Abstract
Human factor H (HufH), a key inhibitor of the alternative pathway of complement, binds to Neisseria gonorrhoeae and constitutes an important mechanism of human-specific complement evasion. The C-terminal domain 20 of HufH contains the binding site for sialylated gonococci. We exploited differences in amino acid sequences between human and non-binding chimpanzee fH domain 20 to create cross-species mutations to define amino acids important for binding to sialylated gonococci. We used fH/Fc fusion constructs that contained contiguous fH domains 18–20 fused to Fc fragments of murine IgG2a. The Fc region was used both as a tag for detection of each fusion molecule on the bacterial surface and as an indicator for complement-dependent killing. Arg-1203 was critical for binding to both porin (Por) B.1A and PorB.1B strains. Modeling of the R1203N human-to-chimpanzee mutation using the crystal structure of HufH19–20 as a template showed a loss of positive charge that protrudes at the C terminus of domain 20. We tested the functional importance of Arg-1203 by incubating sialylated gonococci with normal human serum, in the presence of wild-type HufH18–20/Fc or its R1203A mutant. Gonococci bound and were killed by wild-type HufH18–20/Fc but not by the R1203A mutant. A recombinant fH/Fc molecule that contained chimpanzee domain 20, humanized only at amino acid 1203 (N1203R) also bound to sialylated gonococci and restored killing. These findings provide further insights into the species specificity of gonococcal infections and proof-of-concept of a novel therapeutic approach against gonorrhea, a disease rapidly becoming resistant to conventional antibiotics.
Keywords: Amino Acid, Bacteria, Complement, Crystal Structure, Site-directed Mutagenesis, Neiseria gonorrhoeae, Complement, Factor H, Sialylation, Species Specificity
Introduction
Neisseria gonorrhoeae, the causative agent of gonorrhea, is the second most commonly reported bacterial sexually transmitted infection in the United States. Approximately 301,000 new cases were reported in the U.S.A. in 2009 (1). More than 60 million cases are estimated to occur annually worldwide (2). The complement system is a crucial innate immune defense mechanism against invading pathogens; N. gonorrhoeae interacts specifically with certain human complement components that are present in the genital tract (3), the most common portal of entry of this organism.
Gonococci have developed numerous mechanisms to evade complement-mediated killing. Most gonococci that cause bacteremic or disseminated infection are resistant to killing by nonimmune normal human serum (NHS),2 a property termed “stable” serum resistance (4). Stable serum resistance may in part be mediated by the ability of gonococcal porin B (PorB) to bind plasma complement inhibitory proteins such as factor H (fH) (5) and/or C4b-binding protein (C4BP) (6). PorB molecules are 34–37-kDa proteins, which exist as trimers in their native configurations and function as selective ion channels (7). N. gonorrhoeae porB exists as two alleles, which encode either PorB.1A or PorB.1B (formerly called Por1A and Por1B, respectively) (8). PorB.1A gonococci frequently cause disseminated disease, whereas PorB.1B strains usually cause local urogenital disease and pelvic inflammatory disease in women (8, 9). Most gonococcal PorB.1A strains can bind fH and/or C4BP (hence they exhibit stable serum resistance), whereas PorB.1B strains bind fH weakly (5) unless their lipooligosaccharide (LOS) is sialylated (5, 10). Some PorB.1B strains can bind C4BP independently of LOS sialylation (6).
Factor H (fH) is an important fluid-phase alternative complement pathway inhibitor. It inhibits the assembly of an active C3 convertase by competing with factor B for C3b binding, accelerates the decay of the alternative pathway C3 convertase (C3bBb), and acts as a cofactor in factor I-mediated cleavage of C3b to iC3b (11–15). fH is composed of 20 short consensus repeat (also called complement control protein (CCP)) domains (16), each comprising ∼60 amino acids (17). The structural integrity of each domain is maintained by two disulfide bonds (18).
Gonococcal LOS that contains lacto-N-neotetraose (19) becomes sialylated when the bacteria are grown in media supplemented with 5′-cytidinemonophospho-N-acetylneuraminic acid (CMP-NANA) (19, 20). Sialylation of LOS also occurs in vivo (21) and renders those strains that are otherwise susceptible to complement-mediated killing, resistant to killing by NHS (22). This is termed “unstable” serum resistance because it is a property that is lost when organisms are grown in media that lack CMP-NANA (23). Sialylation of gonococci enhances their ability to bind human fH (HufH) (10). The primary binding site for sialylated gonococci (both PorB.1A- and PorB.1B-expressing strains) resides in a region spanned by the three C-terminal fH domains, 18–20 (24).
Natural infection with N. gonorrhoeae is restricted to humans. HufH, but not fH from other primates such as chimpanzee (Pan troglotydes) and rhesus macaque (Macaca mulatta), bind directly and strongly to gonococci (24). In this study, we used fH/Fc fusion constructs that contained contiguous fH domains 18–20 from human (Hu) or chimpanzee (Ch), fused in frame at their C-terminal ends to the N terminus of the Fc fragment of murine IgG2a, to examine the effects of amino acid mutations in fH domain 20 on the binding of fH to sialylated gonococci. The Fc region served as a tag for detection of each fusion molecule and also enabled complement-dependent bacterial killing. We exploited the differences in the amino acid sequence between human and chimpanzee fH located in the C-terminal fH domains to define the amino acids that are important for binding of HufH to sialylated N. gonorrhoeae.
EXPERIMENTAL PROCEDURES
Bacterial Strains
N. gonorrhoeae serotype PorB.1B strain F62 (25) and serotype PorB.1A strain 252 (5) have been described previously. Strain 252 was chosen in this case as an example of a PorB.1A strain that binds to HufH but fails to bind the classical complement pathway inhibitor, C4BP (5), thereby eliminating confounding by C4BP in bactericidal assays. Bacteria that had been grown on chocolate agar supplemented with Isovitalex® equivalent (26) in 5% CO2 for 10–12 h at 37 °C were suspended in gonococcal liquid media (26) and grown to the mid-log phase. Sialylation of gonococcal LOS was achieved by adding CMP-NANA to a final concentration of 2 μg/ml to the growth media. Bacteria were washed and resuspended in Hanks' balanced salt solution (HBSS) containing 0.15 mm CaCl2 and 1 mm MgCl2 (HBSS2+) for use in binding assays. Escherichia coli (Invitrogen) used for site-directed mutagenesis were cultured in Luria Bertani (LB) broth or on LB agar.
Serum and Complement Reagents
Serum was obtained from seven normal healthy adult volunteers with no history of gonococcal infection who provided written informed consent. Participation was approved by the University of Massachusetts Medical School's Human Subjects/IRB, Office of Research (IRB protocol 11732). Sera were pooled and stored at −70 °C. For some experiments, complement was inactivated by heating serum at 56 °C for 30 min. HufH was purchased from Complement Technologies (Tyler, TX). Depletion of IgG and IgM from serum, testing for the adequacy of depletion and hemolytic activity of the depleted serum were all performed exactly as described previously (27). Heparin, sodium salt (ICN Biomedicals), was isolated from porcine intestinal mucosa and has most chains in the range of 17,000–19,000 Da. Anti-thrombin III activity was ∼100 United States Pharmacopoeias units/mg. The amount of heparin needed to prevent coagulation was between 20 and 50 unit/ml of whole blood.
Sequence Comparison
Amino acid sequences of domains 18, 19, and 20 of human and chimpanzee factor H were obtained from the GenBankTM database (accession nos. Y00716 and XP001136531, respectively) and were aligned using the BIOEDIT alignment program.
Recombinant fH Fragment/Fc Fusion Proteins (fH/Fc)
FH/Fc fusion constructs were used to examine the effects of amino acid mutations in fH domain 20 on the binding of fH to sialylated gonococci. These constructs contained contiguous fH domains 18–20 from human (Hu) or chimpanzee (Ch), fused in frame at their C-terminal ends to the N terminus of the Fc fragment of murine IgG2a (fH/Fc fusion proteins). The predicted amino acid sequence of the wild-type Hu18–20/Fc fusion protein is shown in supplemental Fig. S1. Construction of these chimeric molecules and estimation of the concentration of the fH/Fc fusion proteins in concentrated cell culture supernatants have been described previously (24). Briefly, Chinese hamster ovary cells were transfected with each of the fH/Fc constructs using Lipofectin (Invitrogen), according to the manufacturer's instructions. Media from transfected cells were collected over a 2-day period, culture supernatants were concentrated using Amicon Ultra (Millipore), and fH/Fc protein concentrations were determined by ELISA. Recombinant proteins that were used in bactericidal assays were purified over HiTrap Protein A HP (GE Healthcare); their concentrations were determined by absorbance at 280 nm.
Site-directed Mutagenesis of fH Domain 20
To map the amino acids in HufH domain 20 and to assess systematically their importance for binding to sialylated N. gonorrhoeae, we created ten HufH18–20/Fc mutants by replacing one or two amino acid residues in domain 20 with the corresponding chimpanzee residue(s). A list of primers used to create the recombinant fH/Fc proteins and those used for site-directed mutagenesis are listed in supplemental Table S1.
Single amino acid mutations were also generated in the reverse direction (chimpanzee-to-human) in the recombinant fH/Fc molecule, HufH18–19/ChfH20/Fc. N1203R or A1217T substitutions were made in chimpanzee domain 20. Point mutations were created using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Construct sequences were validated by DNA sequencing.
Flow Cytometry
Bacteria grown in gonococcal liquid media were washed and resuspended in HBSS2+; 108 organisms were used in each FACS assay. To detect the binding of recombinant fH/Fc fusion proteins, 108 bacteria, suspended in HBSS2+ containing 1% BSA (pH 7.4), were incubated with concentrated supernatants that contained 0.5 μg (∼48 nm) of recombinant fH/Fc protein (as determined by ELISA) in a final reaction volume of 100 μl for 30 min at 37 °C. After washing, FITC-labeled goat anti-mouse IgG (Sigma) diluted 1:100 in 1% BSA-HBSS2+ was used to detect bacteria-bound fH/Fc fusion proteins.
To examine whether select fH/Fc fusion proteins inhibited binding of full-length fH to bacteria, 108 organisms, suspended in 1% BSA-HBSS2+ (pH 7.4) were incubated with increasing concentrations of purified fH/Fc proteins together with 0.5 μg (32 nm) of purified HufH in a final reaction volume of 100 μl for 30 min at 37 °C. Full-length fH that bound to organisms (not inhibited from binding by fH/Fc proteins) was detected with mAb 90X (28) used at a concentration of 10 μg/ml in 1% BSA-HBSS2+, followed by FITC-labeled anti-mouse IgG (Fab-specific) diluted 1:100 in 1% BSA- HBSS2+. Thus, only bound mAb 90X, a complete antibody containing Fab, was detected with this conjugate; fH/Fc fusion proteins, lacking Fab were not recognized in this experiment. Flow cytometry was performed using a LSRII flow cytometer (Becton Dickinson); data analysis was performed using the FlowJo data analysis software package (Tree Star, Inc.).
Inhibition of Binding of HufH18–20/Fc to Sialylated Gonococci by Heparin
To examine whether heparin inhibited binding of HufH18–20/Fc to bacteria (heparin-binding sites on fH are located in domain 20 (29, 30)), 108 organisms, suspended in 1% BSA-HBSS2+ (pH 7.4) were incubated with heparin (sodium salt) at either of two concentrations (∼56 or ∼222 μm) plus 0.5 μg (48 nm) of purified HufH18–20/Fc in a final volume of 100 μl, for 30 min at 37 °C. HufH18–20/Fc that bound to bacteria was detected with FITC-labeled anti-mouse IgG.
Modeling of HufH19–20 R1203N
The tertiary structure of the fH19–20 protein with the R1203N mutation was generated by homology-based molecular modeling using the previously solved crystal structure of wild type fH19–20 as a template (31). The generated structure was energy minimized using the Steepest descent algorithm in the Discover module of the Insight II software package (Accelrys Inc., San Diego, CA).
Electrostatic potentials of the fH19–20 wild-type and the R1203N mutant structures were calculated after addition of hydrogens using the Adaptive Poisson-Boltzmann Solver (32) that was run using the PyMOL graphic interface (version 1.3, Schrödinger, LLC). The program PDB 2PQR (33) was used to generate the PQR file required by the Adaptive Poisson-Boltzmann Solver.
Serum Bactericidal Assay
Serum bactericidal assays were used to test whether fH/Fc fusion proteins that inhibited binding of full-length HufH to bacteria resulted in killing of gonococci by NHS. In certain experiments, IgG- and IgM-depleted NHS was used as a source of active complement. Serum bactericidal assays were performed as described previously (26) that also included the use heat inactivated serum as negative controls. Briefly, ∼2000 cfu of gonococci (sialylated) were incubated with purified fH/Fc fusion proteins at concentrations ranging from 0.5 to 33 μg/ml followed by the addition of 10% NHS (v/v). The final volume of reaction mixtures was 150 μl. Aliquots of 25 μl were immediately removed from reaction mixtures and plated in duplicate (t0) and again after 30 min. of incubation at 37 °C (t30). Following overnight incubation of the plates at 37 °C in 5% CO2, colonies were counted, and the percent killing calculated as the ratio of the number of colonies at t0 versus the number at t30.
RESULTS
HufH Domain 20 Contains Binding Site for Sialylated N. gonorrhoeae
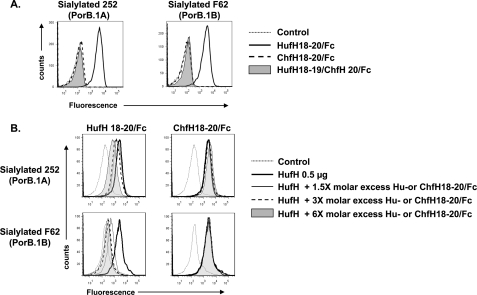
Domains 18–20 of HufH contain the binding site for sialylated gonococci (24). Our goal was to determine the amino acid(s) in these domains that contribute to human-specific binding of fH to sialylated gonococci. We used chimpanzee fH (ChfH), which does not bind to sialylated gonococci (24), as a basis to compare the amino acid sequences in fH domains 18–20 and then substituted individual amino acids. As a prelude to the subsequent experiments in this study, we first ascertained that only HufH18–20/Fc, but not ChfH18–20/Fc, bound to each of the two sialylated strains of gonococci (Fig. 1A). We also confirmed that HufH18–20/Fc, but not ChfH18–20/Fc, inhibited binding of full-length fH to sialylated gonococci in a dose-dependent manner (Fig. 1B). The inhibition by HufH18–20/Fc was more efficient with the PorB.1B strain F62 than with the PorB1.A strain 252 (Fig. 1B, left panels); ∼4-fold more HufH/18–20 was required to achieve similar levels of inhibition of binding to sialylated strain 252 compared with sialylated strain F62.
FIGURE 1.
A, binding of human and chimpanzee fH domains 18–20/Fc fusion proteins to sialylated N. gonorrhoeae. Binding of HufH18–20/Fc is indicated by the solid line and (lack of) binding of ChfH18–20/Fc by the dashed line. Lack of binding of protein HufH18–19/ChfH20/Fc (HufH20 replaced by ChfH20) is shown by the shaded gray histogram. In all graphs, the x axis represents fluorescence on a log10 scale, and the y axis represents the number of events. The control graph (no fH/Fc protein added) is depicted by the faint dotted line. One representative experiment of three independently performed experiments is shown. B, inhibition of binding of full-length HufH to sialylated N. gonorrhoeae by HufH18–20/Fc and ChfH18–20/Fc. Sialylated N. gonorrhoeae strains 252 (PorB.1A) and F62 (PorB.1B) were incubated with 0.5 μg of full-length HufH and either ≈ 1.5, 3, or 6× molar excess (compared with full-length HufH) of purified HufH18–20/Fc (left panel) or ChfH18–20/Fc (right panel). Full-length HufH that bound to bacteria was detected with mAb 90× (28), followed by anti-mouse IgG (Fab-specific) FITC. In all graphs, the x axis represents fluorescence on a log10 scale, and the y axis represents the number of events. No HufH or fusion protein was contained in the reaction mixtures represented by the “Control” histograms. One representative experiment is shown of two independently performed experiments.
Domain 18 of HufH and ChfH are identical and domain 19 differs by only two amino acids; in domain 20, there are a total of 11 amino acid differences between human and chimpanzee (Fig. 2A). Therefore, we speculated that the C-terminal domain 20 was likely to harbor the gonococcal binding site. This was confirmed using a fusion protein that contained HufH domains 18–19 and ChfH domain 20 fused to murine Fc (HufH18–19/ChfH20/Fc), which did not bind to sialylated PorB.1A or PorB.1B strains of N. gonorrhoeae (Fig. 1A). Next, we sought to define the amino acids in domain 20 that were important for HufH interactions with sialylated gonococci.
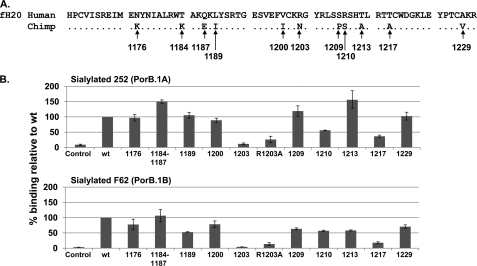
FIGURE 2.
Binding of HufH18–20/Fc to sialylated N. gonorrhoeae: effect of human-to-chimpanzee mutations in HufH domain 20. A, amino acid sequence comparison of human and chimpanzee fH domains 20 (fH20). Identical amino acid residues are denoted by dots. B, binding to sialylated gonococcal strains 252 and F62 by HufH18–20/Fc constructs that separately contained the indicated chimpanzee fH domain 20 point mutations relative to HufH18–20/Fc wild-type. The bar graph labeled Control contains no fusion proteins. In all bar graphs, the x axis represents each mutant, and the y axis represents the percentage of binding relative to WT using median fluorescence. Values represent the mean calculated from two or more independently performed experiments ± S.E.
Arginine 1203 in Domain 20 of HufH Is Critical for Binding to Sialylated N. gonorrhoeae
The 11 amino acids in HufH domain 20 that are unique to humans were changed one at a time (in one instance, two were simultaneously changed (T1184K/Q1187E)) to the corresponding residues in ChfH domain 20 using site-directed mutagenesis (Fig. 2A). Ten HufH 18–20/Fc mutant proteins were constructed initially. Western blot analysis of the recombinant HufH18–20/Fc proteins in concentrated supernatants is shown in supplemental Fig. S2. We compared HufH18–20/Fc mutant protein binding (using median fluorescence) to sialylated gonococci against wild type HufH18–20/Fc binding. Eight of the mutant proteins (N1176K, T1184K/Q1187E, L1189I, V1200I, S1209P, R1210S, T1213A, or A1229V) bound to both sialylated PorB.1A and PorB.1B strains ≥50% relative to the wild-type (Fig. 2B). The R1203N mutation resulted in a significant decrease in binding to both strains (Fig. 2B). Based on this observation, an additional mutant that substituted alanine instead of asparagine (R1203A), was used to verify that the observed effect of the R1203N mutation was not based on a conformational change in the region of the mutation (34) but rather that the residue Arg-1203 itself was important for HufH binding to sialylated gonococci (Fig. 2B). Similar to the R1203N mutant, binding of the R1203A mutant to sialylated gonococci was also decreased significantly (Fig. 2B), suggesting that the Arg-1203 residue played a key role in the interaction of HufH with sialylated gonococci. In addition, the T1217A mutant showed 36 and 18% binding to sialylated PorB.1A and PorB.1B, respectively, relative to the binding of WT HufH18–20/Fc (Fig. 2B).
Most gonococci express the lacto-N-neotetraose LOS species that becomes sialylated in vivo, the rationale behind the use of sialylated bacteria in these studies. To understand whether LOS sialylation influenced the amino acid residues of fH18–20 that determined species-specificity of fH binding to N. gonorrhoeae, we compared the binding profiles of HufH18–20/Fc and its mutant proteins both to sialylated and unsialylated gonococci. Whereas unsialylated PorB.1B strain F62 binds barely detectable amounts of HufH by FACS (35), PorB.1A strain 252 binds to HufH even in the unsialylated state (24), and therefore, a comparison between sialylated and unsialylated bacteria was carried out using both forms of strain 252 only. As shown in supplemental Fig. S3, binding of HufH18–20/Fc and its mutant proteins was similar, with the expected increases in levels of binding seen throughout when LOS was sialylated.
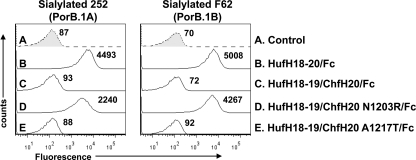
”Humanizing” Chimpanzee Domain 20 Selectively at Position 1203 Restores fH Binding to Gonococci
Having shown that Arg-1203 was important for binding of HufH to sialylated gonococci, we next considered whether replacement of Asn-1203 in ChfH20 with arginine, the human counterpart, was sufficient to restore binding of fH to sialylated gonococci. This single substitution (HufH18–19/ChfH20 N1203R/Fc) was sufficient to enable binding of this construct to both sialylated gonococcal strains (Fig. 3). Because the Thr-1217 residue in HufH20 also appeared to be important in binding to gonococci (Fig. 2B), we also examined whether binding of a mutant construct that possessed A1217T (called HufH18–19/ChfH20 A1217T/Fc) in the back-ground of HufH18–19/ChfH20/Fc could restore binding to gonococci. As seen in Fig. 3, this mutant did not bind to the sialylated gonococcal strains.
FIGURE 3.
Binding of HufH18–19/ChfH20/Fc (HufH20 replaced by ChfH20) constructs to sialylated N. gonorrhoeae. Amino acid residues at positions 1203 and 1217 in ChfH20 were individually substituted by their human counterparts. In all graphs, the x axis represents fluorescence on a log10 scale, and the y axis represents the number of events. The graph labeled HufH18–20/Fc serves as the positive control, and HufH18–19/ChfH20/Fc and Control, the latter containing no fusion proteins, serve as negative controls. One representative experiment of two independently performed experiments is shown.
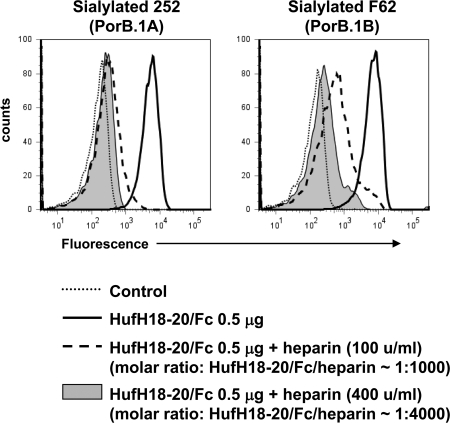
Heparin Inhibits Binding of HufH18–20/Fc to Sialylated Gonococci
Heparin is a highly sulfated negatively charged carbohydrate that is often used as a surrogate for host glycosaminoglycans (29, 30, 36) that bind fH to down-regulate complement activation on human cell surfaces (36, 37). A heparin-binding site is located in domain 20 of fH (29, 30). Consistent with the ionic nature of the heparin-HufH20 bond, positively charged residues such as Arg-1182, Arg-1203, Arg-1206, and Lys-1230 in HufH20 are involved in the interaction with heparin (31, 36, 38). Because Arg-1203 is involved in binding of fH to heparin (39, 40) as well as to sialylated gonococci, we hypothesized that heparin would inhibit binding of HufH to sialylated gonococci. Indeed, heparin inhibited the binding of HufH18–20/Fc to sialylated PorB.1A and PorB.1B strains of N. gonorrhoeae in a dose-responsive manner (Fig. 4), indicating that binding sites for heparin and sialylated gonococci in HufH domain 20 overlap.
FIGURE 4.
Heparin inhibits binding of HufH18–20/Fc to sialylated gonococci. Binding of HufH18–20/Fc to sialylated 252 (PorB.1A) and F62 (PorB.1B) strains was measured by flow cytometry in the presence of heparin (final concentrations of 100 or 400 units (u)/ml; denoted by the dashed line and shaded gray histograms, respectively). Binding of HufH18–20/Fc in the absence of heparin is indicated by the solid line and Control (no HufH18–20/Fc added) by the dotted line. The x axis represents fluorescence on a log10 scale, and the y axis represents the number of events. One representative experiment of three independently performed experiments is shown.
Modeling of R1203N Mutant Structure
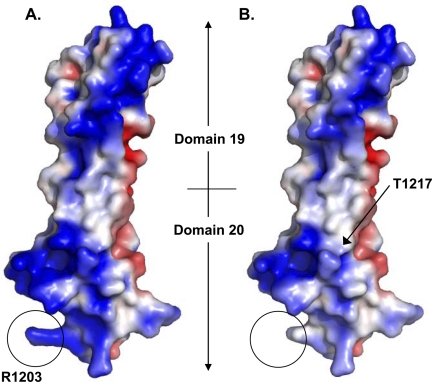
Point mutations may affect structural integrity of a protein. However, it is unlikely that mutating the amino acids in HufH to their ChfH counterparts would destabilize the tertiary structure of the molecule. Because the R1203N mutation in HufH 18–20 abolished binding to gonococci, we modeled the effect of the mutation by homology-based molecular modeling of the R1203N mutant using the crystal structure of fH domains 19–20 (31) as a template. In the crystal structure, the Arg-1203 residue is exposed on the surface of the tip of domain 20 (31). In the model, it was apparent that the mutation, R1203N, resulted in a dramatic local change by removal of the positive charge on a protrusion that is formed by the side chain of the residue (Fig. 5, A and B). The side chain of Arg-1203 is directed away from cysteine 1201 (supplemental Fig. S4); mutating this residue to an alanine does not affect the integrity of the disulfide bond or the overall domain structure as evidenced by x-ray crystallography (41).
FIGURE 5.
Comparison of the crystal structure of HufH domains 19–20 (31) (A) with a molecular model of fH domains 19–20 with the R1203N mutation (B). In both A and B, surface representation of charge is shown with colors; red indicates electronegative, and blue indicates electropositive regions (range from −3 to +3 kT/e). The site of the R1203N mutation is circled. The model (B) indicates that the effect of the R1203N mutation upon surface charge is limited to the immediate area surrounding the mutated residue, i.e. the encircled area only. Location of Thr-1217 is indicated in the model structure with a black arrow.
The residue Thr-1217 had also appeared to be involved in binding of HufH18–20 to N. gonorrhoeae (Fig. 2B), but this finding was not corroborated by mutating the residues on ChfH to the corresponding human residues (Fig. 3). Although the Thr-1217 residue is surface exposed in the crystal structure of fH19–20 (Fig. 5B), Thr-1217 is located on the side surface of domain 20, ∼24 Å from the Arg-1203 residue.
Complement-dependent Killing of Sialylated Gonococci by HufH18–20/Fc
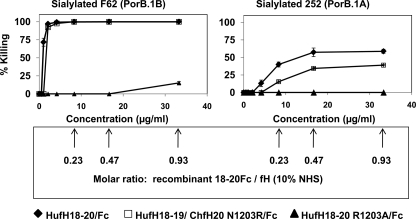
The ability of HufH18–20/Fc to bind to sialylated gonococci at a location where full-length fH binds (Fig. 1B) in combination with the ability of Fc to activate the classical pathway of complement (supplemental Fig. S5) prompted us to investigate whether HufH 18–20/Fc and the derived HufH18–19/ChfH 20 N1203R/Fc mutant, which also bound to sialylated gonococcal strains F62 and 252, supported complement-dependent killing of N. gonorrhoeae. Adding HufH18–20/Fc to NHS resulted in killing of sialylated strain F62 (PorB.1B) in a dose-dependent manner (Fig. 6, left panel). Seventy two percent killing was seen at concentrations as low as 1 μg/ml. Adding HufH18–19/ChfH 20 N1203R/Fc to NHS showed similar results. As a control, we used HufH18–20 R1203A/Fc that did not bind to gonococci; as expected, addition of this mutant molecule did not diminish survival of the sialylated F62 strain, except at the highest concentration used (33.2 μg/ml yielded 15% killing). Sialylated PorB.1A strain 252 was killed 13% by complement in NHS in the presence of HufH18–20/Fc at concentrations (4.2 μg/ml; Fig. 6, right panel) that had resulted in complete killing of sialylated strain F62. Increasing the concentration of HufH18–20/Fc to 8.3, 16.6, and 33.2 μg/ml enhanced killing to 40, 57, and 59% respectively. At a concentration of 33.2 μg/ml, HufH18–19/ChfH20 N1203R/Fc resulted in 39% killing of sialylated 252. As expected, HufH18–20 R1203A/Fc did not kill sialylated 252. Addition of HufH18–20/Fc to heat inactivated NHS resulted in no killing of either sialylated F62 and 252 strains (data not shown) confirming that killing was complement-dependent.
FIGURE 6.
Inhibition of binding of HufH to sialylated gonococci results in reversal of inhibition (killing) of N. gonorrhoeae by the alternative pathway of complement in NHS. Killing of sialylated gonococcal strains F62 (PorB.1B; left panel) and 252 (PorB.1A; right panel) in 10% NHS containing increasing amounts of HufH18–20/Fc or HufH18–20/Fc mutant proteins: HufH18–20/Fc (filled diamonds), HufH18–19/ChfH20 N1203R/Fc (open squares) and HufH18–20 R1203A/Fc (filled triangles). The x axis represents the concentration of purified recombinant HufH18–20/Fc proteins added to reaction mixtures that contained 10% NHS; the y axis indicates the percent (%) killing. The numbers in the box represent the molar ratio of recombinant fH domains 18–20/Fc to endogenous fH in NHS (10%) at different concentrations of the recombinant domains used. Results represent the mean killing calculated from three or more independently performed experiments ± S.E.
NHS contains natural antibodies directed against pathogenic Neisseriae (42–45). To address whether anti-Neisserial antibodies present in NHS played a role in modulating killing of gonococci by HufH18–20/Fc, we compared killing of recombinant fH/Fc protein in NHS versus IgG and IgM-depleted NHS, each used as an source of complement. As shown in supplemental Fig. S6, both sialylated F62 and 252 were killed equally by HufH18–20/Fc in either NHS or IgG- and IgM-depleted NHS that contained equal amounts of active complement.
DISCUSSION
To survive the milieu of the genital tract and establish infection, N. gonorrhoeae must evade the innate immune system at this location. Naturally occurring antibodies and complement are important effector arms of the immune response in the lower genital tract, particularly in women (3). Gonococci have developed numerous mechanisms to evade complement-mediated killing in vivo; one such mechanism is sialylation of lacto-N-neotetraose LOS. Mechanisms of serum resistance conferred by gonococcal lacto-N-neotetraose LOS sialylation include decreased binding of antibodies (46) and enhanced binding of HufH (10).
The ability of gonococci to bind HufH selectively may contribute an explanation of why gonorrhea occurs naturally only in humans (24). HufH and ChfH domain 20 differ by 11 amino acid residues. Nine of these 11 amino acid residues (except Val-1200 and Ala-1229) are surface exposed (31). Although the other amino acids common to both human and chimpanzee fH are undoubtedly involved in the binding between fH and sialylated gonococci, we have shown that Arg-1203 is an important residue that dictates the species specificity of fH binding to sialylated gonococci. R1203N (or R1203A) mutations alone were sufficient to abrogate binding of fH to both sialylated PorB.1A and PorB.1B strains of N. gonorrhoeae. Complementing this observation, a chimpanzee-to-human mutation, N1203R, in ChfH domain 20 resulted in a gain of function, that is, binding to gonococci. Crystal data of HufH domains 19–20 (31) and our current modeling data of HufH20 with an R1203N mutation (Fig. 5), indicates that this surface exposed positively charged residue located 29 amino acids upstream of the C terminus of HufH is essential for binding to sialylated gonococci enabling organisms to evade complement-mediated killing. Our findings provide additional understanding of the molecular basis of human-specific binding of fH to sialylated gonococci.
Arg-1203 is a component of the heparin-binding site in HufH 20 (39, 40), and in accordance with these previous studies, the HufH18–20/Fc R1203N mutant showed decreased binding to heparin by ELISA when compared with HufH18–20/Fc (supplemental Fig. S7). A model of the R1203N mutation shows that loss of a positive charge at this location disrupts a positively charged belt, which is presumed to be the heparin-binding site in HufH 20 (31, 36, 38). In addition, heparin inhibited binding of HufH18–20/Fc (and full-length HufH) to sialylated PorB.1A and PorB.1B gonococci, suggesting that the point(s) of contact between fH and sialylated gonococci overlap with the heparin binding site(s) in fH. The 1203 residue is also involved in the binding of factor H and complement C3b and C3d ((47) and supplemental Fig. S8).
In earlier studies, we demonstrated that LOS sialic acid increases HufH binding to PorB.1B-bearing strains (10); subsequently, we have also shown that gonococcal Por is essential for the interaction between HufH and sialylated PorB.1B-bearing gonococci (35). However, it is possible that sialylated LOS, gonococcal Por and HufH form a ternary complex. It is noteworthy that subtle variations are seen in PorB.1A and PorB.1B interactions with recombinant HufH18–20/Fc mutants (Fig. 2B). Variations observed in the interactions of the two PorB types with HufH18–20/Fc are not entirely unexpected because the predicted surface-exposed loop sequences of the two PorB molecules differ substantially (48).
The mutation T1217A appeared to have had an effect on the binding of HufH to sialylated gonococci. However, the reverse mutation created in HufH18–19/ChfH20/Fc did not restore binding to sialylated gonococci. The structure of HufH19–20 (31) indicates that residue Arg-1203 is on a different surface of the fH19–20 protein than the Thr-1217 residue. Although the location of the Thr-1217 residue on the structure of FH19–20 does not exclude the possibility that it was involved in the interaction, it also possible that the T1217A mutation itself (independently) may have resulted in decreased binding efficiency between the recombinant (HufH18–20 T1217A/Fc) and sialylated gonococci due to, for example, (small) aliphatic changes at this site that might have resulted in diminished solubility of the recombinant protein and resultant diminished binding in the reaction mixtures (49, 50). Failure to corroborate this finding when humanization of position 1217 in HufH18–19/ChfH20 A1217T/Fc did not result in enhanced binding as it had when position 1203 was humanized, suggests that residue Thr-1217 is not directly involved in the interaction with gonococci.
In this study, we have shown that binding of HufH18–20/Fc to both PorB.1A and PorB.1B strains enabled complement-dependent killing of these gonococcal strains (Fig. 6). Sialylated PorB.1B strain F62 was killed completely by HufH18–20/Fc at 2 μg/ml. However, sialylated PorB.1A strain 252 required a higher concentration of HufH18–20/Fc to kill it and complete killing of this strain by HufH18–20/Fc was not achieved by concentrations that exceeded by nearly 10-fold those required to kill sialylated PorB.1B strain F62. We have reported that sialylated PorB.1B strain F62 binds HufH via domains 18–20 exclusively, whereas PorB.1A strain 252 binds HufH via domains 18–20 and separately, by domain 6 (24, 51). A proposed functional structure of fH suggests that monomeric HufH adopts a folded-back conformation (52) where domains, 12–14 contain the site of a bend or hinge (30) that increases proximity of short consensus repeat 6 and 20 domains to binding sites on N. gonorrhoeae. This may result in enhanced binding (and resultant function) of factor H. A requirement for engagement of fH via two closely spaced binding sites on this molecule for the sialylated PorB.1A strain might explain the inability of HufH18–20/Fc to completely kill this strain.
In conclusion, we have defined amino acids in human fH that are involved in binding on the surface of N. gonorrhoeae. Our strategy and findings may aid in the development of molecules that not only bind to gonococci but that also activate the classical pathway through the Fc at vulnerable sites on N. gonorrhoeae otherwise occupied by factor H. We acknowledge that such an approach will require further refinement of the molecule such that it binds only to the pathogen and not to eukaryotic cells to minimize damage to the host. This may provide novel therapeutic options against this pathogen that is rapidly becoming resistant to multiple antibiotics (53). In addition, a deeper molecular understanding of species-specific binding of fH to gonococci has been elucidated, which may shed additional light on the basis for the restriction of gonococcal infection to humans.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI32725, AI054544, AI084048, and AI069084. This work was also supported by the Academy of Finland (Projects 201506 and 202529) and The Sigrid Jusélius Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S8.
- NHS
- normal human serum
- fH
- factor H
- HufH
- human fH
- Por
- porin
- LOS
- lipo-oligosaccharide
- CMP-NANA
- 5′-cytidinemonophospho-N-acetylneuraminic acid
- ChfH
- chimpanzee fH
- HBSS
- Hanks' balanced salt solution.
REFERENCES
- 1. Centers for Disease Control and Prevention (2010) Sexually Transmitted Disease Surveillance 2009, U. S. Department of Health and Human Services, Atlanta, GA [Google Scholar]
- 2. Gerbase A. C., Rowley J. T., Heymann D. H., Berkley S. F., Piot P. (1998) Sex Transm. Infect. 74, S12–16 [PubMed] [Google Scholar]
- 3. Price R. J., Boettcher B. (1979) Fertil. Steril. 32, 61–66 [DOI] [PubMed] [Google Scholar]
- 4. O'Brien J. P., Goldenberg D. L., Rice P. A. (1983) Medicine 62, 395–406 [PubMed] [Google Scholar]
- 5. Ram S., McQuillen D. P., Gulati S., Elkins C., Pangburn M. K., Rice P. A. (1998) J. Exp. Med. 188, 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ram S., Cullinane M., Blom A. M., Gulati S., McQuillen D. P., Monks B. G., O'Connell C., Boden R., Elkins C., Pangburn M. K., Dahlbäck B., Rice P. A. (2001) J. Exp. Med. 193, 281–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blake M. S., Gotschlich E. C. (1986) in Bacterial Outer Membrane as Model Systems (Inouye M. ed.) pp. 377–400, John Wiley, New York [Google Scholar]
- 8. Cannon J. G., Buchanan T. M., Sparling P. F. (1983) Infect. Immun. 40, 816–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brunham R. C., Plummer F., Slaney L., Rand F., DeWitt W. (1985) J. Infect. Dis. 152, 339–343 [DOI] [PubMed] [Google Scholar]
- 10. Ram S., Sharma A. K., Simpson S. D., Gulati S., McQuillen D. P., Pangburn M. K., Rice P. A. (1998) J. Exp. Med. 187, 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fearon D. T., Austen K. F. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 1683–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pangburn M. K., Schreiber R. D., Müller-Eberhard H. J. (1977) J. Exp. Med. 146, 257–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sim R. B., Twose T. M., Paterson D. S., Sim E. (1981) Biochem. J. 193, 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weiler J. M., Daha M. R., Austen K. F., Fearon D. T. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 3268–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whaley K., Ruddy S. (1976) J. Exp. Med. 144, 1147–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ripoche J., Day A. J., Harris T. J., Sim R. B. (1988) Biochem. J. 249, 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kristensen T., Tack B. F. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Norman D. G., Barlow P. N., Baron M., Day A. J., Sim R. B., Campbell I. D. (1991) J. Mol. Biol. 219, 717–725 [DOI] [PubMed] [Google Scholar]
- 19. Mandrell R. E., Griffiss J. M., Macher B. A. (1988) J. Exp. Med. 168, 107–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parsons N. J., Patel P. V., Tan E. L., Andrade J. R., Nairn C. A., Goldner M., Cole J. A., Smith H. (1988) Microb. Pathog. 5, 303–309 [DOI] [PubMed] [Google Scholar]
- 21. Apicella M. A., Mandrell R. E., Shero M., Wilson M. E., Griffiss J. M., Brooks G. F., Lammel C., Breen J. F., Rice P. A. (1990) J. Infect. Dis. 162, 506–512 [DOI] [PubMed] [Google Scholar]
- 22. Nairn C. A., Cole J. A., Patel P. V., Parsons N. J., Fox J. E., Smith H. (1988) J. Gen. Microbiol. 134, 3295–3306 [DOI] [PubMed] [Google Scholar]
- 23. Ward M. E., Watt P. J., Glynn A. A. (1970) Nature 227, 382–384 [DOI] [PubMed] [Google Scholar]
- 24. Ngampasutadol J., Ram S., Gulati S., Agarwal S., Li C., Visintin A., Monks B., Madico G., Rice P. A. (2008) J. Immunol. 180, 3426–3435 [DOI] [PubMed] [Google Scholar]
- 25. Schneider H., Griffiss J. M., Williams G. D., Pier G. B. (1982) J. Gen. Microbiol. 128, 13–22 [DOI] [PubMed] [Google Scholar]
- 26. McQuillen D. P., Gulati S., Rice P. A. (1994) Methods Enzymol. 236, 137–147 [DOI] [PubMed] [Google Scholar]
- 27. Dutta Ray T., Lewis L. A., Gulati S., Rice P. A., Ram S. (2011) J. Immunol. 186, 4881–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jokiranta T. S., Zipfel P. F., Hakulinen J., Kühn S., Pangburn M. K., Tamerius J. D., Meri S. (1996) FEBS Lett. 393, 297–302 [DOI] [PubMed] [Google Scholar]
- 29. Pangburn M. K., Atkinson M. A., Meri S. (1991) J. Biol. Chem. 266, 16847–16853 [PubMed] [Google Scholar]
- 30. Schmidt C. Q., Herbert A. P., Kavanagh D., Gandy C., Fenton C. J., Blaum B. S., Lyon M., Uhrín D., Barlow P. N. (2008) J. Immunol. 181, 2610–2619 [DOI] [PubMed] [Google Scholar]
- 31. Jokiranta T. S., Jaakola V. P., Lehtinen M. J., Pärepalo M., Meri S., Goldman A. (2006) EMBO J. 25, 1784–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) Nucleic Acids Res. 35, W522–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cunningham B. C., Wells J. A. (1989) Science 244, 1081–1085 [DOI] [PubMed] [Google Scholar]
- 35. Madico G., Ngampasutadol J., Gulati S., Vogel U., Rice P. A., Ram S. (2007) J. Immunol. 178, 4489–4497 [DOI] [PubMed] [Google Scholar]
- 36. Jokiranta T. S., Cheng Z. Z., Seeberger H., Jòzsi M., Heinen S., Noris M., Remuzzi G., Ormsby R., Gordon D. L., Meri S., Hellwage J., Zipfel P. F. (2005) Am. J. Pathol. 167, 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ferreira V. P., Herbert A. P., Hocking H. G., Barlow P. N., Pangburn M. K. (2006) J. Immunol. 177, 6308–6316 [DOI] [PubMed] [Google Scholar]
- 38. Manuelian T., Hellwage J., Meri S., Caprioli J., Noris M., Heinen S., Jozsi M., Neumann H. P., Remuzzi G., Zipfel P. F. (2003) J. Clin. Invest. 111, 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ferreira V. P., Herbert A. P., Cortés C., McKee K. A., Blaum B. S., Esswein S. T., Uhrín D., Barlow P. N., Pangburn M. K., Kavanagh D. (2009) J. Immunol. 182, 7009–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herbert A. P., Uhrín D., Lyon M., Pangburn M. K., Barlow P. N. (2006) J. Biol. Chem. 281, 16512–16520 [DOI] [PubMed] [Google Scholar]
- 41. Bhattacharjee A., Lehtinen M. J., Kajander T., Goldman A., Jokiranta T. S. (2008) Mol. Immunol. 45, 4125. [DOI] [PubMed] [Google Scholar]
- 42. Griffiss J. M., Jarvis G. A., O'Brien J. P., Eads M. M., Schneider H. (1991) J. Immunol. 147, 298–305 [PubMed] [Google Scholar]
- 43. Rice P. A., McCormack W. M., Kasper D. L. (1980) J. Immunol. 124, 2105–2109 [PubMed] [Google Scholar]
- 44. Schoolnik G. K., Ochs H. D., Buchanan T. M. (1979) J. Immunol. 122, 1771–1779 [PubMed] [Google Scholar]
- 45. Yamasaki R., Maruyama T., Yabe U., Asuka S. (2005) J. Biochem. 137, 487–494 [DOI] [PubMed] [Google Scholar]
- 46. Vogel U., Weinberger A., Frank R., Müller A., Köhl J., Atkinson J. P., Frosch M. (1997) Infect. Immun. 65, 4022–4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bhattacharjee A., Lehtinen M. J., Kajander T., Goldman A., Jokiranta T. S. (2010) Mol. Immunol. 47, 1686–1691 [DOI] [PubMed] [Google Scholar]
- 48. Swanson J., Dorward D., Lubke L., Kao D. (1997) J. Bacteriol. 179, 3541–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Clackson T., Ultsch M. H., Wells J. A., de Vos A. M. (1998) J. Mol. Biol. 277, 1111–1128 [DOI] [PubMed] [Google Scholar]
- 50. Moreira I. S., Fernandes P. A., Ramos M. J. (2007) Proteins 68, 803–812 [DOI] [PubMed] [Google Scholar]
- 51. Agarwal S., Ram S., Ngampasutadol J., Gulati S., Zipfel P. F., Rice P. A. (2010) J. Immunol. 185, 4344–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aslam M., Perkins S. J. (2001) J. Mol. Biol. 309, 1117–1138 [DOI] [PubMed] [Google Scholar]
- 53. Tapsall J. W., Ndowa F., Lewis D. A., Unemo M. (2009) Expert. Rev. Anti. Infect. Ther. 7, 821–834 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.