are put into close proximity such that disulfide linkage can be formed. Indeed, it has been documented that the local protein microenvironment can greatly influence the ionization and reactivity of the thiolate groups of cysteines at physiological pH (48). In any case, the fact that CD38 can form such extensive disulfides makes it, perhaps, a unique example and challenges the well accepted belief that cytosolic proteins should be devoid of disulfide.
The approach demonstrated in this study should be generally applicable to any cells that can be transfected with the construct. We have further shown that the effectiveness of the construct in altering cellular cADPR levels can be modulated by specific mutations: decreased by mutating the catalytic residue Glu-226 and increased by the double mutation E146A/T221F. Further fine-tuning could be achieved with other mutations. We have systematically mutagenized various residues in the active site of CD38 and documented the effects on its enzymatic activities (13, 14, 49, 50). These mutations can be applied to produce various constructs suitable for specific purposes.
Alternatively, a cell-permeant inhibitor of CD38 can be used to modulate the cADPR-synthesizing activity of EGFP-sCD38 expressed in cells. Nicotinamide, a water-soluble vitamin and part of the vitamin B group, is one such inhibitor. We have previously shown that it is innocuous and well tolerated at high concentrations by cells; it is effective in inhibiting cADPR production by endogenous CD38 (51).
Further control was achieved using a tetracycline-inducible system, allowing cADPR production to be turned on and off within a few hours. The cellular cADPR content changes with a time course that lags behind that of the expression of the engineered CD38 during both the turn-on and turn-off processes, consistent with the causal role of the enzyme. It could be argued that the method would not allow fast enough elevation of cADPR to mimic some hormonal actions. There are, however, many physiological functions mediated by cADPR that are slower processes, such as cell proliferation (52), differentiation (53), chemotaxis (11, 54), and secretion (5). These processes should be suitable for investigation using this new approach.
Most importantly, the approach we described here sets the stage for making transgenic mice for tissue-specific and inducible expression of EGFP-sCD38. The EGFP tag would serve as a marker for identifying the expressing cells or tissues. This should ultimately allow direct assessment of the physiological roles of the cADPR/CD38 pathway in live animals and the investigation of how the pathway may regulate behavior (5).
Acknowledgments
We thank J. Marchant for the gift of pDsRed-ER and T. He for help in constructing pEGFP-sCD38(DM).
This work was supported by Hong Kong General Research Fund Grants 769107, 768408, 769309, and 770610 (to H. C. L.), 765909 and 766510 (to Q. H.), and 785110 (to H. M. Z.) and National Natural Science Foundation of China/Research Grants Council Joint Research Scheme Grant N_HKU 722/08 (to H. C. L.).
The atomic coordinates and structure factors (code 3RAJ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ER
- endoplasmic reticulum
- cADPR
- cyclic ADP-ribose
- EGFP
- enhanced green fluorescent protein
- sCD38
- soluble CD38
- RFP
- red fluorescent protein.
REFERENCES
- 1. Clapper D. L., Walseth T. F., Dargie P. J., Lee H. C. (1987) J. Biol. Chem. 262, 9561–9568 [PubMed] [Google Scholar]
- 2. Lee H. C., Walseth T. F., Bratt G. T., Hayes R. N., Clapper D. L. (1989) J. Biol. Chem. 264, 1608–1615 [PubMed] [Google Scholar]
- 3. Wu Y., Kuzma J., Maréchal E., Graeff R., Lee H. C., Foster R., Chua N. H. (1997) Science 278, 2126–2130 [DOI] [PubMed] [Google Scholar]
- 4. Zocchi E., Carpaneto A., Cerrano C., Bavestrello G., Giovine M., Brozzone S., Guida L., Franco L., Usai C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14859–14864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jin D., Liu H. X., Hirai H., Torashima T., Nagai T., Lopatina O., Shnayder N. A., Yamada K., Noda M., Seike T., Fujita K., Takasawa S., Yokoyama S., Koizumi K., Shiraishi Y., Tanaka S., Hashii M., Yoshihara T., Higashida K., Islam M. S., Yamada N., Hayashi K., Noguchi N., Kato I., Okamoto H., Matsushima A., Salmina A., Munesue T., Shimizu N., Mochida S., Asano M., Higashida H. (2007) Nature 446, 41–45 [DOI] [PubMed] [Google Scholar]
- 6. Lee H. C. (2002) Cyclic ADP-ribose and NAADP: Structures, Metabolism and Functions, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 7. Lee H. C. (2004) Curr. Mol. Med. 4, 227–237 [DOI] [PubMed] [Google Scholar]
- 8. Howard M., Grimaldi J. C., Bazan J. F., Lund F. E., Santos-Argumedo L., Parkhouse R. M., Walseth T. F., Lee H. C. (1993) Science 262, 1056–1059 [DOI] [PubMed] [Google Scholar]
- 9. Lee H. C. (2006) Mol. Med. 12, 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kato I., Yamamoto Y., Fujimura M., Noguchi N., Takasawa S., Okamoto H. (1999) J. Biol. Chem. 274, 1869–1872 [DOI] [PubMed] [Google Scholar]
- 11. Partida-Sánchez S., Cockayne D. A., Monard S., Jacobson E. L., Oppenheimer N., Garvy B., Kusser K., Goodrich S., Howard M., Harmsen A., Randall T. D., Lund F. E. (2001) Nat. Med. 7, 1209–1216 [DOI] [PubMed] [Google Scholar]
- 12. Jackson D. G., Bell J. I. (1990) J. Immunol. 144, 2811–2815 [PubMed] [Google Scholar]
- 13. Munshi C., Aarhus R., Graeff R., Walseth T. F., Levitt D., Lee H. C. (2000) J. Biol. Chem. 275, 21566–21571 [DOI] [PubMed] [Google Scholar]
- 14. Graeff R., Liu Q., Kriksunov I. A., Kotaka M., Oppenheimer N., Hao Q., Lee H. C. (2009) J. Biol. Chem. 284, 27629–27636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Q., Graeff R., Kriksunov I. A., Jiang H., Zhang B., Oppenheimer N., Lin H., Potter B. V., Lee H. C., Hao Q. (2009) J. Biol. Chem. 284, 27637–27645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Q., Kriksunov I. A., Graeff R., Munshi C., Lee H. C., Hao Q. (2005) Structure 13, 1331–1339 [DOI] [PubMed] [Google Scholar]
- 17. Lee H. C. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 317–345 [DOI] [PubMed] [Google Scholar]
- 18. Ogunbayo O. A., Zhu Y., Rossi D., Sorrentino V., Ma J., Zhu M. X., Evans A. M. (2011) J. Biol. Chem. 286, 9136–9140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Flora A., Zocchi E., Guida L., Franco L., Bruzzone S. (2004) Ann. N.Y. Acad. Sci. 1028, 176–191 [DOI] [PubMed] [Google Scholar]
- 20. Bruzzone S., Guida L., Zocchi E., Franco L., De Flora A. (2001) FASEB J. 15, 10–12 [DOI] [PubMed] [Google Scholar]
- 21. Franco L., Zocchi E., Usai C., Guida L., Bruzzone S., Costa A., De Flora A. (2001) J. Biol. Chem. 276, 21642–21648 [DOI] [PubMed] [Google Scholar]
- 22. Guida L., Bruzzone S., Sturla L., Franco L., Zocchi E., De Flora A. (2002) J. Biol. Chem. 277, 47097–47105 [DOI] [PubMed] [Google Scholar]
- 23. Adebanjo O. A., Anandatheerthavarada H. K., Koval A. P., Moonga B. S., Biswas G., Sun L., Sodam B. R., Bevis P. J., Huang C. L., Epstein S., Lai F. A., Avadhani N. G., Zaidi M. (1999) Nat. Cell Biol. 1, 409–414 [DOI] [PubMed] [Google Scholar]
- 24. Khoo K. M., Han M. K., Park J. B., Chae S. W., Kim U. H., Lee H. C., Bay B. H., Chang C. F. (2000) J. Biol. Chem. 275, 24807–24817 [DOI] [PubMed] [Google Scholar]
- 25. Kou W., Banerjee S., Eudy J., Smith L. M., Persidsky R., Borgmann K., Wu L., Sakhuja N., Deshpande M. S., Walseth T. F., Ghorpade A. (2009) J. Neurosci. Res. 87, 2326–2339 [DOI] [PubMed] [Google Scholar]
- 26. Yalcintepe L., Albeniz I., Adin-Cinar S., Tiryaki D., Bermek E., Graeff R. M., Lee H. C. (2005) Exp. Cell Res. 303, 14–21 [DOI] [PubMed] [Google Scholar]
- 27. Yamada M., Mizuguchi M., Otsuka N., Ikeda K., Takahashi H. (1997) Brain Res. 756, 52–60 [DOI] [PubMed] [Google Scholar]
- 28. Hegde R. S., Mastrianni J. A., Scott M. R., DeFea K. A., Tremblay P., Torchia M., DeArmond S. J., Prusiner S. B., Lingappa V. R. (1998) Science 279, 827–834 [DOI] [PubMed] [Google Scholar]
- 29. Hegde R. S., Voigt S., Lingappa V. R. (1998) Mol. Cell 2, 85–91 [DOI] [PubMed] [Google Scholar]
- 30. Stewart R. S., Harris D. A. (2003) J. Biol. Chem. 278, 45960–45968 [DOI] [PubMed] [Google Scholar]
- 31. Seppälä S., Slusky J. S., Lloris-Garcerá P., Rapp M., von Heijne G. (2010) Science 328, 1698–1700 [DOI] [PubMed] [Google Scholar]
- 32. Tate C. G. (2010) Science 328, 1644–1645 [DOI] [PubMed] [Google Scholar]
- 33. Tohgo A., Takasawa S., Noguchi N., Koguma T., Nata K., Sugimoto T., Furuya Y., Yonekura H., Okamoto H. (1994) J. Biol. Chem. 269, 28555–28557 [PubMed] [Google Scholar]
- 34. Centlivre M., Zhou X., Pouw S. M., Weijer K., Kleibeuker W., Das A. T., Blom B., Seppen J., Berkhout B., Legrand N. (2010) Gene Ther. 17, 14–25 [DOI] [PubMed] [Google Scholar]
- 35. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 36. Graeff R., Lee H. C. (2002) Biochem. J. 361, 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–325 [DOI] [PubMed] [Google Scholar]
- 38. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 40. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 42. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robinson L. C., Marchant J. S. (2008) Biochem. Biophys. Res. Commun. 368, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cumming R. C., Andon N. L., Haynes P. A., Park M., Fischer W. H., Schubert D. (2004) J. Biol. Chem. 279, 21749–21758 [DOI] [PubMed] [Google Scholar]
- 45. Brennan J. P., Wait R., Begum S., Bell J. R., Dunn M. J., Eaton P. (2004) J. Biol. Chem. 279, 41352–41360 [DOI] [PubMed] [Google Scholar]
- 46. Stewart E. J., Aslund F., Beckwith J. (1998) EMBO J. 17, 5543–5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Markusic D., Oude-Elferink R., Das A. T., Berkhout B., Seppen J. (2005) Nucleic Acids Res. 33, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bulaj G., Kortemme T., Goldenberg D. P. (1998) Biochemistry 37, 8965–8972 [DOI] [PubMed] [Google Scholar]
- 49. Graeff R., Liu Q., Kriksunov I. A., Hao Q., Lee H. C. (2006) J. Biol. Chem. 281, 28951–28957 [DOI] [PubMed] [Google Scholar]
- 50. Graeff R., Munshi C., Aarhus R., Johns M., Lee H. C. (2001) J. Biol. Chem. 276, 12169–12173 [DOI] [PubMed] [Google Scholar]
- 51. Munshi C. B., Graeff R., Lee H. C. (2002) J. Biol. Chem. 277, 49453–49458 [DOI] [PubMed] [Google Scholar]
- 52. Zocchi E., Podestà M., Pitto A., Usai C., Bruzzone S., Franco L., Guida L., Bacigalupo A., De Flora A. (2001) FASEB J. 15, 1610–1612 [DOI] [PubMed] [Google Scholar]
- 53. Yue J., Wei W., Lam C. M., Zhao Y. J., Dong M., Zhang L. R., Zhang L. H., Lee H. C. (2009) J. Biol. Chem. 284, 29335–29342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bruzzone S., Moreschi I., Usai C., Guida L., Damonte G., Salis A., Scarfì S., Millo E., De Flora A., Zocchi E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5759–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]