Abstract
Deacetylation of histone proteins at the HIV type 1 (HIV-1) long terminal repeat (LTR) by histone deactylases (HDACs) can promote transcriptional repression and virus latency. As such, HDAC inhibitors (HDACI) could be used to deplete reservoirs of persistent, quiescent HIV-1 proviral infection. However, the development of HDACI to purge latent HIV-1 requires knowledge of the HDAC isoforms contributing to viral latency and the development of inhibitors specific to these isoforms. In this study, we identify the HDACs responsible for HIV-1 latency in Jurkat J89GFP cells using a chemical approach that correlates HDACI isoform specificity with their ability to reactivate latent HIV-1 expression. We demonstrate that potent inhibition or knockdown of HDAC1, an HDAC isoform reported to drive HIV-1 into latency, was not sufficient to de-repress the viral LTR. Instead, we found that inhibition of HDAC3 was necessary to activate latent HIV-1. Consistent with this finding, we identified HDAC3 at the HIV-1 LTR by chromatin immunoprecipitation. Interestingly, we show that valproic acid is a weak inhibitor of HDAC3 (IC50 = 5.5 mm) relative to HDAC1 (IC50 = 170 μm). Because the total therapeutic concentration of valproic acid ranges from 275 to 700 μm in adults, these data may explain why this inhibitor has no effect on the decay of latent HIV reservoirs in patients. Taken together, our study suggests an important role for HDAC3 in HIV-1 latency and, importantly, describes a chemical approach that can readily be used to identify the HDAC isoforms that contribute to HIV-1 latency in other cell types.
Keywords: Antiviral Agents, Epigenetics, Histone Deacetylase, HIV, Viral Transcription, HIV Latency
Introduction
Combination antiretroviral therapy (cART)3 can effectively reduce plasma HIV-1 to undetectable levels. However, upon its interruption, there is usually a rapid rebound of viremia (1). This viremia is thought to arise from latently infected reservoirs such as memory CD4(+) T cells or CD34(+) multipotent hematopoietic progenitor cells (2–5). Therefore, any long term therapeutic strategy targeted toward eliminating HIV-1 infection must include compounds that purge the latent viral reservoirs thereby rendering them susceptible to cART.
HIV-1 can be maintained in a latent state by multiple different mechanisms that inhibit virus gene expression after integration into the cellular DNA (6–8). For example, epigenetic modifications at or near the HIV-1 5′-long terminal repeat (LTR) can induce chromatin condensation that diminishes the accessibility of the HIV-1 promoter to transcription factors. In this regard, it has been well documented that different transcription factors can recruit histone deacetylase (HDAC) enzymes to the HIV-1 LTR where they promote chromatin condensation by deacetylating the ϵ-amino groups of lysine residues in histone tails (9–14). Eleven distinct zinc-dependent HDAC isoforms have been identified in humans. These can be classified into four families, namely class I (HDAC1–3 and -8), IIa (HDAC4, -5, -7, and -9), IIb (HDAC6 and -10), and IV (HDAC11), which differ in structure, enzymatic function, subcellular localization, and expression patterns (15). To date, multiple studies have demonstrated that recruitment of HDAC1 to the HIV-1 LTR by different DNA-binding complexes is sufficient to induce viral latency (9–14). However, HDAC2 and HDAC3 can also bind to the HIV-1 LTR and may also play an important role in viral latency (12, 16, 17).
Treatment of latently infected HIV-1 cell lines and/or CD4(+) T cells from aviremic HIV-1-infected individuals on cART with HDACI can lead to chromatin relaxation and induction of viral transcription (reviewed in Ref. 6). Therefore, HDACIs are considered as potential therapeutic agents for purging the latent viral reservoir in HIV-1-infected individuals. However, the active site structures of the HDAC family are largely conserved, and many HDACIs exhibit activity against multiple HDAC isoforms. For example, suberoylanilide hydroxamic acid (SAHA, vorinostat), an activator of latent HIV-1 expression (18–20), is a nonselective HDACI that inhibits both class I and class II HDAC isoforms (21). Because HDACs exert crucial roles in numerous biological processes, including cell cycle, cell differentiation, and survival (15), simultaneous inhibition of multiple HDAC isoforms will likely reduce their therapeutic window by promoting undesirable side effects and/or toxicity. Accordingly, the development of HDACI for an HIV-1 curative strategy requires knowledge of the HDAC isoforms contributing to viral latency and the development of inhibitors targeting these isoforms. In this study, we use a chemical approach that correlates the isoform specificities of HDACI with their abilities to reactivate latent HIV-1 expression to identify the HDAC isoforms responsible for HIV-1 latency in Jurkat J89GFP cells. The results from this study suggest that potent inhibition of HDAC3 may be important for reactivation of latent HIV-1.
EXPERIMENTAL PROCEDURES
Materials
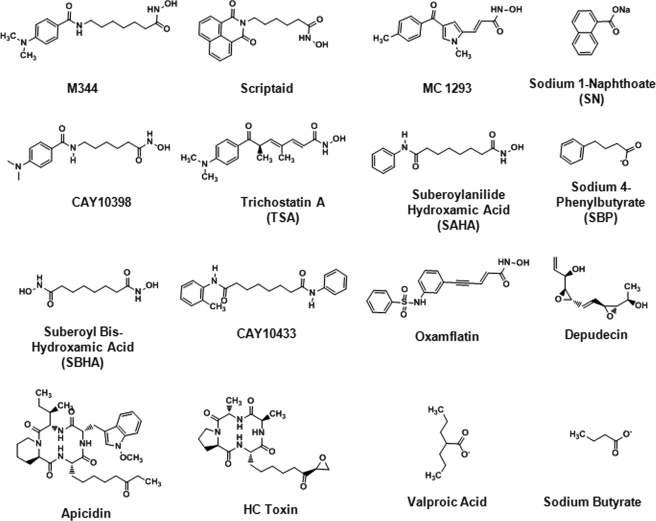
The HDACI 4,5:8,9-dianhydro-1,2,6,7,11-pentadeoxy-d-threo-d-ido-undeca-1,6-dienitol (depudecin), suberoyl bis-hydroxamic acid, cyclo[(2S)-2-amino-8-oxodecanoyl-1-methoxy-l-tryptophyl-l-isoleucyl-(2R)-2-piperidinecarbonyl] (apicidin), cyclo-(d-Pro-l-Ala-d-Ala-l-2-amino-8-oxo-9,10-epoxydecanoic acid) (HC toxin), (2E)-5-[3-(phenylsulfonylamino)phenyl]-pent-2-en-4-ynohydroxamic acid (oxamflatin), 6-(1,3-dioxo-1H,3H-benzo[de]isoquinolin-2-yl)-hexanoic acid hydroxyamide (scriptaid), sodium butyrate, sodium 4-phenylbutyrate, SAHA, valproic acid, and [(R)-(E,E)]-7-[4-(dimethylamino)phenyl]-N-hydroxy-4,6-dimethyl-7-oxo-2,4-heptadienamide] (trichostatin A) were obtained from Enzo Life Sciences (Plymouth Meeting, PA). 4-Dimethylamino-N-(6-hydroxyamino)-6-(oxohexyl]-benzamide (CAY10398) and N-phenyl-N′-(2-aminophenyl) hexamethylenediamide (CAY10433) were obtained from Cayman Chemical Co. (Ann Arbor, MI). Sodium 1-naphthoate was obtained from TCI America (Portland, OR). Droxinostat was purchased from Sigma. Wortmannin was obtained from Sigma. The AKT inhibitor IV was obtained from EMD Biosciences (Gibbstown, NJ). The phospho-AKT antibody and AKT antibody were obtained from Cell Signaling Technology (Boston). The β-actin antibody was obtained from Abcam (Cambridge, MA). DNA oligonucleotide primers were synthesized by Integrated DNA Technologies (San Diego). The recombinant purified HDAC isoforms, the Fluorogenic HDAC assay kit, and the HDAC assay substrates were purchased from BPS Bioscience (San Diego). The J89GFP cells were a kind gift from Dr David Levy.
HDAC Activity Assays
The lysine deacetylase activity of HDAC1–9 was assessed using the fluorogenic HDAC assay (BPS Bioscience) according to the manufacturer's instructions. The HDAC3 used in this assay was complexed with human nuclear receptor co-repressor 2 (NCOR2; amino acids 395–489), which is an activating co-factor of this HDAC isoform (31). All assays were carried out under steady-state conditions, and the assay read-out was optimized for linearity both as a function of time and enzyme concentration. Inhibition assays were carried out in 384-well plates. The assay volume was 25 μl and contained 0.1 mg/ml BSA, 20 μm substrate, and varying concentrations of the HDACI. All HDACI were dissolved in DMSO. The final concentration of DMSO did not exceed 5.0% (v/v). The formation of the fluorescent product was measured using a SpectraMax M2 plate reader (Molecular Devices). The excitation and emission wavelengths were 360 and 450 nm, respectively. The concentrations of HDACI required to inhibit 50% of the deacetylase activity of an HDAC isoform (i.e. IC50) were calculated by regression analysis using SigmaPlot software (Systat Software, Inc., San Jose, CA).
HDACI Cytotoxicity
Jurkat cells were maintained in RMPI 1640 medium supplemented with 10% FBS (Atlanta Biologicals), 0.3 mg/ml l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. HeLa and 293T cells were maintained in DMEM medium supplemented with 10% FBS, 0.3 mg/ml l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. CD8(+)-depleted peripheral blood mononuclear cells were isolated from fresh whole blood (100 ml) of HIV-negative individuals, as described previously (38). To determine HDACI cytotoxicity, 1 × 104 cells were plated in 96-well plates with varying concentrations of drug. Following a 24-h incubation period, cell viability was measured using either the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Roche Applied Science) or CellTiter 96 proliferation (Promega, Madison, WI) assay. The concentration of HDACI that decreased cell viability by 50% (i.e. 50% cytotoxic concentration (CC50)) was calculated by regression analysis using SigmaPlot software.
Reactivation of Latent HIV-1 by HDACI
J89GFP cells are a Jurkat T-cell line that contains a stably integrated, full-length HIV-1 provirus (strain 89.6) with an enhanced green fluorescent protein (EFGP) reporter incorporated into the viral genome (22). The viral genome in these cells is transcriptionally silent. However, upon stimulation with tumor necrosis factor α or HDACI, viral transcription was activated, and viral expression can be measured by EGFP production. We chose this cell line model of HIV-1 latency because the absence of viral expression was not due to mutations in either the Tat-TAR axis (e.g. the ACH2 cell line (34) and the U1 promonocytic cell line (36)) or in the 5′-LTR (e.g. the JΔK cell line (35)). The J89GFP cells were maintained in RMPI 1640 medium supplemented with 10% FBS, 0.3 mg/ml l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. 5 × 105 cells/ml cells were plated in 6-well plates with varying concentrations of HDACI for 6–72 h. The PI3K and Akt inhibitors wortmannin and Akt inhibitor IV (AI4) were used at concentrations of 100 nm and 10 μm, respectively. The cells were then washed in PBS, fixed in 4% paraformaldehyde, and stored at 4 °C until analysis. Reactivation of latent HIV-1 was determined by quantifying the percentage of EGFP-positive cells using a FACScan flow cytometer with FACSDiva software (BD Biosciences).
DNA Microarray Analyses
5 × 105 J89GFP cells were treated with 200 nm SAHA, oxamflatin, scriptaid, and apicidin for 24 h. Control experiments included J89GFP cells grown in the absence of HDACI and Jurkat cells infected with HIV-1 (multiplicity of infection of 1) for 24 h. Total cellular RNA was extracted from these cells using the RNeasy Plus RNA extraction kit (Qiagen Inc.) according to the manufacturer's protocol. RNA quantification, quality assessment, and DNA microarray analyses were carried out by PhalanxBio, Inc. (Palo Alto, CA), using the Human Whole Genome OneArrayTM microarray. Each treatment condition and control were assessed in duplicate biological replicates, and all samples were run in duplicate technical replicates on the arrays. Data analysis was performed by PhalanxBio, Inc., using Rosetta Resolver software.
siRNA Knockdown
siRNAs targeting HDAC1, HDAC2, and HDAC3, as well as a control scrambled sequence control siRNA, were purchased from Qiagen (SA Biosciences). The J89GFP cells were transfected with 60 nm siRNA using the Neon Transfection System from Invitrogen, according to the manufacturer's protocol. The efficiency of gene knockdown was assessed by determining mRNA levels (described below) and by Western blot analyses of protein expression.
Quantitative Analysis of Gene Transcripts
RNA was extracted from treated cells using RNeasy Plus RNA extraction kit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. RNA was quantified using a Nanodrop 2000, and 200–400 ng of total RNA was used in each reaction. RNA was amplified using the QuantiTect SYBR Green RT-PCR kit (Qiagen Inc., Valencia, CA) and the DNA Engine Opticon® system (Bio-Rad). Initiated HIV-1 transcripts were detected using primers TAR-FW (5′-GTTAGACCAGATCTGAGCCT-3′) and TAR-Rev (5′-GTGGGTTCCCTAGTTAGCCA-3′). Elongated HIV-1 transcripts were detected using primers TAT-FW (5′-ACTCGACAGAGGAGAGCAAG-3′) and TAT-REV (5′-GAGTCTGACTGTTCTGATGA-3′). HDAC1 was quantified using primers HDAC1-FW (5′-CCAGTATTCGATGGCCTGTT-3′) and HDAC1-REV (5′-TGTACAGCACCCTCTGGTGA-3′). HDAC2 was quantified using primers HDAC2-FW (5′-ATAAAGCCACTGCCGAAGAA-3′) and HDAC2-REV (5′-TCCTCCAGCCCAATTAACAG-3′). HDAC3 was quantified using primers HDAC3-FW (5′-TGGCTTCTGCTATGTCAACG-3′) and HDAC3-REV (5′-TCTCTGCCCCGACTTCATAC-3′). β-Actin mRNA copies were used as the normalization control (10) and were quantified using the primers β-actin-FW (5′-GTCGACAACGGCTCCGGC-3′) and β-actin-REV (5′-GGTGTGGTGCCAGATTTTCT-3′). Relative gene expression levels were calculated using the ΔΔC(T) method (23).
Chromatin Immunoprecipitation (ChIP) Assays
2 × 106 J89GFP cells were fixed with 1% formaldehyde for 10 min at room temperature. ChIP assays were carried out using the EZ-ChIP assay kit (Millipore Billerica, MA). Immunoprecipitation was performed using 5 μg of antibody (Invitrogen) against HDAC1–3 or -8. Rabbit immunoglobulin G serum (5 μg, Santa Cruz Biotechnology Inc., Santa Cruz, CA) was used to control for nonspecific immunoprecipitation of DNA. Forward (LTRκB primer 5, 5′-AGGTTTGACAGCCGCCTA-3′) and reverse (LTRκB primer 3, 5′-AGAGACCCAGTACAGGCAAAA-3′) primers specific for a 203-bp region in the HIV-1 LTR that encompasses the NF-κB-binding site LTR were used to detect a specific interaction between an HDAC isoform and HIV-1 DNA, as described previously (10).
RESULTS AND DISCUSSION
Potent Inhibition of HDAC1 Is Not Sufficient to Reactivate Latent HIV-1
Previous studies have demonstrated that recruitment of HDAC1 to the HIV-1 LTR by different DNA-binding complexes is sufficient to induce viral latency (9–11, 13, 14). Accordingly, we hypothesized that a potent inhibitor of HDAC1 would reactivate HIV-1 expression in the J89GFP cell line model of viral latency. Initially, we screened 16 structurally diverse HDACI (Fig. 1) for the following: (i) their inhibitory activity against recombinant purified HDAC1; (ii) their cytotoxicity (CC50) in Jurkat cells; and (iii) their ability to reactivate HIV-1 expression in J89GFP cells (Table 1). For point iii, the highest possible sub-cytotoxic concentration of HDACI (determined from the cytotoxicity assessments) was used. Of the 16 HDACI tested, 6 (apicidin, HC toxin, scriptaid, oxamflatin, SAHA, and trichostatin A) exhibited potent activity (IC50 <100 nm) against purified HDAC1. Of these, only three (oxamflatin, apicidin, and trichostatin A) were able to stimulate HIV-1 expression by more than 5% in the J89GFP cells, as measured by flow cytometry analysis of EGFP expression. In this regard, it should be noted that apicidin, scriptaid, oxamflatin, and SAHA exhibited similar CC50 values and were tested in the J89GFP cells at identical concentrations, therefore allowing for a direct comparison of their ability to reactivate latent HIV-1 expression. Interestingly, valproic acid and sodium butyrate were found to be relatively weak inhibitors of HDAC1 (IC50 ∼175 μm), but each elicited a different effect in the J89GFP cells as follows: 1 mm valproic acid reactivated HIV-1 expression in only 4.3% of cells; 1 mm sodium butyrate reactivated HIV-1 expression in 66.4% of cells. Taken together, these HDACI screening studies show that inhibition of HDAC1 is not sufficient to reactivate HIV-1 expression in J89GFP cells.
FIGURE 1.
Chemical structures of HDACI used in this study.
TABLE 1.
In vitro activity against HDAC1, cytotoxicity in Jurkat cells, and reactivation of latent HIV-1 in J89GFP cells by structurally diverse HDACI
| HDACI | IC50 against HDAC1 | Cytotoxicity CC50 | Reactivation dosea | % inhibition of HDAC1 at reactivation doseb | % EGFP-positive J89GFP cells (after 24 h) |
|---|---|---|---|---|---|
| μm | μm | μm | |||
| HC toxin | 0.000154 | 0.05 | 0.005 | 99.7 | 1.3 |
| Apicidin | 0.000299 | 10.0 | 0.5 | 99.7 | 40.6 |
| Oxamflatin | 0.003959 | 6.0 | 0.5 | 99.2 | 31.2 |
| Scriptaid | 0.006421 | 6.0 | 0.5 | 98.7 | 3.2 |
| SAHA | 0.0137 | 10.0 | 0.5 | 97.3 | 2.5 |
| TSA | 0.0169 | 0.10 | 0.05 | 74.7 | 17.2 |
| M344 | 0.0941 | 0.5 | 0.1 | 51.5 | 3.1 |
| CAY10398 | 1.7780 | 1.0 | 0.5 | 21.9 | 0.9 |
| MC1293 | 4.245 | 10.0 | 5 | 54.1 | 2.7 |
| CAY10433 | 9.36 | 1000 | 10 | 51.6 | 5.2 |
| SBHA | 4.54 | 100 | 100 | 95.6 | 57.5 |
| Depudecin | 25.33 | 5.0 | 1 | 3.8 | 1.3 |
| Sodium 1-naphthoate | 200.6 | 10 | 10 | 4.75 | 1 |
| Valproic acid | 171 | 10,000 | 1000 | 85.4 | 4.3 |
| Sodium butyrate | 175 | >10,000 | 1000 | 85.1 | 66.4 |
| Sodium 4-phenylbutyrate | 162 | 10,000 | 1000 | 86.1 | 2.5 |
a The highest nontoxic concentration of HDACI (determined from the cytotoxicity assays in Jurkat cells) was administered to the J89GFP cells to determine the inhibitor's ability to reactivate latent HIV-1.
b Maximum possible inhibition of HDAC1 at the concentration of inhibitor used in the reactivation experiments in J89GFP cells is shown (the actual inhibition in the J89GFP cells is likely to be significantly less due to inefficient cellular uptake and nonspecific protein binding).
Inhibition of HDAC3 Correlates with the Reactivation of Latent HIV-1
Based on the data described above, we next carried out in-depth analyses on the HDACIs apicidin, oxamflatin, scriptaid, and SAHA (Fig. 2). Each of these HDACIs exhibit similar potency against purified HDAC1 and similar cytotoxicity (CC50) values in Jurkat cells (Table 1). However, at concentrations of inhibitor ranging from 0 to 500 nm, apicidin and oxamflatin reactivated HIV-1 expression in the J89GFP cells in a dose-dependent manner, whereas SAHA and scriptaid elicited no effect (Fig. 2A). A time course experiment demonstrated that this lack of activity was not due to an early or late EGFP peak that was missed at the 24-h time point used in the dose-response experiments (Fig. 2B). Because the EGFP expression quantitated in Fig. 2, A and B, only provides information on the translated protein, we also used quantitative RT-PCR to assess the formation of HIV-1 RNA transcripts (Fig. 2C). Consistent with the flow cytometry analyses, oxamflatin and apicidin significantly increased the abundance of elongated HIV-1 transcripts, but not initiated transcripts, compared with untreated J89GFP cells. By contrast, scriptaid and SAHA did not significantly increase the formation of either initiated or elongated HIV-1 transcripts.
FIGURE 2.
Reactivation of latent HIV-1 in J89GFP cells by oxamflatin, apicidin, scriptaid, and SAHA. A, J89GFP cells were treated with varying concentrations of HDACI (0–500 nm), and the percentage of cells expressing EGFP was quantitated after 24 h by FACs. Error bars represent S.E. from at least three independent experiments. B, J89GFP cells were treated with 200 nm HDACI, and the percentage of cells expressing EGFP was quantitated at different time intervals (0–75 h) by FACs. C, increase in HIV-1 RNA transcripts in cells treated with 200 nm HDACI for 24 h as measured by quantitative RT-PCR. Error bars represent S.E. from at least three independent experiments. The fold increase in transcript relative to untreated cells is indicated above each bar for all four HDACI. D, J89GFP cells were treated with varying concentrations of HDACI (0–2 μm), and the percentage of cells expressing EGFP was quantitated after 24 h by FACs. Error bars represent S.E. from at least three independent experiments.
We also carried out cDNA microarray analyses to determine the magnitude and the extent of global gene expression changes observed in the J89GFP cells after 24 h of treatment with 200 nm oxamflatin, scriptaid, SAHA, or apicidin. Control experiments included untreated J89GFP cells and Jurkat cells infected with HIV-1 for 24 h. The cDNA microarray data are provided in supplemental Tables 1 and 2 and Fig. 1. SAHA and scriptaid were found to significantly (p < 0.01) up- or down-regulate 3 and 1% of all genes compared with untreated cells, respectively. One study reported that SAHA altered regulation in at least 22% of genes in CEM cells; however, a much higher concentration of drug (2.5 μm) was used which caused ∼50% of cell death after 24 h (30). Oxamflatin and apicidin resulted in gene expression changes in ∼11 and 8% of all genes compared with untreated J89GFP, respectively. These higher gene expression levels compared with scriptaid and SAHA could be due to inhibition of different HDAC isoforms (see below) and/or the expression of HIV-1 proteins in the J89GFP cells. Microarray DNA analyses revealed that 2.7% of all genes displayed altered expression changes in HIV-1-infected Jurkat cells compared with uninfected cells. Nevertheless, these data indicate that SAHA and scriptaid were taken up into the J89GFP cells and induced gene expression changes. However, at the concentrations tested, they lacked the ability to induce expression of latent HIV-1.
To gain insight into the mechanisms by which oxamflatin and apicidin, but not scriptaid and SAHA, reactivated latent HIV-1, we determined the in vitro inhibitory activity of each of the HDACI against recombinant purified HDAC1 and -2, the 3-NCOR2 complex, and -4–9 (Table 2). In general, each of the HDACI exhibited little or no activity against the class II HDAC isoforms, although scriptaid exhibited excellent activity against HDAC6 (IC50 = 34 nm) and oxamflatin activity against HDAC6 (IC50 = 390 nm) and HDAC7 (IC50 = 840 nm). By contrast, all four of the HDACI were found to be very potent inhibitors of the class I HDAC isoforms 1, 2, and 8 (IC50 <20 nm). Interestingly, oxamflatin and apicidin, both of which induced HIV-1 outgrowth in the J89GFP cells, also potently inhibited the HDAC3-NCOR2 complex (IC50 <10 nm). By contrast, scriptaid (IC50 = 320 nm) and SAHA (IC50 = 600 nm) were ∼100-fold less active against the HDAC3-NCOR2 complex. Based on our IC50 calculations, a 200 nm dose of apicidin and oxamflatin would inhibit >95% of the deacetylase activity of HDAC1–3 and -8. (In the J89GFP cells, these values would likely be significantly less due to inefficient inhibitor uptake and nonspecific protein binding.) By contrast, a 200 nm dose of scriptaid and SAHA would inhibit >95% of HDAC1, -2, and -8 but would only inhibit 25 and 18% of the deacetylase activity of HDAC3, respectively. These values for HDAC3 would increase to 45 and 36%, respectively, at a dose of 500 nm. Taken together, these data provide strong evidence that potent inhibition of HDAC3 is required to reactivate the expression of HIV-1 in the J89GFP cells. The data also suggested that increasing the concentrations of scriptaid and SAHA to allow inhibition of HDAC3 would result in the activation of latent HIV-1 in the J89GFP cells. Indeed, in Fig. 2D we show that 2 μm scriptaid and SAHA promote activation of latent HIV-1 in the J89GFP. At this concentration, the total inhibition of HDAC3 approaches 76 and 70% for scriptaid and SAHA, respectively. To further assess the importance of HDAC3 inhibition in the reactivation of latent HIV-1 infection, we assessed the ability of droxinostat to reactivate latent HIV-1 expression in the J89GFP cells. Previous studies reported that this HDACI selectively inhibited HDAC3 and -8 but not HDAC1 and -2 (37). Indeed, we found that droxinostat is a reasonably potent inhibitor of recombinant HDAC3-NCOR2 complex and HDAC8 (IC50 = 2.0 and 3.0 μm, respectively) but shows only weak activity against HDAC1 and -2 (IC50 = 63 and 250 μm, respectively) (Table 2). Of note, droxinostat was found to reactivate latent HIV-1 expression in a dose-dependent manner (Fig. 3). At a 40 μm concentration of droxinostat, HDAC3 and -8 would be inhibited by >95%, whereas there would be only partial inhibition of HDAC1 (38%) and HDAC2 (14%). Taken together, these studies provide additional evidence that HDACI with specificity toward HDAC3 can reactivate latent HIV-1 expression in J89GFP cells.
TABLE 2.
In vitro activity of HDACI against class I and class II HDAC isoforms
| HDAC isoform | IC50 against HDAC isoforms (nm) |
|||||
|---|---|---|---|---|---|---|
| Apicidin | Oxamflatin | Scriptaid | SAHA | Droxinostat | Valproic acid | |
| Class I | ||||||
| HDAC1 | 0.30 ± 0.15a1 | 3.96 ± 0.87 | 0.64 ± 0.09 | 13.7 ± 0.15 | 63,000 | 171,000 |
| HDAC2 | 1.2 ± 0.80 | 0.16 ± 0.11 | 1.4 ± 0.74 | 62.0 ± 0.15 | 250,000 | 634,000 |
| HDAC3 | 0.98 ± 0.22 | 10.3 ± 1.2 | 607 ± 93 | 869 ± 0.15 | 2000 | 5,500,000 |
| HDAC8 | 0.26 ± 0.09 | 0.37 ± 0.15 | 14.5 ± 1.1 | 6.8 ± 0.15 | 5000 | 756,000 |
| Class II | ||||||
| HDAC4 | >50,000 | 3800 ± 1100 | 14,000 ± 1500 | >50,000 | NDb | ND |
| HDAC5 | >50,000 | >50,000 | >50,000 | >50,000 | ND | ND |
| HDAC6 | >50,000 | 390 ± 73 | 34 ± 9 | 5500 ± 760 | ND | ND |
| HDAC7 | >50,000 | 840 ± 39 | 2200 ± 350 | >50,000 | ND | ND |
| HDAC9 | >50,000 | >50,000 | >50,000 | >50,000 | ND | ND |
a Data represent the mean ± S.D. from three replicate experiments.
b ND, not determined.
FIGURE 3.
Reactivation of latent HIV-1 in J89GFP cells by the HDAC3-specific inhibitor droxinostat. The chemical structure of droxinostat is shown. The number of J89GFP cells expressing EGFP was quantitated after 24 h by FACs. Error bars represent S.E. from two independent experiments.
Several studies have recently demonstrated that oxamflatin, apicidin, scriptaid, and SAHA can reactivate latent HIV-1 in different cell lines and/or in resting CD4(+) T cells from aviremic patients (18, 19, 20, 32). In each of these studies relatively high concentrations (>500 nm) of inhibitor were used. Of note, each of these HDACI display significant toxicity in cell lines (CC50 values range from 5 to 10 μm) and in CD8(+)-depleted peripheral blood mononuclear cells (CC50 values range from 0.1 to 7.5 μm) (Table 3). In this regard, the small therapeutic window of these HDACI highlights one potential limitation for their inclusion in therapeutic combinations targeted toward the eradication of HIV-1.
TABLE 3.
Cytotoxicity of HDACI in different cells
| HDACI | CC50 (μm) |
|||
|---|---|---|---|---|
| Jurkata | HeLaa | 293Ta | CD8(+)-depleted PBMCb | |
| Apicidin | 10.0 ± 0.1 | 11.3 ± 1.7 | 12.2 ± 1.4 | 0.1 |
| Oxamflatin | 5.9 ± 0.1 | 9.4 ± 0.8 | 6.0 ± 1.0 | 0.3 |
| SAHA | 10.0 ± 0.1 | 11.3 ± 0.3 | 14.1 ± 0.2 | 7.5 |
| Scriptaid | 6.1 ± 0.3 | 8.9 ± 1.8 | 5.8 ± 0.2 | 0.7 |
a Data represent the mean ± S.D. from three replicate experiments.
b Data represent the mean from two independent replicate experiments.
HDAC3 Resides at the HIV-1 LTR in J89GFP Cells
The data described above provide strong evidence that inhibition of HDAC3 is important for the activation of latent HIV-1. However, only two studies have identified this HDAC isoform at the HIV-1 LTR (16, 17). Accordingly, we performed ChIP assays in the J89GFP cells using antibodies specific for the class I HDAC isoforms (Fig. 4A). We detected a strong signal for HDAC1 and -3 indicating that both of these HDAC isoforms resided at the HIV-1 LTR. We also detected a weak signal for HDAC2 suggesting that this isoform may also be present at the HIV-1 LTR in J89GFP cells. HDAC8 did not associate with the HIV-1 LTR. These findings are consistent with a recent study by Keedy et al. (17), who also reported that HDAC1–3 resided at the HIV-1 LTR in J89GFP cells. Of note, this study also reported that HDAC8 is primarily sequestered in the cytoplasm and not the nucleus in J89GFP cells (17). Quantitative real time PCR experiments confirmed the presence of HDAC1 and -3, but not HDAC2, at the HIV-1 LTR (Fig. 4B). Importantly, the occupancy of HDAC1 and -3 at the HIV-1 LTR in the J89GFP cells is lost upon treatment with apicidin (Fig. 4B) or oxamflatin (data not shown). To further assess the role of HDAC1–3 in maintaining HIV-1 latency, we knocked down their gene expression by siRNA (Fig. 4C). The magnitude of the siRNA-mediated gene silencing was confirmed by quantitative PCR analyses of mRNA levels (Fig. 4C) and by Western blot analysis (data not shown). Knockdown of HDAC3 resulted in significant and substantial cell death that prevented subsequent analyses of HIV-1 latency. Interestingly, the knockdown of either HDAC1 or -2, or a combination of HDAC1 and -2, did not result in the reactivation of latent HIV-1 expression in the J89GFP cells (Fig. 4D). These data provide strong supporting evidence that inhibition of HDAC1 and -2 is insufficient to reactivate latent HIV-1 expression.
FIGURE 4.
Role of HDAC1–3 in maintaining HIV-1 latency. A, ChIP assays identify HDAC1–3, but not HDAC8, at the HIV-1 LTR in J89GFP cells. Two different HDAC1 antibodies (a and b) were used in the ChIP assays. B, quantitative real time PCR experiments show enrichment (versus rabbit IgG) of HDAC1 and -3 at the HIV-1 LTR in J89GFP cells that is lost upon treatment with apicidin. PCR data were normalized by quantification of the GAPDH promoter in the input samples. C, mRNA expression of HDAC1–3 after knockdown by siRNA. HDAC1* and HDAC2* reports on the mRNA levels in experiments in which both HDAC1 and -2 were knocked down simultaneously. An siRNA with a scrambled sequence was used as a control. D, reactivation of latent HIV-1 in J89GFP cells after knockdown of HDAC1 or -2 or HDAC1 and -2. The number of J89GFP cells expressing EGFP was quantitated after 24 h by FACs. Error bars represent S.E. from three independent experiments.
Reactivation of Latent HIV-1 Expression by Apicidin and Oxamflatin Is Partially Dependent on the PI3K/Akt Signaling Pathway
Peterlin and co-workers (20) have shown that SAHA activates HIV-1 expression in latently infected cells via the PI3K/Akt pathway. To determine whether this pathway is also activated by apicidin and oxamflatin, we first assessed Akt phosphorylation levels in J89GFP cells treated with TNF-α (a positive control) or with 500 nm apicidin, oxamflatin, SAHA, or scriptaid (Fig. 5A). Increased Akt phosporylation was observed following treatment with apicidin but not oxamflatin, SAHA, or scriptaid. Because the concentration of SAHA used in this experiment (500 nm) is not sufficient to reactivate latent HIV-1 expression in the J89GFP cells, it was not unexpected that Akt was not activated. To further determine the impact of apicidin and oxamflatin on activation of the PI3K/Akt signaling pathway, we used PI3K (wortmannin) and Akt (Akt inhibitor IV (AI4)) inhibitors. Indeed, Akt and PI3K inhibitors decreased but did not completely eliminate both apicidin and oxamflatin-induced viral replication (Fig. 5B). Importantly, these inhibitors had no significant effect on the basal levels of HIV-1 production (Fig. 5B). These results suggest that the latent HIV-1 reactivation activity of both apicidin and oxamflatin is intertwined with activation of the PI3K/Akt signaling pathway.
FIGURE 5.
Activation of the PI3K/Akt signaling pathway by apicidin and oxamflatin. A, Western blot analysis of Akt phosphorylation in J89GFP cells treated with TNF-α (positive control) or with 500 nm apicidin, oxamflatin, SAHA, or scriptaid. Phosphorylated Akt and β-actin were detected using monoclonal antibodies specific for these proteins. B, J89GFP cells were treated with 500 nm HDACI with or without wortmannin (100 nm) or the Akt inhibitor IV (AI 4, 10 μm). The number of J89GFP cells expressing EGFP was quantitated after 24 h by FACs. Error bars represent S.E. from three independent experiments.
Valproic Acid Is a Weak Inhibitor of HDAC3
In 2004, Ylisastigui et al. (24) demonstrated that treatment of resting CD4(+) T cells of aviremic patients with valproic acid induced histone acetylation and promoted virus outgrowth. An initial proof-of-concept study in which four volunteers infected with HIV added oral valproic acid to their cART regimen for 3 months reported a significant decline in the frequency of resting CD(+) T-cell infection (25). However, several follow-up studies found that valproic acid does not reduce the size of latent HIV reservoir (26–29). Interestingly, we find that valproic acid is a weak inhibitor of the HDAC3-NCOR2 complex (IC50 = 5.5 mm, Table 2) and that high concentrations of inhibitor (>1 mm) are required in J89GFP cells to activate HIV-1 expression (Fig. 6). The total and free therapeutic concentrations of valproic acid in adults range from 275 to 700 μm and from 27 to 100 μm, respectively. These therapeutic concentrations would be insufficient to inhibit HDAC3 in vivo. Accordingly, our data may explain why valproic acid has no effect on the decay of latent HIV reservoirs in patients.
FIGURE 6.
Reactivation of latent HIV-1 in J89GFP cells by varying concentrations (0–10 mm) of valproic acid. The percentage of J89GFP cells expressing EGFP was quantitated after 24 h by FACs.
Conclusions
This study demonstrates that HDAC3 resides at the HIV-1 LTR in J89GFP cells and that inhibition of this HDAC isoform is required for the activation of latent HIV-1 expression by HDACI. Interestingly, Archin et al. (33) recently demonstrated that an HDACI (MRK12) specific for HDAC1 and -2 was unable to reactivate latent HIV-1 in J89GFP cells and in resting CD4(+) T-cells from aviremic patients. By contrast, an inhibitor (MRK13) specific for HDAC1–3 promoted virus outgrowth in both assay systems (33). Taken together, these studies suggest that potent inhibition of HDAC3 should be an important criterion in the development of HDACI for HIV-1 curative strategies. Unfortunately, neither our study nor that of Archin et al. (33) could address whether inhibition of HDAC3 alone is sufficient to induce virus outgrowth or if inhibition of HDAC1 and/or -2 is also required. The identification and development of inhibitors specific for HDAC3 may address this question.
Finally, it is likely that there are several reservoirs of latent HIV-1 infection in aviremic patients on cART. For example, resting CD4(+) T-cells and CD34(+) multipotent hematopoietic progenitor cells have both been identified as reservoirs of latent HIV-1 infection (2–5). The HDAC isoforms recruited to the HIV-1 LTRs in these different cell types may be different. In this regard, the chemical approach described in this study can readily be used to identify the HDAC isoforms that contribute to HIV-1 latency in other cell types. The primary advantages of this approach include the ability to rapidly conduct studies in cell types that cannot be easily transfected with siRNA (or shRNA) molecules or in patient-derived tissues or cells where there may be insufficient material to carry out genetic studies. Importantly, our chemical approach also provides an immediate assessment of the therapeutic potential of HDACI to reactivate latent HIV-1 expression in different cell types and/or tissues.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Tables 1 and 2.
- cART
- combination antiretroviral therapy
- HDAC
- histone deacetylase
- HDACI
- HDAC inhibitor
- SAHA
- suberoylanilide hydroxamic acid
- EGFP
- enhanced green fluorescent protein
- FW
- forward
- REV
- reverse
- HIV-1
- HIV type 1.
REFERENCES
- 1. Davey R. T., Jr., Bhat N., Yoder C., Chun T. W., Metcalf J. A., Dewar R., Natarajan V., Lempicki R. A., Adelsberger J. W., Miller K. D., Kovacs J. A., Polis M. A., Walker R. E., Falloon J., Masur H., Gee D., Baseler M., Dimitrov D. S., Fauci A. S., Lane H. C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 15109–15114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chun T. W., Stuyver L., Mizell S. B., Ehler L. A., Mican J. A., Baseler M., Lloyd A. L., Nowak M. A., Fauci A. S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13193–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Finzi D., Hermankova M., Pierson T., Carruth L. M., Buck C., Chaisson R. E., Quinn T. C., Chadwick K., Margolick J., Brookmeyer R., Gallant J., Markowitz M., Ho D. D., Richman D. D., Siliciano R. F. (1997) Science 278, 1295–1300 [DOI] [PubMed] [Google Scholar]
- 4. Wong J. K., Hezareh M., Günthard H. F., Havlir D. V., Ignacio C. C., Spina C. A., Richman D. D. (1997) Science 278, 1291–1295 [DOI] [PubMed] [Google Scholar]
- 5. Carter C. C., Onafuwa-Nuga A., McNamara L. A., Riddell J., 4th, Bixby D., Savona M. R., Collins K. L. (2010) Nat. Med. 16, 446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colin L., Van Lint C. (2009) Retrovirology 6, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Margolis D. M. (2010) Curr. HIV/AIDS Rep. 7, 37–43 [DOI] [PubMed] [Google Scholar]
- 8. Trono D., Van Lint C., Rouzioux C., Verdin E., Barré-Sinoussi F., Chun T. W., Chomont N. (2010) Science 329, 174–180 [DOI] [PubMed] [Google Scholar]
- 9. Coull J. J., Romerio F., Sun J. M., Volker J. L., Galvin K. M., Davie J. R., Shi Y., Hansen U., Margolis D. M. (2000) J. Virol. 74, 6790–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams S. A., Chen L. F., Kwon H., Ruiz-Jarabo C. M., Verdin E., Greene W. C. (2006) EMBO J. 25, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Imai K., Okamoto T. (2006) J. Biol. Chem. 281, 12495–12505 [DOI] [PubMed] [Google Scholar]
- 12. Marban C., Suzanne S., Dequiedt F., de Walque S., Redel L., Van Lint C., Aunis D., Rohr O. (2007) EMBO J. 26, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang G., Espeseth A., Hazuda D. J., Margolis D. M. (2007) J. Virol. 81, 10914–10923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tyagi M., Karn J. (2007) EMBO J. 26, 4985–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haberland M., Montgomery R. L., Olson E. N. (2009) Nat. Rev. Genet. 10, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malcolm T., Chen J., Chang C., Sadowski I. (2007) Virus Genes 35, 215–223 [DOI] [PubMed] [Google Scholar]
- 17. Keedy K. S., Archin N. M., Gates A. T., Espeseth A., Hazuda D. J., Margolis D. M. (2009) J. Virol. 83, 4749–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edelstein L. C., Micheva-Viteva S., Phelan B. D., Dougherty J. P. (2009) AIDS Res. Hum. Retroviruses 25, 883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Archin N. M., Espeseth A., Parker D., Cheema M., Hazuda D., Margolis D. M. (2009) AIDS Res. Hum. Retroviruses 25, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Contreras X., Schweneker M., Chen C. S., McCune J. M., Deeks S. G., Martin J., Peterlin B. M. (2009) J. Biol. Chem. 284, 6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Savarino A., Mai A., Norelli S., El Daker S., Valente S., Rotili D., Altucci L., Palamara A. T., Garaci E. (2009) Retrovirology 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kutsch O., Benveniste E. N., Shaw G. M., Levy D. N. (2002) J. Virol. 76, 8776–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 24. Ylisastigui L., Archin N. M., Lehrman G., Bosch R. J., Margolis D. M. (2004) AIDS 18, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 25. Lehrman G., Hogue I. B., Palmer S., Jennings C., Spina C. A., Wiegand A., Landay A. L., Coombs R. W., Richman D. D., Mellors J. W., Coffin J. M., Bosch R. J., Margolis D. M. (2005) Lancet 366, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Archin N. M., Cheema M., Parker D., Wiegand A., Bosch R. J., Coffin J. M., Eron J., Cohen M., Margolis D. M. (2010) PLoS One 5, e9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Archin N. M., Eron J. J., Palmer S., Hartmann-Duff A., Martinson J. A., Wiegand A., Bandarenko N., Schmitz J. L., Bosch R. J., Landay A. L., Coffin J. M., Margolis D. M. (2008) AIDS 22, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sagot-Lerolle N., Lamine A., Chaix M. L., Boufassa F., Aboulker J. P., Costagliola D., Goujard C., Pallier C., Paller C., Delfraissy J. F., Lambotte O. (2008) AIDS 22, 1125–1129 [DOI] [PubMed] [Google Scholar]
- 29. Siliciano J. D., Lai J., Callender M., Pitt E., Zhang H., Margolick J. B., Gallant J. E., Cofrancesco J., Jr., Moore R. D., Gange S. J., Siliciano R. F. (2007) J. Infect. Dis. 195, 833–836 [DOI] [PubMed] [Google Scholar]
- 30. Peart M. J., Smyth G. K., van Laar R. K., Bowtell D. D., Richon V. M., Marks P. A., Holloway A. J., Johnstone R. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3697–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guenther M. G., Barak O., Lazar M. A. (2001) Mol. Cell. Biol. 21, 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ying H., Zhang Y., Lin S., Han Y., Zhu H. Z. (2010) Int. J. Mol. Med. 26, 265–272 [DOI] [PubMed] [Google Scholar]
- 33. Archin N. M., Keedy K. S., Espeseth A., Dang H., Hazuda D. J., Margolis D. M. (2009) AIDS 23, 1799–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emiliani S., Van Lint C., Fischle W., Paras P., Jr., Ott M., Brady J., Verdin E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6377–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Antoni B. A, Rabson A. B., Kinter A., Bodkin M., Poli G. (1994) Virology 202, 684–694 [DOI] [PubMed] [Google Scholar]
- 36. Emiliani S., Fischle W., Ott M., Van Lint C., Amella C. A., Verdin E. (1998) J. Virol. 72, 1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wood T. E., Dalili S., Simpson C. D., Sukhai M. A., Hurren R., Anyiwe K., Mao X., Suarez Saiz F., Gronda M., Eberhard Y., MacLean N., Ketela T., Reed J. C., Moffat J., Minden M. D., Batey R. A., Schimmer A. D. (2010) Mol. Cancer Ther. 9, 246–256 [DOI] [PubMed] [Google Scholar]
- 38. Reuse S., Calao M., Kabeya K., Guiguen A., Gatot J. S., Quivy V., Vanhulle C., Lamine A., Vaira D., Demonte D., Martinelli V., Veithen E., Cherrier T., Avettand V., Poutrel S., Piette J., de Launoit Y., Moutschen M., Burny A., Rouzioux C., De Wit S., Herbein G., Rohr O., Collette Y., Lambotte O., Clumeck N., Van Lint C. (2009) PLoS One 4, e6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.