Abstract
The integration of metabolic signals required for the regulation of hepatic lipid homeostasis is complex. Previously, we showed that mice lacking expression of the mitogen-activated protein kinase (MAPK) phosphatase-1 (MKP-1) have increased fatty acid oxidation and are protected from the development of hepatic steatosis. Here, we show that leptin receptor-deficient (db/db) mice lacking MKP-1 are also resistant to the development of hepatic steatosis. Microarray analyses of livers from db/db mice lacking MKP-1 showed suppression of peroxisome proliferator-activated receptor γ (PPARγ) target genes. We identified the fat-specific protein 27 (Fsp27), which promotes PPARγ-mediated hepatic steatosis, as repressed in livers of both db/db and high fat diet-fed mice lacking MKP-1. Hepatocytes from MKP-1-deficient mice exhibited reduced PPARγ-induced lipid droplet formation. Mechanistically, loss of MKP-1 inhibited PPARγ function by increasing MAPK-dependent phosphorylation on PPARγ at its inhibitory residue of serine 112. These results demonstrate that in addition to inhibiting hepatic fatty acid oxidation, MKP-1 promotes hepatic lipogenic gene expression through PPARγ. Hence, MKP-1 plays an important role in MAPK-mediated control of hepatic lipid homeostasis.
Keywords: Cell Metabolism, Dual Specificity Phosphoprotein Phosphatase, Liver Metabolism, MAP Kinases (MAPKs), Protein Phosphatase
Introduction
The liver represents a tissue in which multiple signals are sensed and subsequently integrated to maintain metabolic homeostasis. An imbalance in how these metabolic cues are relayed through signaling pathways that lead to the expression of genes involved in hepatic metabolism can result in excess accumulation of fat in the liver (steatosis). A steatotic liver can progress to steatohepatitis, cirrhosis, fibrosis, and ultimately, either liver failure or hepatocellular carcinoma. Therefore, defining the molecular events surrounding the regulation of hepatic lipid homeostasis is of paramount importance, and elucidating the contributing facets of hepatic lipid regulation will provide insight in to both physiological and pathophysiological aspects of liver function (1).
The mitogen-activated protein kinase (MAPK) family includes the growth factor-responsive extracellular signal-regulated kinases 1 and 2 (ERK1/2), and the stress-responsive MAPKs, p38 MAPK and c-jun NH2-terminal kinase (JNK) (2, 3). MAPK activation is mediated by phosphorylation on a threonine and tyrosine residue in the activation loop by upstream MAP kinase kinases in response to diverse stimuli (2, 3). The MAPKs have been implicated in the regulation of hepatic lipid deposition. Mice lacking expression of JNK1, but not JNK2, have an improved steatotic phenotype in a model of methionine and choline deficiency (4), whereas mice lacking JNK1 specifically in the liver are insulin-resistant and develop hepatic steatosis (5). p38 MAPK promotes fatty acid β-oxidation by direct phosphorylation of the nuclear receptor, peroxisome proliferator-activated receptor (PPAR)3 α, which plays an important role in hepatic lipid metabolism (6), and in cultured hepatocytes p38 MAPK activity has been linked to decreased hepatic lipogenesis (7). These data provide evidence that the stress-responsive MAPKs are involved in regulating hepatic lipid metabolism.
The MAPKs are directly inactivated by the MAPK phosphatases (MKPs) through dephosphorylation of the regulatory threonine and tyrosine residues on the MAPKs (8, 9). MKPs display overlapping substrate preference to the MAPKs and achieve specificity through differences in tissue distribution, subcellular localization, and inducibility of expression (10). Largely through genetic efforts, it has been established that MKPs play unique and essential roles in multiple physiological systems (11–15). Previously, we described mice lacking MKP-1 to be resistant to the development of age- and high fat diet-induced obesity and hepatic steatosis (13, 15). We found that in hepatocytes derived from MKP-1-deficient mice, ligand-induced activation of PPARα is increased (13). These results suggested that MKP-1 negatively regulates hepatic fatty acid oxidation. However, whether MKP-1 regulates hepatic lipid metabolism solely through its effect on PPARα has yet to be investigated.
Here, we use the leptin-resistant (db/db) genetic mouse model of gross obesity to elucidate additional mechanisms through which MKP-1 is in involved in hepatic lipid management. We demonstrate that db/db mice lacking MKP-1 are resistant to the development of hepatic steatosis. We have uncovered a novel role for MKP-1 in the control of hepatic lipogenic gene expression. We found that MAPK-mediated phosphorylation of PPARγ at a site that negatively regulates its activity is regulated by MKP-1 in the liver. These data delineate an essential role for MKP-1 in PPARγ-mediated lipogenic gene expression and further suggest that the effects of MKP-1 in the liver occur through its actions on MAPK-mediated phosphorylation of the PPARs.
EXPERIMENTAL PROCEDURES
Maintenance of Mice
These studies were reviewed and approved by the Institutional Animal Care and Use Committee of Yale University School of Medicine. Female C57BL6/J mkp-1+/− mice as described previously (16) were crossed to BKS.Cg-m+/+ Leprdb/J breeder pairs (Jackson Laboratories) until both gray and obese pups were obtained in the same litter, indicating the presence of both db and m alleles. db/m;mkp-1+/− mice were intercrossed to obtain db/db;mkp-1+/+ mice and db/db;mkp-1−/− mice. Mice lacking the expression of mkp-1 (mkp-1−/−) were bred and subjected to high fat feeding as described previously (15).
Primary Cell Cultures and Tissue Analysis
Primary hepatocytes were isolated by collagenase digestion (17) and cultured as described previously (13). Livers were prepared for Oil Red O and hemotoxylin staining as described (13).
Transfection and Immunoblotting
SkHep cells were grown in DMEM (Invitrogen) containing 10% FBS (Gemini), 1% sodium pyruvate, and 1% penicillin/streptomycin (Invitrogen). Hepatocytes were transfected with pcDNA3, pcDNA3-FLAG-PPARγ (Addgene plasmid 8895) (18), pcDNA3-FLAG-PPARγ S112A, and pcDNA3-MKP-1 with Lipofectamine 2000 (Invitrogen). Cells were lysed and immunoblotting was performed as described (15).
RNA Isolation and Analysis
RNA isolation and reverse transcription were as previously described (15). RT-PCR was performed on ABI 7500 Fast Real Time PCR System using premade probes (Applied Biosystems). RNA for the microarray was additionally purified using RNeasy columns (Qiagen). Microarray analysis was performed using 430 whole genome array chips (Affymetrix).
Lipid Extraction and Analysis
Lipids were extracted with a 2:1 chloroform:methanol mixture and the organic layer dried overnight and reconstituted (60% butanol, 40% of a 2:1 mixture of Triton-X114 and methanol). Triglycerides were measured using a colorimetric triglyceride assay kit (Wako Diagnostics). Cholesterol was measured using a colorimetric cholesterol assay kit (Pointe Scientific). For oleate studies, primary hepatocytes were loaded overnight with complete Williams Medium E (Invitrogen) containing 500 μm oleic acid conjugated to 0.5% free fatty acid-free BSA (Sigma Aldrich) and 1 μCi of [9,10-3H]oleic acid (PerkinElmer Life Science). Uptake, export, β-oxidation, and triglyceride turnover assays were performed as described (19). For imaging and gene expression studies, primary hepatocytes were treated overnight with dimethyl sulfoxide, 100 μm rosiglitazone, or 1 mm oleic acid (Cayman Chemical) conjugated to 1% free fatty acid-free BSA. Cells were processed for RNA or fixed with 4% paraformaldehyde, stained with BODIPY 493/503 (Invitrogen), and mounted in DAPI-containing medium (Vector Laboratories). Cells were imaged (Zeiss AxioCam) and lipid content analyzed with BioPix software.
Statistics
Statistics were calculated by unpaired two-way t tests assuming equal variances (Microsoft Excel). Growth curves were analyzed by one-way ANOVA with Bonferroni post test using GraphPad InStat (Version 3.05 for Windows, Graph Pad Software). Triglyceride turnover rates were compared by linear regression with Graph Pad Prism Software.
RESULTS
db/db Mice Lacking MKP-1 Are Smaller and Resistant to Hepatic Steatosis
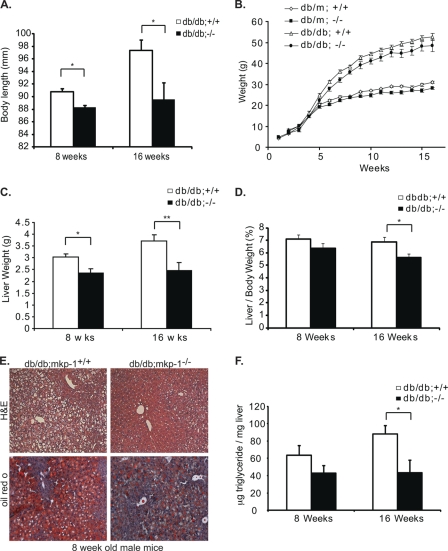
To understand how MKP-1 regulates metabolic processes involved in the development of metabolic disease, we intercrossed mkp-1−/− mice to a genetic model of gross obesity, the leptin receptor-deficient (db/db) mice (20). We found that 16-week-old db/db;mkp-1−/− mice displayed an 8% decrease in overall body length compared with db/db;mkp-1+/+ mice (Fig. 1A). At 8 weeks of age db/db;mkp-1−/− mice weighed 17% less and by 16 weeks 21% less, compared with db/db;mkp-1+/+ mice (Fig. 1B). This weight difference was not attributed to changes in adiposity or skeletal muscle mass (Table 1). Eight-week-old db/db;mkp-1−/− mice displayed a 23% decrease in liver mass compared with db/db;mkp-1+/+ mice (Fig. 1C) that progressed to 34% by 16 weeks of age (Fig. 1C). Liver mass was also significantly decreased even when normalized to body weight at 16 weeks of age (Fig. 1D). Histological examination of the liver showed that 8-week-old male db/db;mkp-1+/+ mice were highly steatotic (Fig. 1E). In contrast, the architecture of livers isolated from db/db;mkp-1−/− mice exhibited markedly less lipid accumulation (Fig. 1E). Oil Red O staining confirmed that livers derived from db/db;mkp-1−/− mice were less steatotic compared with db/db;mkp-1+/+ livers (Fig. 1E). Importantly, hepatic triglyceride content was significantly decreased by 51% after 16 weeks of age in db/db;mkp-1−/− mice (Fig. 1F). Hence, loss of MKP-1 in db/db mice, where the actions of leptin are absent, results in a decrease in body length, body weight, and a marked resistance to hepatic steatosis, similar to high fat-fed mkp-1−/− mice (13).
FIGURE 1.
Resistance to hepatic steatosis in db/db;mkp-1−/− mice. A, stature of db/db;mkp-1+/+ and db/db;mkp-1 mice. db/db;mkp-1−/− are shorter than db/db;mkp-1+/+ mice. Data are mean ± S.E. (error bars; n = 3–9; *, p < 0.05). B, growth curves for chow-fed db/m and db/db male mice that were either wild type for MKP-1 or lacking MKP-1. Weights were monitored for 16 weeks. Data are mean ± S.E. (n = 3–28). C, liver weights of randomly fed 8- and 16-week-old male db/db;mkp-1+/+ and db/db;mkp-1−/− mice. Data are mean ± S.E. (n = 11–16; *, p < 0.05; **, p < 0.005). D, liver to body weight ratio percentage from data shown in C (*, p < 0.05). E, H&E staining (upper) and Oil Red O staining (lower) of liver sections from 8-week-old male db/db;mkp-1+/+ and db/db;mkp-1−/− mice. F, hepatic triglyceride content measured in 8- and 16-week-old male db/db;mkp-1+/+ and db/db;mkp-1−/− mice. Data are mean ± S.E. (n = 9–15; *, p < 0.05).
TABLE 1.
Body and tissue weights, liver cholesterol, and serum metabolites of male db/db;mkp-1+/+ and db/db;mkp-1−/− mice were collected at the indicated ages
n = 3–15; *, p < 0.05; **, p < 0.005. NEFA, nonesterified fatty acids; ND, not determined; TA, tibialis anterior.
| Parameters | 8 weeks |
16 weeks |
||
|---|---|---|---|---|
| db/db;mkp-1+/+ | db/db;mkp-1−/− | db/db;mkp-1+/+ | db/db;mkp-1−/− | |
| Body weight (g) | 43.0 ± 0.6 | 35.7 ± 1.7** | 53.8 ± 1.3 | 42.5 ± 3.8** |
| Epididymal fat (g) | 2.67 ± 0.15 | 2.46 ± 0.42 | 2.89 ± 0.16 | 2.65 ± 0.25 |
| Heart (g) | 0.135 ± 0.003 | 0.135 ± 0.006 | 0.154 ± 0.004 | 0.141 ± 0.016 |
| Gastrocnemius (g) | 0.087 ± 0.003 | 0.078 ± 0.004 | 0.102 ± 0.004 | 0.105 ± 0.019 |
| TA (g) | 0.028 ± 0.004 | 0.030 ± 0.005 | 0.037 ± 0.004 | 0.042 ± 0.009 |
| Liver cholesterol (μg/mg) | 1.67 ± 0.08 | 1.93 ± 0.1 | 1.91 ± 0.11 | 2.22 ± 0.16 |
| Glucose (mg/dl) | 461.1 ± 26.5 | 684.2 ± 84.3* | ND | ND |
| Insulin (ng/ml) | 1.38 ± 0.12 | 0.99 ± 0.17 | ND | ND |
| Serum triglycerides (mg/dL) | 45.7 ± 10.6 | 39.42 ± 6.4 | ND | ND |
| Serum cholesterol (mg/dL) | 193.0 ± 18.1 | 202.8 ± 22.1 | ND | ND |
| NEFA (mmol/liter) | 0.622 ± 0.063 | 0.513 ± 0.031 | ND | ND |
| β-Hydroxybutyrate (mmol/liter) | 0.724 ± 0.268 | 0.452 ± 0.116 | ND | ND |
Increased Hepatic Triglyceride Turnover in db/db;mkp-1−/− Mice Liver
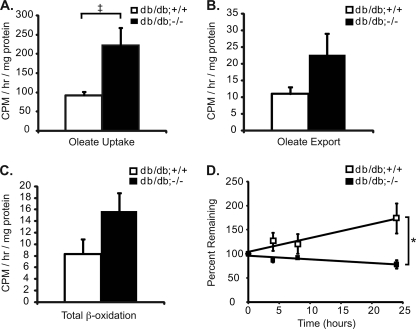
Multiple metabolic defects contribute to the development of hepatic steatosis including defects in lipid import, lipid export, fatty acid β-oxidation, de novo lipogenesis, and triglyceride turnover (1). To determine which of these metabolic effects are altered in db/db;mkp-1−/− mice, we isolated primary hepatocytes from 7–9-week-old male db/db;mkp-1+/+ and db/db;mkp-1−/− mice and performed lipid-loading experiments with [9,10–3H]oleic acid as a precursor of triglyceride accumulation. We found a >2-fold increase in oleic acid import (Fig. 2A) and a 2-fold increase in oleic acid export (Fig. 2B) in db/db;mkp-1−/− compared with db/db;mkp-1+/+ hepatocytes. Additionally, there was an 89% increase in β-oxidation in db/db;mkp-1−/− hepatocytes compared with those derived from db/db;mkp-1+/+ animals (Fig. 2C). The increase in β-oxidation was consistent with previous observations where we showed that MKP-1-deficient mice exhibited increased PPARα activation (13). Next, we measured triglyceride turnover in hepatocytes labeled with [9,10-3H]oleic acid overnight, which was removed and replaced with medium containing triacsin C, a fatty acyl-CoA synthetase inhibitor, to inhibit triglyceride synthesis (19). The triglyceride content in db/db;mkp-1−/− hepatocytes decreased to 78% over a period of 24 h, whereas db/db;mkp-1+/+ hepatocytes did not (Fig. 2D). These results demonstrate that lipid turnover is increased in db/db;mkp-1−/− animals, which in addition to increased β-oxidation, likely contributes to the resistance to hepatic steatosis.
FIGURE 2.
Increased triglyceride turnover and β-oxidation in livers of db/db;mkp-1−/− mice. Primary hepatocytes were isolated from db/db;mkp-1+/+ and db/db;mkp-1−/− mice, loaded with [9,10-3H]oleic acid, and lipid content was measured. A, uptake into hepatocytes. B, export into media. C, β-oxidation. D, triglyceride turnover. Data are mean ± S.E. (error bars; n = 3 or 4; ‡, p = 0.05; *, p < 0.05).
Regulation of PPARγ Target Genes in db/db;mkp-1−/− Mice
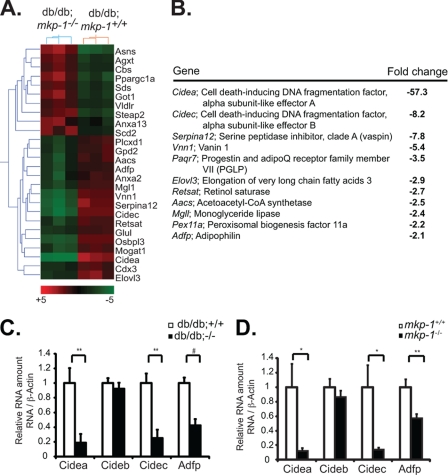
To elucidate additional mechanisms by which db/db;mkp-1−/− mice were resistant to the development of hepatic steatosis, we performed a genome-wide microarray experiment from livers of 8-week-old db/db;mkp-1+/+ and db/db;mkp-1−/− mice. The microarray results revealed >150 transcripts that were either induced or repressed by >2-fold in db/db;mkp-1−/− livers (Fig. 3A). Genes were grouped by ontogeny for those known to be involved in glucose metabolism, lipid metabolism, lipid transport, or electron transport to identify target candidates involved in hepatic lipid metabolism. A subset of highly repressed genes were identified that are known transcriptional targets of the nuclear receptor PPARγ, a master regulator of lipid metabolism that is involved in promoting hepatic steatosis and is a known MAPK substrate (21–23) (Fig. 3, A and B). The two most highly repressed genes were the cell death-inducing DNA fragmentation factor A-like effector A (cidea) and CIDEC/fat-specific protein 27 (FSP27), which were repressed by 57- and 8-fold, respectively (Fig. 3, A and B). cidec is a PPARγ target gene that has been shown to be necessary and sufficient for the development of hepatic steatosis in the ob/ob mouse (19). Other PPARγ target genes that were found to be repressed in the livers of db/db;mkp-1−/− mice included Serpina12/vaspin (24), vanin (vnn1) (25), elongation of very long chain fatty acids (Elovl3) (26), acetoacetyl-CoA synthetase (Aacs) (27), monoglyceride lipase (mgll) (28), peroxisomal biogenesis factor 11a (pex11a) (29), and adipose differentiation-related protein/adipophilin (adfp) (30) (Fig. 3B).
FIGURE 3.
Decreased expression of PPARγ target genes in db/db;mkp-1−/− mice. A, heat map of genes involved in lipid metabolism, lipid transport, or electron transport that was >2-fold induced or repressed in db/db;mkp-1−/− mice (n = 3, false discovery rate < 0.1). B, genes regulated by PPARγ repressed in db/db;mkp-1−/− mice. C and D, quantitative RT-PCR from livers of 8-week-old male db/db;mkp-1+/+ and db/db;mkp-1−/− mice (C) and male mkp-1+/+ and mkp-1−/− mice (D) fed a high fat diet for 16 weeks. Data are mean ± S.E. (error bars; n = 8 or 9, *, p < 0.05; **, p < 0.005; #, p < 0.0005).
We focused our analysis on the cide gene family, as both cidea and cidec were the two most highly repressed genes identified in the microarray experiments (Fig. 3A). As a control, we included cideb the other cide family member (31). To verify the microarray results, quantitative RT-PCR was performed in livers of db/db;mkp-1+/+ and db/db;mkp-1−/− mice. We found an 81% decrease in cidea expression and a 75% decrease in cidec expression (Fig. 3C). No changes in cideb expression in livers of db/db;mkp-1−/− mice were found (Fig. 3C). We also confirmed that the lipid-binding protein, adfp, was also significantly repressed (Fig. 3C). These results indicate that the expression of key lipid-binding proteins are repressed in the livers of db/db;mkp-1−/− mice which is consistent with the reduced lipid load observed in the livers of these mice.
We demonstrated previously that high fat diet-fed mkp-1−/− mice are resistant to the development of hepatic steatosis (13). To determine whether the effects of MKP-1 on PPARγ target genes are not solely a function of the lack of leptin receptor expression, we measured cidea, cideb, cidec, and adfp levels in the livers of wild-type and mkp-1−/− mice fed a high fat diet for 8 weeks. We found that although cideb was unaffected, cidea and cidec mRNA levels were significantly repressed in the livers of high fat-fed mkp-1−/− mice compared with wild-type controls (Fig. 3D). In addition, adfp was also significantly reduced in the livers of high fat-fed mkp-1−/− mice compared with wild-type controls (Fig. 3D). These observations demonstrate that MKP-1 is required for the activation of PPARγ target genes independently of the effects of leptin resistance.
Reduced Lipid Droplet Formation in db/db;mkp-1−/− Hepatocytes
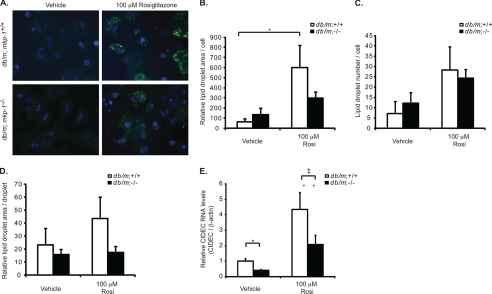
To determine whether the resistance to hepatic steatosis in db/db;mkp-1−/− mice was related to a defect in PPARγ-dependent lipid droplet formation, we isolated primary hepatocytes from nonobese db/m;mkp-1+/+ and db/m;mkp-1−/− mice and induced lipid droplet formation with either oleate or the PPARγ agonist, rosiglitazone. Cells were fixed and stained with BODIPY 493/503, and lipid droplet number and size were quantitated. Treatment with oleate increased lipid droplet area, number, and size to a similar extent in both db/m;mkp-1+/+ and db/m;mkp-1−/− hepatocytes (data not shown), indicating that the capacity of db/m;mkp-1−/− cells to form lipid droplets through exogenous pathways is equivalent to wild type. However, treatment of db/m;mkp-1+/+ and db/m;mkp-1−/− hepatocytes with rosiglitazone to induce PPARγ activity resulted in a 40% reduction of relative lipid droplet area per cell and a 60% reduction in lipid droplet area per droplet (Fig. 4, A, B, and D). These changes were observed in the absence of a change in lipid droplet number per cell (Fig. 4C). Gene expression changes in these primary hepatocytes were also assessed. In untreated hepatocytes, cidec expression was reduced in db/m;mkp-1−/− compared with db/m;mkp-1+/+ hepatocytes, and rosiglitazone-induced cidec expression was also impaired (Fig. 4E). These results indicate that hepatocytes derived from db/m;mkp-1−/− mice are defective in lipid droplet formation, consistent with the interpretation that MKP-1 is required for the expression of lipid droplet-forming genes cidea and cidec.
FIGURE 4.
PPARγ-mediated lipid accumulation and CIDEC expression in db/db;mkp-1−/− mice. Primary hepatocytes were isolated from db/m;mkp-1+/+ and db/m;mkp-1−/− mice. Cells were treated overnight with dimethyl sulfoxide (Vehicle) or 100 μm rosiglitazone (Rosi). A, lipids stained with BODIPY 493/503 and nuclei with DAPI. B, area of lipid droplets/cell. C, no. of lipid droplets/cell. D, area of individual lipid droplets. E, quantitative RT-PCR for CIDEC from hepatocytes treated as above. Data are mean ± S.E. (error bars; n = 5–10; ‡, p = 0.05; *, p < 0.05; **, p < 0.005).
Enhanced MAPK and PPARγ Phosphorylation in db/db;mkp-1−/− Hepatocytes
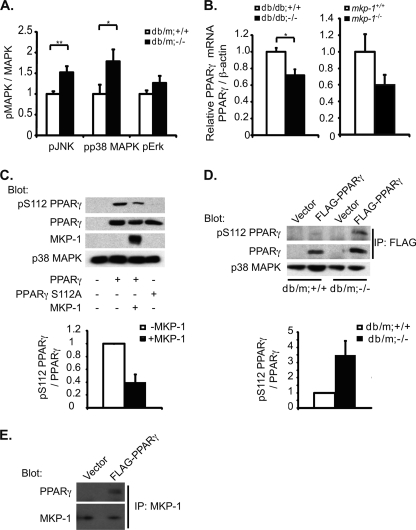
Loss of MKP-1 in the db/db background likely results in enhanced MAPK signaling. We determined whether there was enhanced MAPK activity in mkp-1−/− mice on the db/db background. Lysates from db/m;mkp-1+/+ and db/m;mkp-1−/− hepatocytes were prepared and immunoblotted for levels of phosphorylated ERK1/2, JNK, and p38 MAPK. There were significant increases in phosphorylated JNK and p38 MAPK by 50 and 80%, respectively, in db/m;mkp-1−/− compared with db/m;mkp-1+/+ hepatocytes (Fig. 5A). Although the levels of phosphorylated ERK1/2 did not reach statistical significance, their levels were consistently elevated in db/m;mkp-1−/− compared with db/m;mkp-1+/+ hepatocytes (Fig. 3A). These results demonstrate that the resistance to the acquisition of a fatty liver in MKP-1-deficient mice in the db/db background manifests from enhanced activities of the MAPKs. We measured PPARγ mRNA levels in livers of db/db;mkp-1+/+ and db/db;mkp-1−/− mice by quantitative RT-PCR and found approximately a 33% decrease in PPARγ mRNA levels in db/db;mkp-1−/− livers compared with livers from db/db;mkp-1+/+ mice (Fig. 5B). The expression level of PPARγ mRNA was also reduced in high fat diet-fed mkp-1−/− mice (Fig. 5B). Therefore, MKP-1 is involved in promoting the expression of hepatic PPARγ mRNA.
FIGURE 5.
Increased MAPK activity and PPARγ phosphorylation in db/m;mkp-1−/− hepatocytes. A, primary hepatocytes isolated from db/m;mkp-1+/+ and db/m;mkp-1−/− mice and immunoblotted for phospho- and total MAPKs. Phospho-MAPK values were assessed by densitometry and normalized to total MAPK values. Data are mean ± S.E. (error bars; n = 8 or 9; *, p < 0.05, **, p < 0.005). B, quantitative RT-PCR of PPARγ mRNA from livers of 8-week-old male db/db;mkp-1+/+ and db/db;mkp-1−/− (left panel) and male mkp-1+/+ and mkp-1−/− mice fed a high fat diet for 16 weeks (right panel). Data are mean ± S.E. (n = 9; *, p < 0.05). C, SkHep cells transfected with FLAG-PPARγ or FLAG-PPARγ S112A with or without MKP-1 and immunoblotted for Ser(P)112 and total PPARγ. Data are representative of three independent experiments. D, primary hepatocytes from db/m;mkp-1+/+ or db/m;mkp-1−/− mice transfected with pcDNA3 or FLAG-PPARγ. FLAG was immunoprecipitated (IP) and immunoblotted for Ser(P)112 and total PPARγ (n = 3). E, SkHep cells transfected with vector or FLAG-PPARγ. MKP-1 was immunoprecipitated and immunoblotted for MKP-1 or PPARγ.
PPARγ activity is controlled by multiple factors including ligand binding and phosphorylation by the MAPKs (21, 32, 33). Specifically, ERK1/2 and JNK phosphorylate PPARγ1 and PPARγ2 on Ser84 and Ser112, respectively, resulting in decreased ligand binding and transcriptional activity (22, 23, 33). We examined the possibility that phosphorylation of PPARγ at its MAPK site may be increased in the absence of MKP-1, which would impair PPARγ-mediated activation of its target genes. We used SkHep cells to determine whether overexpression of MKP-1 decreased PPARγ phosphorylation on Ser112. Cells were transfected with PPARγ2 along with MKP-1, and the phosphorylation status of PPARγ was assessed using phospho-specific Ser112 PPARγ antibodies. These experiments showed that overexpression of MKP-1 reduced phosphorylation of Ser112 PPARγ by 60% (Fig. 5C).
To determine whether MKP-1 expression was essential for PPARγ phosphorylation on Ser112, we measured PPARγ phosphorylation in hepatocytes derived from db/m;mkp-1+/+ and db/m;mkp-1−/− mice. We transfected FLAG-tagged PPARγ2 into these cells and measured the levels of immunoprecipitated FLAG-tagged Ser(P)112 on PPARγ. More than a 3-fold increase in Ser(P)112 PPARγ in hepatocytes derived from db/m;mkp-1−/− mice compared with those from db/m;mkp-1+/+ mice was detected (Fig. 5D). Finally, we tested whether MKP-1 complexes with PPARγ. SkHep were transfected with FLAG-tagged PPARγ2, MKP-1 was immunoprecipitated, and these complexes were immunoblotted for PPARγ. MKP-1 was detected in a complex with PPARγ in hepatocytes (Fig. 5E). These results indicate that MKP-1 can complex with PPARγ and that it is able to regulate MAPK-mediated phosphorylation of PPARγ in hepatocytes. Together, these results demonstrate that MKP-1 is required for optimal hepatic PPARγ function at both the level of transcription (Fig. 5B) and post-translational modifications through negative regulation of its inhibitory MAPK phosphorylation site (Fig. 5, C and D).
DISCUSSION
The conclusion that MKP-1 promotes hepatic steatosis is based upon the observation that db/db;mkp-1−/− mice were resistant to the accumulation of triglycerides in the liver accompanied by a commensurate protection from the increase in liver mass associated with this genetic model of obesity. The levels of triglyceride content in db/db;mkp-1−/− mice were significantly lower in 16-week-old mice, strongly suggesting that loss of MKP-1 inhibited rather than delayed the development of hepatic steatosis. Previously, we suggested that the ability of MKP-1-deficient mice to resist the development of hepatic steatosis was due, at least in part, to the fact that p38 MAPK-dependent phosphorylation on PPARα was enhanced resulting in increased ligand-induced activation of hepatic β-oxidation (6). However, whether MKP-1 loss resulted in increased β-oxidation was not formally tested. Here, we show that hepatocytes derived from db/db;mkp-1−/− mice exhibited increased levels of hepatic β-oxidation, demonstrating that enhanced fatty acid oxidation contributes to the resistance to hepatic steatosis in these mice.
In this study, we have explored additional mechanisms through which MKP-1 might regulate hepatic lipid content. Both triglyceride export and import were elevated by comparable levels in db/db;mkp-1−/− livers, suggesting that an imbalance in lipid export and import exists, but this net outcome would otherwise negate the effect of these pathways influencing overall triglyceride content. However, triglyceride turnover was significantly increased in hepatocytes from db/db;mkp-1−/− mice. Therefore, loss of MKP-1 protects against the development of hepatic steatosis also as a result of increased rates of triglyceride turnover.
It was conceivable that both increased PPARα-induced β-oxidation and triglyceride turnover could account entirely for the resistance to hepatic steatosis in db/db;mkp-1−/− mice. In fact, we previously demonstrated that MKP-1 negatively regulates PPARα (13). Consistent with that mechanism, we found that there are increased levels of fatty acid oxidation in db/db;mkp-1−/− mice. However, an unbiased analysis of gene expression changes revealed the possibility of additional mechanisms. We discovered that db/db;mkp-1−/− displayed an enriched subset of suppressed PPARγ target genes. The second most highly repressed gene identified in the liver of db/db;mkp-1−/− mice (8-fold repressed compared with wild type) was cidec/Fsp27. cidec is an interesting candidate because it is both necessary and sufficient to promote hepatic steatosis in ob/ob mice and negatively regulates hepatic triglyceride turnover and facilitates triglyceride storage in multiple cell types (19, 34, 35). Mouse knock-out models of cidec demonstrate that it is responsible for unilocular lipid droplet formation (36, 37). Consistent with these observations, hepatocytes derived from db/db;mkp-1−/− mice contained smaller lipid droplets. It is thought that multilocular droplets increase lipid surface presentation to circulating lipases, providing greater access to fatty acid availability for mitochondrial β-oxidation (37). This too may contribute to the increased β-oxidation in the livers of db/db;mkp-1−/− mice. Finally, CIDEC inhibits hepatic triglyceride turnover, and consistent with this, hepatocytes derived from db/db;mkp-1−/− mice displayed increased hepatic triglyceride turnover. Unlike cidec, the expression of cidea was not significantly different in rosiglitazone-stimulated db/db;mkp-1+/+ and db/db;mkp-1−/− hepatocytes. Nevertheless, CIDEA has been implicated in lipid metabolism, and genetic deletion of cidea results in increased lipolysis, energy expenditure, elevated levels of thermogenesis, and resistance to diet-induced obesity (38). The reduced expression of cidea in the liver is consistent with attenuated hepatic steatosis, resistance to diet-induced obesity, and increased energy expenditure in MKP-1-deficient mice (13, 39). Collectively, these results identify a link among MKP-1, hepatic triglyceride turnover, and the machinery involved in lipid droplet formation.
The mechanism engaged by MKP-1 to regulate PPARγ-mediated target gene activity is related to the enhanced levels of either JNK and/or p38 MAPK activity in the livers db/db;mkp-1−/− mice. It has been suggested that p38 MAPK can affect PPARγ mRNA expression (7). However, those experiments demonstrated that p38 MAPK positively, rather than negatively, regulated PPARγ mRNA in the liver of high fat diet-fed mice (7). Moreover, overexpression of MKP-4 in the liver, which inhibits JNK and ERK1/2, was shown to prevent hepatic steatosis (40). Because ERK1/2 and JNK inhibit PPARγ activity by phosphorylation on Ser112 (22, 32, 33, 41) we speculated that MKP-1 might be exerting its effects on PPARγ activity by negatively regulating MAPK-dependent phosphorylation at this site. We found that Ser112 phosphorylation on PPARγ was attenuated when MKP-1 was overexpressed. Conversely, hepatocytes derived from db/m mice lacking MKP-1 exhibited increased PPARγ Ser112 phosphorylation. These data argue that MKP-1 plays an essential role in negatively regulating Ser112 phosphorylation on PPARγ. The functional effects of Ser112 phosphorylation on PPARγ in mice have been investigated (42). In those experiments, the mutant PPARγ that fails to become phosphorylated at Ser112 did not affect weight gain in response to high fat feeding (42). However, the status of the liver in those mice was not investigated. Regardless, our results demonstrate a link between MKP-1 and the regulatory pathway controlled by PPARγ in hepatic lipogenic gene expression.
The levels of PPARγ are increased in steatotic livers (43). Under conditions of high fat feeding, MKP-1 expression is elevated in the liver (44). It is conceivable that increased MKP-1 expression in the liver under conditions of high fat feeding attenuates nuclear MAPK-mediated phosphorylation of PPARγ. Interestingly, liver-specific JNK knock-out mice exhibit increased hepatic steatosis (5), suggesting that enhanced JNK activity prevents hepatic steatosis. MKP-1 can function to control JNK substrates (13, 45), and it has been demonstrated that JNK can phosphorylate PPARγ (46). Therefore, it is possible that MKP-1 acts through JNK to mediate PPARγ phosphorylation at Ser112. MKP-1 was also found to be in a complex with PPARγ, suggesting that it is physically positioned to dephosphorylate the MAPKs that phosphorylate PPARγ. Further experiments will be required to define the molecular interactions between MKP-1 and PPARγ.
In summary, the results presented here show that in a genetic mouse model of obesity MKP-1 plays an essential role in the maintenance of hepatic lipid homeostasis. We have identified that MKP-1 regulates PPARγ function in the liver which reveals a link between MKP-1 and hepatic lipid droplet formation and storage. Hence, MKP-1 represents a potential pharmacological target for the treatment of hepatic steatosis in obesity.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK75776 and DK57751 (to A. M. B.). This work was also supported by Yale Liver Center Grant P30 DK34989.
- PPAR
- peroxisome proliferator-activated receptor
- CIDE
- cell death-inducing DNA fragmentation factor A-like effector
- FSP27
- fat-specific protein 27
- MKP-1
- MAPK phosphatase-1.
REFERENCES
- 1. Anderson N., Borlak J. (2008) Pharmacol. Rev. 60, 311–357 [DOI] [PubMed] [Google Scholar]
- 2. Raman M., Chen W., Cobb M. H. (2007) Oncogene 26, 3100–3112 [DOI] [PubMed] [Google Scholar]
- 3. Weston C. R., Davis R. J. (2007) Curr. Opin. Cell Biol. 19, 142–149 [DOI] [PubMed] [Google Scholar]
- 4. Schattenberg J. M., Singh R., Wang Y., Lefkowitch J. H., Rigoli R. M., Scherer P. E., Czaja M. J. (2006) Hepatology 43, 163–172 [DOI] [PubMed] [Google Scholar]
- 5. Sabio G., Cavanagh-Kyros J., Ko H. J., Jung D. Y., Gray S., Jun J. Y., Barrett T., Mora A., Kim J. K., Davis R. J. (2009) Cell Metab. 10, 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barger P. M., Browning A. C., Garner A. N., Kelly D. P. (2001) J. Biol. Chem. 276, 44495–44501 [DOI] [PubMed] [Google Scholar]
- 7. Xiong Y., Collins Q. F., An J., Lupo E., Jr., Liu H. Y., Liu D., Robidoux J., Liu Z., Cao W. (2007) J. Biol. Chem. 282, 4975–4982 [DOI] [PubMed] [Google Scholar]
- 8. Dickinson R. J., Keyse S. M. (2006) J. Cell Sci. 119, 4607–4615 [DOI] [PubMed] [Google Scholar]
- 9. Boutros T., Chevet E., Metrakos P. (2008) Pharmacol. Rev. 60, 261–310 [DOI] [PubMed] [Google Scholar]
- 10. Owens D. M., Keyse S. M. (2007) Oncogene 26, 3203–3213 [DOI] [PubMed] [Google Scholar]
- 11. Maillet M., Purcell N. H., Sargent M. A., York A. J., Bueno O. F., Molkentin J. D. (2008) J. Biol. Chem. 283, 31246–31255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y., Blattman J. N., Kennedy N. J., Duong J., Nguyen T., Wang Y., Davis R. J., Greenberg P. D., Flavell R. A., Dong C. (2004) Nature 430, 793–797 [DOI] [PubMed] [Google Scholar]
- 13. Wu J. J., Roth R. J., Anderson E. J., Hong E. G., Lee M. K., Choi C. S., Neufer P. D., Shulman G. I., Kim J. K., Bennett A. M. (2006) Cell Metabolism 4, 61–73 [DOI] [PubMed] [Google Scholar]
- 14. Christie G. R., Williams D. J., Macisaac F., Dickinson R. J., Rosewell I., Keyse S. M. (2005) Mol. Cell. Biol. 25, 8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roth R. J., Le A. M., Zhang L., Kahn M., Samuel V. T., Shulman G. I., Bennett A. M. (2009) J. Clin. Invest. 119, 3817–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu J. J., Bennett A. M. (2005) J. Biol. Chem. 280, 16461–16466 [DOI] [PubMed] [Google Scholar]
- 17. Boyer J. L., Phillips J. M., Graf J. (1990) Methods Enzymol. 192, 501–516 [DOI] [PubMed] [Google Scholar]
- 18. Hauser S., Adelmant G., Sarraf P., Wright H. M., Mueller E., Spiegelman B. M. (2000) J. Biol. Chem. 275, 18527–18533 [DOI] [PubMed] [Google Scholar]
- 19. Matsusue K., Kusakabe T., Noguchi T., Takiguchi S., Suzuki T., Yamano S., Gonzalez F. J. (2008) Cell Metab. 7, 302–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anstee Q. M., Goldin R. D. (2006) Int. J. Exp. Pathol. 87, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Desvergne B., Michalik L., Wahli W. (2006) Physiol. Rev. 86, 465–514 [DOI] [PubMed] [Google Scholar]
- 22. Hu E., Kim J. B., Sarraf P., Spiegelman B. M. (1996) Science 274, 2100–2103 [DOI] [PubMed] [Google Scholar]
- 23. Adams M., Reginato M. J., Shao D., Lazar M. A., Chatterjee V. K. (1997) J. Biol. Chem. 272, 5128–5132 [DOI] [PubMed] [Google Scholar]
- 24. Hida K., Wada J., Eguchi J., Zhang H., Baba M., Seida A., Hashimoto I., Okada T., Yasuhara A., Nakatsuka A., Shikata K., Hourai S., Futami J., Watanabe E., Matsuki Y., Hiramatsu R., Akagi S., Makino H., Kanwar Y. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10610–10615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu S., Matsusue K., Kashireddy P., Cao W. Q., Yeldandi V., Yeldandi A. V., Rao M. S., Gonzalez F. J., Reddy J. K. (2003) J. Biol. Chem. 278, 498–505 [DOI] [PubMed] [Google Scholar]
- 26. Petrovic N., Walden T. B., Shabalina I. G., Timmons J. A., Cannon B., Nedergaard J. (2010) J. Biol. Chem. 285, 7153–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aguiló F., Camarero N., Relat J., Marrero P. F., Haro D. (2010) Biochem. J. 427, 255–264 [DOI] [PubMed] [Google Scholar]
- 28. Festuccia W. T., Laplante M., Berthiaume M., Gélinas Y., Deshaies Y. (2006) Diabetologia 49, 2427–2436 [DOI] [PubMed] [Google Scholar]
- 29. Welch J. S., Ricote M., Akiyama T. E., Gonzalez F. J., Glass C. K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schadinger S. E., Bucher N. L., Schreiber B. M., Farmer S. R. (2005) Am. J. Physiol. Endocrinol. Metab. 288, E1195–1205 [DOI] [PubMed] [Google Scholar]
- 31. Inohara N., Koseki T., Chen S., Wu X., Núñez G. (1998) EMBO J. 17, 2526–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burns K. A., Vanden Heuvel J. P. (2007) Biochim. Biophys. Acta 1771, 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shao D., Rangwala S. M., Bailey S. T., Krakow S. L., Reginato M. J., Lazar M. A. (1998) Nature 396, 377–380 [DOI] [PubMed] [Google Scholar]
- 34. Keller P., Petrie J. T., De Rose P., Gerin I., Wright W. S., Chiang S. H., Nielsen A. R., Fischer C. P., Pedersen B. K., MacDougald O. A. (2008) J. Biol. Chem. 283, 14355–14365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Puri V., Konda S., Ranjit S., Aouadi M., Chawla A., Chouinard M., Chakladar A., Czech M. P. (2007) J. Biol. Chem. 282, 34213–34218 [DOI] [PubMed] [Google Scholar]
- 36. Toh S. Y., Gong J., Du G., Li J. Z., Yang S., Ye J., Yao H., Zhang Y., Xue B., Li Q., Yang H., Wen Z., Li P. (2008) PloS One 3, e2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishino N., Tamori Y., Tateya S., Kawaguchi T., Shibakusa T., Mizunoya W., Inoue K., Kitazawa R., Kitazawa S., Matsuki Y., Hiramatsu R., Masubuchi S., Omachi A., Kimura K., Saito M., Amo T., Ohta S., Yamaguchi T., Osumi T., Cheng J., Fujimoto T., Nakao H., Nakao K., Aiba A., Okamura H., Fushiki T., Kasuga M. (2008) J. Clin. Invest. 118, 2808–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou Z., Yon Toh S., Chen Z., Guo K., Ng C. P., Ponniah S., Lin S. C., Hong W., Li P. (2003) Nat. Genet. 35, 49–56 [DOI] [PubMed] [Google Scholar]
- 39. Roth Flach R. J., Bennett A. M. (2010) Aging 2, 170–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emanuelli B., Eberlé D., Suzuki R., Kahn C. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3545–3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Camp H. S., Tafuri S. R. (1997) J. Biol. Chem. 272, 10811–10816 [DOI] [PubMed] [Google Scholar]
- 42. Rangwala S. M., Rhoades B., Shapiro J. S., Rich A. S., Kim J. K., Shulman G. I., Kaestner K. H., Lazar M. A. (2003) Dev. Cell 5, 657–663 [DOI] [PubMed] [Google Scholar]
- 43. Matsusue K., Haluzik M., Lambert G., Yim S. H., Gavrilova O., Ward J. M., Brewer B., Jr., Reitman M. L., Gonzalez F. J. (2003) J. Clin. Invest. 111, 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reddy S. T., Nguyen J. T., Grijalva V., Hough G., Hama S., Navab M., Fogelman A. M. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 1676–1681 [DOI] [PubMed] [Google Scholar]
- 45. Jeanneteau F., Deinhardt K., Miyoshi G., Bennett A. M., Chao M. V. (2010) Nat. Neurosci. 13, 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yin R., Dong Y. G., Li H. L. (2006) Acta Pharmacol. Sin. 27, 1146–1152 [DOI] [PubMed] [Google Scholar]