Abstract
SIRT1 is involved in the pathogenesis of obesity, diabetes, and aging. However, it is not clear how SIRT1 activity is regulated by intracellular kinases in cells. In this study, we investigated SIRT1 phosphorylation and protein degradation in response to JNK1 activation in obese mice. Mouse SIRT1 is phosphorylated by JNK1 at Ser-46 (Ser-47 in human SIRT1), which is one of the four potential residues targeted by JNK1. The phosphorylation induces a brief activation of SIRT1 function and degradation of SIRT1 thereafter by the proteasome. Ubiquitination occurs in SIRT1 protein after the phosphorylation. Mutation of Ser-46 to alanine prevents the phosphorylation, ubiquitination, and degradation. In vivo, SIRT1 undergoes an extensive degradation in hepatocytes in obesity as a consequence of persistent activation of JNK1. The degradation leads to inhibition of SIRT1 function, which contributes to development of hepatic steatosis. The degradation disappears in obesity when JNK1 is inactivated in mice. JNK2 exhibits an opposite activity in the regulation of SIRT1 degradation. The JNK1-SIRT1 pathway provides a new molecular mechanism for the pathogenesis of hepatic steatosis in obesity.
Keywords: Insulin Resistance, Liver Metabolism, Obesity, SIRT, Ubiquitination
Introduction
SIRT1 (Sirtuin 1) is a member of the sirtuin family of histone deacetylases whose activities are induced by nicotinamide adenine dinucleotide (NAD+) (1). SIRT1 regulates energy metabolism and lifespan through transcriptional silencing (2, 3). SIRT1 activity is reported in many organs/tissues in the regulation of gene transcription. Liver is one of the major organs where SIRT1 is involved in the modification of lipid and glucose metabolism (4–8). Activation of SIRT1 by resveratrol or small molecules promotes fatty acid oxidation, prevents diet-induced fatty liver, and attenuates insulin resistance (9–12). SIRT1 knockdown in liver induces local accumulation of fatty acids and cholesterol (13). SIRT1 inactivation increases the risk of hepatic steatosis in mice with global or organ-specific knock-out (7, 8). Mechanisms of SIRT1 actions involve the regulation of transcriptional activities of peroxisome proliferator-activated receptor γ (14), sterol regulatory element-binding protein (SREBP) (15), HNF4 (16), FoxO1 (17), NF-κB (18), and activator protein-1 (AP-1) (19). SIRT1 also regulates nuclear coactivators, such as PGC-1α and CRTC2 (also known as TORC2), in the control of hepatic gluconeogenesis (4, 5). SIRT1 interaction with those nuclear proteins provides molecular mechanisms for hepatic steatosis and metabolic disorders in obesity.
SIRT1 activity is reduced in liver and adipose tissue in response to obesity (9, 10, 14). The alteration is related to loss of SIRT1 protein or a reduction in intracellular NAD+ levels. The protein reduction is associated with hyperinsulinemia and hyperglycemia in mammalian systems (20). In cells, SIRT1 protein is reduced by insulin or glucose (20). However, the signaling pathway for SIRT1 reduction is unknown.
c-Jun N-terminal kinase 1 (JNK1) activation contributes to the development of steatohepatitis in obesity (21, 22). Suppression of hepatic JNK1 activity by chemical inhibitors or RNAi-mediated gene knockdown attenuates hepatic steatosis (23) and improves insulin sensitivity (24, 25). In obesity, JNK1 is activated by several obesity-associated factors, such as inflammation, endoplasmic reticulum stress, oxidative stress, diglyceride, or ceramide (26–29). JNK1 contributes to insulin resistance by direct phosphorylation of the IRS-1 protein (26, 28, 30). Our recent study of liver-specific JNK1 knock-out mice suggests that JNK1 activity is required for protection of liver from hepatic steatosis (31). The study has raised a concern about the exact role of JNK1 in the regulation of lipid metabolism in liver. It is unknown why JNK1 has opposite activities in the pathogenesis of hepatic steatosis.
In the present study we investigated regulation of SIRT1 protein by JNK1 in cellular and animal models. We present evidence that JNK1 phosphorylates SIRT1 and induces SIRT1 activation, which is followed by ubiquitination and degradation of SIRT1. Persistent JNK1 activation leads to inhibition of SIRT1 activity through extensive protein degradation, which contributes to hepatic steatosis in obesity. The JNK1-SIRT1 pathway provides a new molecular mechanism for fatty liver in obesity.
MATERIALS AND METHODS
Animal Models
Male C57BL/6 (B6) mice were purchased at 4 weeks of age from The Jackson Laboratory (Bar Harbor, ME). The global knock-out mice for JNK1-, JNK2-, and liver-specific knock-out mice for JNK1/2 were from Dr. Roger Davis' laboratory. Age- and gender-matched littermates were used as controls. SIRT1+/− mice on the 129/J background were a gift of Dr. Frederick W. Alt at the Howard Hughes Medical Institute and Department of Genetics, Harvard University Medical School, Boston, MA 02115. SIRT1+/− transgenic mice were backcrossed with C57BL/6 mice for 10 generations to obtain the C57BL/6 gene background. The heterozygous knock-out (KO) (SIRT1+/−) and wild type (WT) littermates were used in the study. All of the mice were housed in the animal facility in group of 4 mice per box with a 12:12-h light-dark cycle and constant temperature (22–24 °C). The mice had free access to water and diet. All procedures were performed in accordance with the National Institutes of Health guidelines and approved by the Institution Animal Care and Use Committee at the Pennington Biomedical Research Center.
High Fat Diet (HFD)2
HFD (D12331, Research Diets, New Brunswick, NJ) contains 58% of kcal from fat. The mice were fed the HFD at 5 weeks of age. Their body weight and composition were examined every 2 weeks.
Cell Culture and Reagents
Mouse embryonic fibroblast (MEF) cell lines including WT and JNK1-KO and JNK2-KO were described elsewhere (32). The cell lines 3T3-L1 (CL-173), rat H4IIE, human HepG2, and HEK293 were purchased from the American Type Culture Collection and maintained in 10% fetal bovine serum, Dulbecco's modified Eagle's medium in a 5% CO2 incubator. The cells were starved in Dulbecco's modified Eagle's medium containing 0.2% fatty acid-free bovine serum albumin overnight before treatment with 200 nm insulin and 50 mm glucose. The JNK activator (anisomycin, ST-102), inhibitor (SP600125, EI305), and proteasome inhibitor (MG-132, PI102) were purchased from Biomol International (Plymouth Meeting, PA). The ERK inhibitor (PD98059, 513000) was purchased from Calbiochem. Cycloheximide (C-7698) was from Sigma.
Nuclear Magnetic Resonance
Body composition was measured using quantitative nuclear magnetic resonance (NMR) as described elsewhere (33). The mouse was placed into a small tube individually and then examined with a Brucker model mq10 NMR analyzer (Brucker, Milton, ON, Canada). The fat and lean mass were measured within 1 min. Each measurement was made in triplicate for each mouse to get a mean value.
Immunoblot
Whole cell lysates were prepared by sonication in lysis buffer and used in Western blots as described elsewhere (34). An antibody to SIRT1 (07-131) was purchased from Upstate Biotechnology (Lake Placid, NY). Antibodies to HA (sc-7392), pJNK (sc-6254), and JNK1 (sc-474) were from Santa Cruz. pSIRT1 Ser-47 (#2314) and pAkt Ser-473 (#9271) were from Cell Signaling. Antibodies to β-actin (ab6276) and ubiquitin (ab122) were purchased from Abcam (Cambridge, MA). To detect multiple signals from one membrane, the membrane was stripped with a stripping buffer.
Immunoprecipitation (IP) and Kinase Assay
IP was carried out using whole cell lysates (400 μg of total protein), 2–4 μg of antibody, and 40 μl of protein G-Sepharose beads (Amersham Biosciences) as described elsewhere. The kinase assay was conducted at room temperature for 30 min in 20 μl of kinase assay buffer containing 5 μCi of [32P]ATP (34). The product was resolved by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane for autoradiography or immunoblotting. The recombinant JNK1 (#14-327) was purchased from Millipore (Temecula, CA), and c-Jun fusion protein beads (#9811) were from Cell Signaling (Beverly, MA).
Plasmid and Transfection
Expression vectors for JNK1, ERK1, and Akt were expressed in HEK293 cells through transient transfection. The plasmids for ERK1 and JNK1 were a gift from Dr. Daniel Hwang (Nutrition Department, University of California at Davis). The expression plasmid for mouse SIRT1 (pCruzHA SIRT1, 10962) was purchased from Addgene (Cambridge, MA) (35). The SIRT1 Ala-46 mutant was made by substituting Ser-46 with alanine in the pCruzHA SIRT1 vector using the QuikChange site-directed mutagenesis kit (Stratagene). The Akt expression plasmid was from Dr. Bin-Hua Jiang (BMC Cancer Center, West Virginia University). Transient transfection was conducted using Lipofectamine as reported previously (36) or electroporation with the Amaxa Nucleofector TM II (Lonza Inc. Allendale, NJ).
Hematoxylin and Eosin Staining
Fresh tissues (liver) were collected and fixed in 4% buffered formalin solution (HT50-1-2; Sigma). Tissue slides were obtained through serial cross-section cutting at 8 μm in thickness and stained with a standard procedure.
Glycerol and Triglyceride (TAG) Assays
Triglycerides were extracted from 50 mg of liver tissues as described elsewhere (37). The extract (2 μl) was used to measure glycerol and TAG with a serum TAG determination kit (Sigma).
SIRT1 Activity Assay
SIRT1 deacetylase activity was measured with a SIRT1 fluorimetric activity assay/drug discovery kit (catalog #AK555, Biomol International LP). Nuclear extract protein (5 μg) was incubated for 10 min at 37 °C with 25 μm concentrations of the indicated fluorogenic acetylated peptide substrate. Reactions were stopped by the addition of 1 mm nicotinamide, and the deacetylation-dependent fluorescent signal was determined using a 360-nm excitation laser and measuring 460-nm emission on a fluorescence plate reader.
Statistical Analysis
Data are presented as the means ± S.E. All in vitro experiments were conducted at least three times. Student's t test was used in the statistical analysis with significance set at p ≤ 0.05.
RESULTS
Activation of JNK1 Leads to SIRT1 Degradation
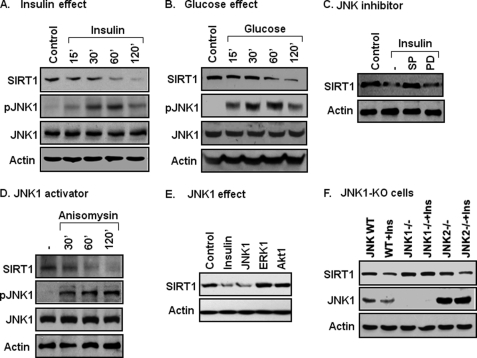
SIRT1 protein is regulated by insulin and glucose in the body. To understand the molecular mechanism, we examined SIRT1 protein in cells after insulin or glucose treatment. Insulin reduced SIRT1 in a time-dependent manner (Fig. 1A). Glucose exhibited a similar activity in the cells (Fig. 1B). We searched for a kinase that is activated in both conditions and then directed our focus on JNK1. JNK1 activity was increased by insulin or glucose as indicated by an increase in its phosphorylation (Fig. 1, A and B). Suppression of JNK1 activity by a chemical inhibitor (SP600125, SP) prevented SIRT1 reduction (Fig. 1C). ERK was also activated by insulin or glucose (data not shown), but ERK inhibition did not influence SIRT1 response to insulin (Fig. 1C). These data suggest that SIRT1 reduction is related to JNK1 activation. To test this possibility, SIRT1 was examined when JNK1 was specifically activated with a chemical activator (anisomycin) or by JNK1 overexpression. In both conditions, SIRT1 protein was reduced (Fig. 1, D and E). In the overexpression study, ERK or Akt1 was used in the control, and SIRT1 protein was not altered by these serine kinases (Fig. 1E).
FIGURE 1.
Association of JNK1 activation and SIRT1 protein reduction. A, insulin effect is shown. SIRT1 protein reduction was induced with insulin (200 nm) in 3T3-L1 cells. SIRT1 protein and pJNK were measured in whole cell lysates in an immunoblot. B, glucose effect is shown. SIRT1 reduction was induced with glucose (50 mm) in 3T3-L1 cells in serum-free DMEM medium (0.25% BSA). C, inhibition of SIRT1 reduction is shown. Cells were pretreated with the JNK inhibitor SP600125 (SP, 50 μm) or MEK inhibitor PD98059 (PD, 30 μm) for 30 min followed by insulin treatment (120 min). D, JNK activator effect is shown. Anisomycin (5 μg/ml) was used to activate JNK, leading to SIRT1 reduction. E, JNK1 overexpression effect is shown. Activities of JNK1, ERK1, and Akt1 were tested for their ability to reduce SIRT1 in a transient transfection of 293 cells with the expression vectors. SIRT1 protein was quantified at 48 h after transfection. F, SIRT1 in JNK-KO cells is shown. SIRT1 protein was measured in an immunoblot after insulin treatment for 2 h. All the experiments were repeated at least three times with consistent results.
JNK has three major isoforms: JNK1, JNK2, and JNK3. In this study, JNK1 and JNK2 were compared in the regulation of SIRT1 protein. JNK3 was not included as it is mainly expressed in the brain. MEF cells of JNK1-KO and JNK2-KO were used to determine SIRT1 response to insulin. In WT cells, SIRT1 was reduced by insulin. The reduction was observed in JNK2-KO cells but not in JNK1-KO cells (Fig. 1F), suggesting that JNK1 is required for SIRT1 reduction. These data suggest that JNK1 activation by insulin or glucose leads to the reduction in SIRT1 protein.
JNK1 Phosphorylates SIRT1 and Induces Protein Degradation
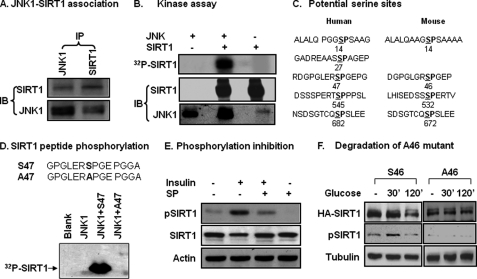
JNK1 is a serine kinase that phosphorylates several downstream proteins in the signaling pathway. It was not known if JNK phosphorylates SIRT1 when this project was started in 2007. We tested the kinase/substrate relationship for JNK and SIRT1. Protein-protein interaction was examined in cells using IP. SIRT1 protein was detected in IP product of JNK1, and JNK1 protein was found in IP product of SIRT1 (Fig. 2A). The protein interaction results provide a basis for the kinase/substrate relationship. In a kinase assay in vitro, recombinant JNK1 was used to phosphorylate SIRT1 protein in the presence of radiolabeled ATP. The phosphorylation was monitored by radioactivity in the SIRT1 protein. JNK1 induced a strong phosphorylation in the SIRT1 protein (Fig. 2B). The phosphorylation was not observed in the controls, in which JNK1 was not included in the reaction.
FIGURE 2.
Phosphorylation of SIRT1 by JNK1. A, JNK1 and SIRT1 association is shown. IP was conducted to detect protein-protein association in the 293 cell lysates (400 μg/point) after anisomycin treatment (5 μg/ml) for 30 min. B, an in vitro kinase assay is shown. The recombinant mouse HA-JNK1 was tested for its ability to phosphorylate the HA-SIRT1 protein in a kinase assay. Assay products were resolved in SDS-PAGE and transferred onto a polyvinylidene difluoride membrane for autoradiography (32P) or immunoblotting (IB). C, shown is a sequence analysis of human and mouse SIRT1. The serine position is indicated by the number. D, a kinase assay with synthesized human SIRT1 peptides is shown. WT and mutant (Ser-Ala) peptides for Ser-47 were used as substrates in the kinase assay. E, SIRT1 phosphorylation was detected by a phospho-specific antibody in cell lysate. Whole cell lysates were made after JNK1 activation by insulin or inhibition by SP600125 (SP) in 3T3-L1 adipocytes. SIRT1 phosphorylation was measured using the phospho-specific antibody to the human SIRT1 Ser-47 (equivalent to mouse Ser-46) in an immunoblot. F, inhibition of SIRT1 reduction by Ser-46 mutation is shown. Reduction of mouse WT (Ser-46) and mutant (Ala-46) SIRT1 proteins were tested in SIRT1−/− MEF cells in a transient transfection. Protein abundance and phosphorylation status were determined after glucose (50 mm) treatment for 2 h. All experiments were repeated at least three times with consistent results.
The JNK1 target motif is characterized by Ser-Pro (SP) residues in the amino acid sequence (38). According to this feature, we analyzed the amino acid sequence of SIRT1 and identified several SP sites (Fig. 2C). There are four SP sites in the mouse and five in the human SIRT1 proteins. The amino acid sequences at some sites are similar to those predicated from mass spectrometry (39). In this study we focused on Ser-46 in the mouse SIRT1 (Ser-47 in human), as the phospho-antibody to this serine residue is available in our laboratory. Mouse SIRT1 was used mostly in this study for JNK1-SIRT1 interaction. Human SIRT1 was tested with a peptide containing the Ser-47 site. In the kinase assay, the human WT peptide (Ser-47) was phosphorylated by JNK1 (Fig. 2D). When the Ser-47 was replaced with alanine (Ala-47), the SIRT1 peptide was no long phosphorylated by JNK1 (Fig. 2D). The data suggest that JNK1 phosphorylates SIRT1 at serine residues in both mouse and human.
The SIRT1 phosphorylation was tested in cells using a phospho-specific antibody, which was raised against phospho-serine 47 of human SIRT1. With this antibody, we observed SIRT1 phosphorylation in mouse 3T3-L1 adipocytes (Fig. 2E). The phosphorylation was detected at Ser-46 in mouse SIRT1 and was inhibited in cells treated with JNK inhibitor (Fig. 2E). When the experiment was repeated in human 293 cells, the phosphorylation was detected in human SIRT1 (data not shown).
The association of phosphorylation and protein reduction suggests that SIRT1 protein may undergo degradation in cells in response to JNK1 activation. To this possibility, we compared WT (Ser-46) and mutant (Ala-46) SIRT1 recombinant protein in response to glucose. The proteins were hemagglutinin (HA)-tagged and expressed in cells through a transient transfection. The wild type (Ser-46) was reduced at 120 min, but the mutant protein (Ala-46) was not reduced in cells treated with glucose (Fig. 2F). These data suggest that SIRT1 serine phosphorylation is required for SIRT1 protein reduction.
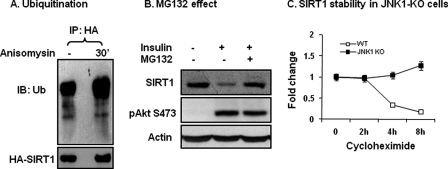
SIRT1 Degradation Is Dependent on Ubiquitination
Phosphorylation-induced protein reduction is often a result of ubiquitination-mediated protein degradation. It is not known if SIRT1 protein is subjected to modification by ubiquitination. To address this issue, we examined SIRT1 ubiquitination using a FLAG-tagged ubiquitin (Ub) in this study. The recombinant proteins for ubiquitin and SIRT1 were expressed together in a transient transfection. SIRT1 was immunoprecipitated with the anti-HA antibody. Ubiquitination was detected with a FLAG antibody in the IP product. The result shows that SIRT1 was ubiquitinated in the basal condition, and the modification was further increased upon JNK1 activation (Fig. 3A). These data suggest that ubiquitination is induced by phosphorylation of SIRT1. An increase in ubiquitination usually induces protein degradation by the proteasome. To test this possibility, we used the proteasome inhibitor MG132. Pretreatment of cells with the proteasome inhibitor prevented SIRT1 protein reduction upon insulin-treatment (Fig. 3B). The inhibition was not a result of blockade of the insulin signaling pathway. The pathway was normal, as insulin-induced Akt phosphorylation was not altered by MG132 (Fig. 3B). The data suggest that SIRT1 ubiquitination is followed by proteasome-mediated protein degradation.
FIGURE 3.
Ubiquitination and proteasome-mediated degradation. A, ubiquitination is shown. HA-SIRT1 was expressed in 293 cells and collected after JNK activation using the anti-HA antibody through IP. Ubiquitination of the SIRT1 protein was determined using the ubiquitin antibody. IB, immunoblot. B, proteasome effect is shown. The proteasome inhibitor MG132 (50 μm) was used to pretreat 3T3-L1 adipocytes for 30 min. SIRT1 degradation was induced by insulin (200 nm, 2 h). Phosphorylation of Akt Ser-473 was used as a control of insulin signaling. C, SIRT1 protein stability is shown. WT and JNK1-KO MEF cells were compared for SIRT1 protein half-life. Protein synthesis was inhibited with cycloheximide. SIRT1 protein abundance was measured in whole cell lysates at multiple time points (h).
To exclude protein synthesis in the SIRT1 reduction, we pretreated cells with cycloheximide, which inhibits protein biosynthesis and activates JNK1 at the same time. In this model SIRT1 exhibited a half-life of 3 h in WT cells (Fig. 3C). In JNK1-KO cells, SIRT1 protein remained unchanged up to 8 h (Fig. 3C), suggesting that SIRT1 degradation, not protein synthesis, contributes to SIRT1 reduction in response to JNK1-mediated phosphorylation. These data provide another line of evidence that JNK1 reduces SIRT1 through protein degradation. JNK1 uses this mechanism to regulate SIRT1 protein abundance.
SIRT1 Reduction Is Associated with Fatty Liver in Obesity
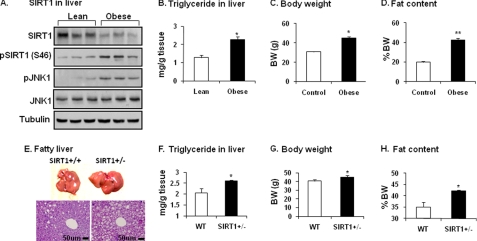
To understand the biological significance of JNK1-SIRT1 interaction, we examined SIRT1 protein in the liver of diet-induced obese (DIO) mice. In this model, blood insulin is increased result in hyperinsulinemia. The liver was examined as it is exposed to the highest level of insulin in the body under obese conditions. Insulin is secreted from β-cells in the pancreas and delivered to the liver by the portal vein. In the obese mice SIRT1 protein was reduced, and JNK1 activity was elevated in the liver (Fig. 4A). SIRT1 reduction was associated with an increase in Ser-46 phosphorylation. In the liver, hepatic steatosis is enhanced as indicated by the elevated triglyceride content (Fig. 4B). Obesity is indicated by the gain in body weight and body fat content in the DIO mice (Fig. 4, C and D). These data suggest that the SIRT1 reduction is associated with JNK1 activation and hepatic steatosis in obese mice.
FIGURE 4.
Decreased SIRT1 in liver of obese mice. A, shown is SIRT1 protein in the liver of DIO mice. SIRT1 protein was measured after tissue homogenization of liver from DIO mice (HFD for 22 weeks). Phosphorylation of SIRT1 and JNK was determined using phospho-specific antibodies. B, TAG in liver is shown. TAG content was determined in liver tissue. C, mouse body weight (BW) at 22 weeks on HFD is shown. D, mouse body fat content at 22 weeks on HFD is shown. Data in panels B–D are presented as the means ± S.E. (n = 10). E, fatty liver in SIRT1+/− mice on HFD is shown. The liver was examined in SIRT1+/− mice at 26 weeks on HFD. Hepatic steatosis is indicated by liver size (picture) and lipid droplets (tissue slide with hematoxylin and eosin staining). F, triglyceride in the livers of mice is shown. G, body weight of the mice is shown. H, body fat content of the mice is shown. Data in panels F–H are presented as the means ± S.E. (n = 8). *, p < 0.05; **, p < 0.001 by Student's t test.
SIRT1 activity is required for protection of liver from steatosis as reported for the pharmacological activator of SIRT1 (10, 12) and SIRT1 inactivation by gene knock-out (7, 8). To understand the JNK1-SIRT1 pathway in the pathogenesis of fatty liver, we compared SIRT1+/− and WT mice in hepatic steatosis. On a HFD, WT mice developed the steatosis as indicated by liver morphology and histology (Fig. 4E). In SIRT1+/− mice, the steatosis was enhanced with a dramatic gain in liver size and lipid droplets in hepatocytes (Fig. 4E). The triglyceride content was increased in liver by 25% over WT (Fig. 4F). The SIRT1-deficient mice had a 10% increase in body weight and a 14% increase in body fat content (Fig. 4, G and H). The genetic model provides in vivo evidence for the JNK1-SIRT1 pathway in the pathogenesis of fatty liver.
JNK1-KO Mice Are Resistant to Liver Steatosis on HFD
JNK1-KO and WT mice were compared for SIRT1 protein in the liver to test the JNK1 activity in vivo. The study was conducted in the mice on HFD. In the JNK1-KO mice, JNK1 was gone, and SIRT1 was significantly higher in liver (Fig. 5A). Liver did not gain as much weight as in the WT mice (Fig. 5B). The hepatocytes did not exhibit as many lipid droplets as observed in the WT mice (Fig. 5B). The liver triglyceride content was 40% less in the JNK1-KO mice (Fig. 5C), although the body weight was only 10% different between the JNK1-KO and WT mice (Fig. 5D). The SIRT1 activity provides a molecular mechanism for protection of JNK1-KO mice from hepatic steatosis. The data provide a line of in vivo evidence for JNK1 regulation of SIRT1.
FIGURE 5.
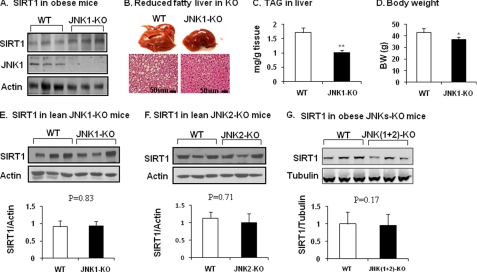
SIRT1 protein in JNK1-KO mice. A, SIRT1 protein in liver tissues is shown. JNK1-KO mice were fed HFD for 22 weeks. SIRT1 protein was determined in the liver tissue homogenate. B, shown is protection of JNK1-KO mice from development of fatty liver. Hepatic steatosis is indicated by liver size (picture) and lipid droplet (slide with hematoxylin and eosin staining). C, triglyceride in liver is shown. D, body weight (BW) is shown. The data in panels C and D are presented as the means ± S.E. (n = 8). E, SIRT1 in lean JNK1-KO mice is shown. SIRT1 protein was determined in the liver of mice on chow diet. F, SIRT1 in lean JNK2-KO mice is shown. SIRT1 protein was determined in the liver of mice on chow diet. G, SIRT1 in obese mice with liver-specific JNK1/2 KO. SIRT1 protein was determined in the liver of double KO mice at 16 weeks on HFD. *, p < 0.05; **, p < 0.001 by Student's t test.
The above data suggest that JNK1 induces SIRT1 protein degradation in obese conditions. Hyperinsulinemia and hyperglycemia may mediate the obesity signals to activate the JNK1-SIRT1 pathway. Does the pathway operate in non-obese conditions? To address this issue, SIRT1 protein was examined in the liver of JNK1 or JNK2 knock-out mice on a regular chow diet. SIRT1 was not altered in either line of the KO mice (Fig. 5, E and F). The data suggest that SIRT1 degradation only occurs when JNK1 is activated in the liver by obesity.
JNK1 and JNK2 differ in substrate affinity and signaling activities. To test JNK2 in the regulation of SIRT1 degradation in obesity, we examined SIRT1 in mice with liver-specific JNK1 and JNK2 double KO. Mice were fed a HFD to induce obesity. SIRT1 protein was determined in the liver at 16 weeks on HFD. Compared with the WT mice, the double KO mice exhibited no difference in SIRT1 protein abundance (Fig. 5G). This is in contrast to the elevated SIRT1 observed in JNK1-KO mice under similar conditions (Fig. 5A). In the JNK1-KO mice, JNK2 activity is enhanced to compensate for the loss of JNK1. In the double KO mice, the JNK2 activity is absent. These data suggest that JNK2 may protect SIRT1 protein from degradation in the obese condition.
Histone Deacetylase Activity of SIRT1
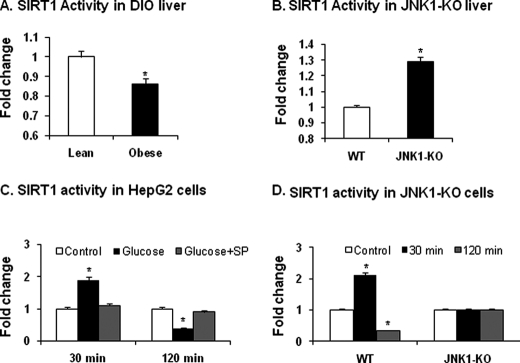
The reduction in SIRT1 protein should lead to a decrease in its enzyme activity. To test this possibility, we measured the catalytic activity of SIRT1 protein using a SIRT1 assay kit. In DIO mice, the enzyme activity was reduced by about 13% in the liver of obese mice (Fig. 6A). When the wild type and global JNK1-KO mice were compared for the enzyme activity, 30% more activity was observed in the KO mice (Fig. 6B). The activity is associated with a higher protein abundance of SIRT1 in the KO mice.
FIGURE 6.
Catalytic activity of SIRT1. A, catalytic activity of SIRT1 in the liver of DIO mice is shown. The test was conducted using the nuclear protein extract from liver tissues of DIO mice fed HFD for 22 weeks. B, catalytic activity of SIRT1 in JNK1-KO mice is shown. The nuclear extract of liver tissues was made from mice at 22 weeks on HFD. C, catalytic activity of SIRT1 is shown. Histone deacetylase activity was examined in the nuclear extract of HepG2 hematoma cells at 30 and 120 min after glucose treatment. JNK inhibitor SP600125 (SP) was used to pretreat the cells for 30 min before glucose treatment. D, catalytic activity of SIRT1 in JNK1-KO MEFs is shown. The assay in panel C was repeated in JNK1-KO MEFs. The data in this figure are presented as the mean ± S.E. (n = 10). *, p < 0.05 by Student's t test.
To examine the enzyme activity of phosphorylated SIRT1 before protein degradation, we determined catalytic activity of SIRT1 in HepG2 cells at two different time points after glucose treatment. At 30 min, the SIRT1 activity was increased by glucose (Fig. 6C). The increase was blocked by the JNK inhibitor SP. At 120 min, the activity was decreased by glucose (Fig. 6C). The decrease was prevented by the JNK inhibitor SP. A similar response was observed in the wild type MEFs (Fig. 6D). However, the time-dependent response disappeared in the JNK1−/− MEFs (Fig. 6D). The results suggest that the catalytic activity of SIRT1 is regulated by both phosphorylation and protein abundance. The phosphorylation leads to an increase first and then a decrease later in the enzyme activity of SIRT1.
DISCUSSION
JNK1-mediated phosphorylation of SIRT1 is a mechanism for the regulation of SIRT1 activity by insulin and glucose. The enzyme activity of SIRT1 is induced by NAD+ (1), natural products (resveratrol) (40, 41), and synthetic small molecules (11). There was not much information regarding SIRT1 regulation by protein phosphorylation. It was not known how SIRT1 protein is reduced by insulin or glucose. In this study we investigated the signaling pathway by which insulin and glucose reduce SIRT1 protein and found that JNK1 phosphorylates SIRT1 at Ser-46 (mouse)/Ser-47 (human). The phosphorylation induces ubiquitination and proteasome-dependent degradation in SIRT1. The JNK1-SIRT1 pathway was investigated using five different approaches in this study; that is, kinase assay, point mutation, JNK1-KO, JNK2-KO, and JNK1/2 double KO. The results consistently support that the pathway operates in vitro and in vivo to control SIRT1 degradation. In this pathway, phosphorylation, activation, ubiquitination, and proteasome degradation form a closely connected sequential in the regulation of SIRT1 activity. The physiological significance of this JNK1-SIRT1 pathway was investigated in the obese mice. Our results support that this pathway plays a critical role in pathogenesis of obesity-associated liver steatosis.
SIRT1 phosphorylation enhances the catalytic activity of SIRT1. Although SIRT1 degradation is a focus in this study, we also examined catalytic activity of SIRT1 after phosphorylation. It was reported that the deacetylase activity of SIRT1 is enhanced by serine/threonine phosphorylation. In the first study, human SIRT1 was examined in a mass spectrometry analysis and found to be phosphorylated at multiple serine/threonine residues in cell cycle progression (39). Cyclin B/Cdk1 was identified as a kinase that phosphorylates threonine 530 and serine 540 in the SIRT1 protein, which enhanced the deacetylase activity of SIRT1. In the second study, JNK1 was found to phosphorylate human SIRT1 at Ser-27, Ser-47, and Thr-530 in response to H2O2 (42). However, the SIRT1 modification was not investigated in the regulation of metabolism in the two studies. Additionally, the conclusions are contradictive in the SIRT1 kinases for Thr-530. In the present study we demonstrated that JNK1 phosphorylates the mouse SIRT1 protein in vitro and in vivo at Ser-46 (Ser-47 in human SIRT1). Phosphorylation induces the enzyme activity and then the protein degradation for SIRT1. In another study, SIRT1 is modified by sumoylation (43).
We compared JNK1 and JNK2 in the regulation of SIRT1 protein stability using isoform-specific JNK-KO MEF cells and knock-out mice as JNK1-mediated phosphorylation induces the protein degradation. Persistent JNK1 activation leads to inhibition of SIRT1 function. This kind of SIRT1 suppression is important for the pathogenesis of liver steatosis in obesity. JNK2 does not have a significant role in the regulation of enzyme activity of SIRT1 but has an opposite activity in the control of SIRT1 degradation. When JNK2 is the only isoform in cells, such as in the JNK1-KO MEFs, the basal SIRT1 protein level is higher relative to that of WT MEFs. In JNK1-KO mice, SIRT1 exhibited a higher protein level in the liver when mice were fed HFD. In the liver of JNK1/2 double-KO mice, the SIRT1 elevation was attenuated. The data suggest that JNK2 may protect SIRT1 from degradation. This conclusion is in agreement with a report by Ford et al. (44), in which JNK2 was found to increase SIRT1 protein in cell culture but not in vivo. In that study JNK2 knockdown by RNAi led to a reduction in SIRT1 protein. In the current study, the JNKs-SIRT1 interaction was examined in vitro and in vivo.
This study provides a new mechanism for the pathogenesis of hepatic steatosis in dietary obese conditions. Hepatic steatosis in obesity, known as nonalcoholic fatty liver disease, is involved in the pathogenesis of insulin resistance. Genetic and pharmacological studies suggest that JNK1 promotes formation of hepatic steatosis in obesity (21–23). In those studies JNK activity is inhibited globally with genetic or pharmacological approaches. When JNK1 was tested in hepatocyte-specific KO mice, we found that liver steatosis was enhanced by the conditional JNK1 inactivation (31). This observation raises a concern about JNK1 activity in the pathogenesis of hepatic steatosis. The current study suggests that the discrepancy is due to lack of a complete understanding of JNK1 activity. We show here that JNK1 has dual roles in the regulation of SIRT1 activity. It induces and then decreases the SIRT1 activity through the phosphorylation-mediated processes. The observations suggest that JNK1 has dual roles in the regulation of SIRT1 activities. In liver, when JNK1 is inactivated, SIRT1 will not act properly in the absence of the phosphorylation. When JNK1 is persistently activated, SIRT1 will be inhibited by extensively protein degradation. In both conditions, the effect is equivalent to SIRT1 inactivation in liver. These possibilities explain the complex JNK1 activities in the pathogenesis of hepatic steatosis.
The JNK1-SIRT1 pathway provides a new mechanism for insulin resistance in obesity. JNK1 is known to induce insulin resistance by direct phosphorylation of IRS-1 protein (26, 28, 30). In this study our data suggest that JNK1 also directly phosphorylates SIRT1. Persistent phosphorylation is observed in liver in obese mice and is involved in inhibition of SIRT1 function. Lack of SIRT1 function contributes to the development of fatty liver, a risk factor for insulin resistance. JNK1 is activated by many factors in obesity. Those include hyperinsulinemia, hyperglycemia, proinflammatory cytokines, endoplasmic reticulum stress, oxidative stress, diglyceride, and ceramide (26–29). All of these factors contribute to the development of insulin resistance. Insulin and glucose were used in our study of the JNK1-SIRT1 relationship. This pathway may apply to other JNK1 inducers in obesity. The multiple factors are responsible for the persistent activation of JNK1 in obesity. The JNK1-mediated SIRT1 degradation contributes to insulin resistance through induction of fatty liver in obesity. In the brain, JNK1 regulates body weight through the hypothalamic-pituitary-thyroid axis (45). This axis may contribute to the attenuation in hepatic steatosis in the global JNK1 knock-out mice, which is observed in this and other studies (21–23).
In summary, we demonstrate that JNK1 phosphorylates SIRT1, which induces a brief SIRT1 activation that is followed by an inhibition from SIRT1 protein degradation. The SIRT1 degradation is mediated by ubiquitination and dependent on proteasome. Persistent JNK1 activation by multiple factors in obesity induces an extensive SIRT1 degradation that leads to SIRT1 inhibition in obesity. This operation of the JNK1-SIRT1 pathway contributes to the pathogenesis of fatty liver and serves as a new mechanism for insulin resistance in obesity.
This work was supported, in whole or in part, by National Institutes of Health Grants DK068036 and DK085495 (to J. Y.).
- HFD
- high fat diet
- MEF
- mouse embryonic fibroblast
- IP
- immunoprecipitation
- TAG
- triglyceride
- Ub
- ubiquitin
- DIO
- diet-induced obese.
REFERENCES
- 1. Blander G., Guarente L. (2004) Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 2. Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 3. Lin S. J., Defossez P. A., Guarente L. (2000) Science 289, 2126–2128 [DOI] [PubMed] [Google Scholar]
- 4. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 5. Liu Y., Dentin R., Chen D., Hedrick S., Ravnskjaer K., Schenk S., Milne J., Meyers D. J., Cole P., Yates J., 3rd, Olefsky J., Guarente L., Montminy M. (2008) Nature 456, 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erion D. M., Yonemitsu S., Nie Y., Nagai Y., Gillum M. P., Hsiao J. J., Iwasaki T., Stark R., Weismann D., Yu X. X., Murray S. F., Bhanot S., Monia B. P., Horvath T. L., Gao Q., Samuel V. T., Shulman G. I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11288–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu F., Gao Z., Zhang J., Rivera C. A., Yin J., Weng J., Ye J. (2010) Endocrinology 151, 2504–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Purushotham A., Schug T. T., Xu Q., Surapureddi S., Guo X., Li X. (2009) Cell Metab. 9, 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. (2006) Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 10. Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., Prabhu V. V., Allard J. S., Lopez-Lluch G., Lewis K., Pistell P. J., Poosala S., Becker K. G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K. W., Spencer R. G., Lakatta E. G., Le Couteur D., Shaw R. J., Navas P., Puigserver P., Ingram D. K., de Cabo R., Sinclair D. A. (2006) Nature 444, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., Jin L., Boss O., Perni R. B., Vu C. B., Bemis J. E., Xie R., Disch J. S., Ng P. Y., Nunes J. J., Lynch A. V., Yang H., Galonek H., Israelian K., Choy W., Iffland A., Lavu S., Medvedik O., Sinclair D. A., Olefsky J. M., Jirousek M. R., Elliott P. J., Westphal C. H. (2007) Nature 450, 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. You M., Cao Q., Liang X., Ajmo J. M., Ness G. C. (2008) J. Nutr. 138, 497–501 [DOI] [PubMed] [Google Scholar]
- 13. Rodgers J. T., Puigserver P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De., Oliveira R., Leid M., McBurney M. W., Guarente L. (2004) Nature 429, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. You M., Liang X., Ajmo J. M., Ness G. C. (2008) Am. J. Physiol. Gastrointest Liver Physiol. 294, G892–G898 [DOI] [PubMed] [Google Scholar]
- 16. Yang J., Kong X., Martins-Santos M. E., Aleman G., Chaco E., Liu G. E., Wu S. Y., Samols D., Hakimi P., Chiang C. M., Hanson R. W. (2009) J. Biol. Chem. 284, 27042–27053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 18. Chen J., Zhou Y., Mueller-Steiner S., Chen L. F., Kwon H., Yi S., Mucke L., Gan L. (2005) J. Biol. Chem. 280, 40364–40374 [DOI] [PubMed] [Google Scholar]
- 19. Gao Z., Ye J. (2008) Biochem. Biophys. Res. Commun. 376, 793–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., Howitz K. T., Gorospe M., de Cabo R., Sinclair D. A. (2004) Science 305, 390–392 [DOI] [PubMed] [Google Scholar]
- 21. Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R. O., Görgün C. Z., Hotamisligil G. S. (2006) Science 313, 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schattenberg J. M., Singh R., Wang Y., Lefkowitch J. H., Rigoli R. M., Scherer P. E., Czaja M. J. (2006) Hepatology 43, 163–172 [DOI] [PubMed] [Google Scholar]
- 23. Singh R., Wang Y., Xiang Y., Tanaka K. E., Gaarde W. A., Czaja M. J. (2009) Hepatology 49, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakatani Y., Kaneto H., Kawamori D., Hatazaki M., Miyatsuka T., Matsuoka T. A., Kajimoto Y., Matsuhisa M., Yamasaki Y., Hori M. (2004) J. Biol. Chem. 279, 45803–45809 [DOI] [PubMed] [Google Scholar]
- 25. Yang R., Wilcox D. M., Haasch D. L., Jung P. M., Nguyen P. T., Voorbach M. J., Doktor S., Brodjian S., Bush E. N., Lin E., Jacobson P. B., Collins C. A., Landschulz K. T., Trevillyan J. M., Rondinone C. M., Surowy T. K. (2007) J. Biol. Chem. 282, 22765–22774 [DOI] [PubMed] [Google Scholar]
- 26. Ozcan U., Cao Q., Yilmaz E., Lee A. H., Iwakoshi N. N., Ozdelen E., Tuncman G., Görgün C., Glimcher L. H., Hotamisligil G. S. (2004) Science 306, 457–461 [DOI] [PubMed] [Google Scholar]
- 27. Samuel V. T., Liu Z. X., Qu X., Elder B. D., Bilz S., Befroy D., Romanelli A. J., Shulman G. I. (2004) J. Biol. Chem. 279, 32345–32353 [DOI] [PubMed] [Google Scholar]
- 28. Gao Z., Zhang X., Zuberi A., Hwang D., Quon M. J., Lefevre M., Ye J. (2004) Mol. Endocrinol. 18, 2024–2034 [DOI] [PubMed] [Google Scholar]
- 29. Chavez J. A., Knotts T. A., Wang L. P., Li G., Dobrowsky R. T., Florant G. L., Summers S. A. (2003) J. Biol. Chem. 278, 10297–10303 [DOI] [PubMed] [Google Scholar]
- 30. Aguirre V., Uchida T., Yenush L., Davis R., White M. F. (2000) J. Biol. Chem. 275, 9047–9054 [DOI] [PubMed] [Google Scholar]
- 31. Sabio G., Cavanagh-Kyros J., Ko H. J., Jung D. Y., Gray S., Jun J. Y., Barrett T., Mora A., Kim J. K., Davis R. J. (2009) Cell Metab. 10, 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao Z., Zuberi A., Quon M. J., Dong Z., Ye J. (2003) J. Biol. Chem. 278, 24944–24950 [DOI] [PubMed] [Google Scholar]
- 33. Gao Z., Wang Z., Zhang X., Butler A. A., Zuberi A., Gawronska-Kozak B., Lefevre M., York D., Ravussin E., Berthoud H. R., McGuinness O., Cefalu W. T., Ye J. (2007) Am. J. Physiol. Endocrinol Metab. 292, E84–E91 [DOI] [PubMed] [Google Scholar]
- 34. Gao Z., Hwang D., Bataille F., Lefevre M., York D., Quon M. J., Ye J. (2002) J. Biol. Chem. 277, 48115–48121 [DOI] [PubMed] [Google Scholar]
- 35. Nemoto S., Fergusson M. M., Finkel T. (2005) J. Biol. Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- 36. Gao Z., He Q., Peng B., Chiao P. J., Ye J. (2006) J. Biol. Chem. 281, 4540–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin J., Yang R., Tarr P. T., Wu P. H., Handschin C., Li S., Yang W., Pei L., Uldry M., Tontonoz P., Newgard C. B., Spiegelman B. M. (2005) Cell 120, 261–273 [DOI] [PubMed] [Google Scholar]
- 38. Dérijard B., Hibi M., Wu I. H., Barrett T., Su B., Deng T., Karin M., Davis R. J. (1994) Cell 76, 1025–1037 [DOI] [PubMed] [Google Scholar]
- 39. Sasaki T., Maier B., Koclega K. D., Chruszcz M., Gluba W., Stukenberg P. T., Minor W., Scrable H. (2008) PLoS One 3, e4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Borra M. T., Smith B. C., Denu J. M. (2005) J. Biol. Chem. 280, 17187–17195 [DOI] [PubMed] [Google Scholar]
- 41. Kaeberlein M., McDonagh T., Heltweg B., Hixon J., Westman E. A., Caldwell S. D., Napper A., Curtis R., DiStefano P. S., Fields S., Bedalov A., Kennedy B. K. (2005) J. Biol. Chem. 280, 17038–17045 [DOI] [PubMed] [Google Scholar]
- 42. Nasrin N., Kaushik V. K., Fortier E., Wall D., Pearson K. J., de Cabo R., Bordone L. (2009) PLoS One 4, e8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang Y., Fu W., Chen J., Olashaw N., Zhang X., Nicosia S. V., Bhalla K., Bai W. (2007) Nat Cell Biol. 9, 1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ford J., Ahmed S., Allison S., Jiang M., Milner J. (2008) Cell Cycle 7, 3091–3097 [DOI] [PubMed] [Google Scholar]
- 45. Sabio G., Cavanagh-Kyros J., Barrett T., Jung D. Y., Ko H. J., Ong H., Morel C., Mora A., Reilly J., Kim J. K., Davis R. J. (2010) Genes Dev. 24, 256–264 [DOI] [PMC free article] [PubMed] [Google Scholar]