Abstract
Cancer cells commonly exhibit increased nonoxidative d-glucose metabolism whereas induction of mitochondrial metabolism may impair malignant growth. We have first used an in silico method called elementary mode analysis to identify inhibition of ALAT (l-alanine aminotransferase) as a putative target to promote mitochondrial metabolism. We then experimentally show that two competitive inhibitors of ALAT, l-cycloserine and β-chloro-l-alanine, inhibit l-alanine production and impair d-glucose uptake of LLC1 Lewis lung carcinoma cells. The latter inhibition is linked to an initial energy deficit, as quantified by decreased ATP content, which is then followed by an activation of AMP-activated protein kinase and subsequently increased respiration rates and mitochondrial production of reactive oxygen species, culminating in ATP replenishment in ALAT-inhibited LLC1 cells. Moreover, we observe altered phosphorylation of p38 MAPK (mitogen-activated protein kinase 14), ERK (extracellular signal-regulated kinase 1/2), and Rb1 (retinoblastoma 1) proteins, as well as decreased expression of Cdc25a (cell decision cycle 25 homolog A) and Cdk4 (cyclin-dependent kinase 4). Importantly, these sequelae of ALAT inhibition culminate in similarly reduced anchorage-dependent and anchorage-independent growth rates of LLC1 cells, together suggesting that inhibition of ALAT efficiently impairs cancer growth by counteracting the Warburg effect due to compensatory activation of mitochondrial metabolism.
Keywords: Amino Acid, Cancer Therapy, Glucose, Glucose Metabolism, Glutamate, Glutamine, Metabolism, Metabolic Regulation, Mitochondria, Mitochondrial Metabolism
Introduction
Compared with nonmalignant entities, cancer cells commonly exhibit increased nonoxidative d-glucose metabolism (glycolysis) (1, 2) whereas mitochondrial activity and in particular respiration rates are severely impaired in malignant cells (3–9). Based on these facts, Otto Warburg proposed that an inappropriate increase in glycolysis due to impaired respiratory capacity may be the cause of malignant growth (1, 2), a concept that was named Warburg's hypothesis in subsequent decades. This effect has been supported on theoretical grounds by the comparably inefficient utilization of available nutrients in cancer cells (10).
Although it is questionable whether the Warburg effect actually causes cancer, impairing d-glucose uptake or d-glucose metabolism in cancer cells unequivocally induces oxidative metabolism and has been shown to effectively impair malignant cell growth in vitro and in vivo (11, 12). In this regard, a typical inhibitor of proximal glycolysis, 2-deoxy-d-glucose (2-DOG),3 has been shown to be particularly effective in impairing cancer growth (13). In later years this effect was used additionally to increase the efficacy of chemotherapeutic agents (14).
Consistent with these promising effects of glycolytic inhibitors like 2-DOG, forcing cancer cells into increased mitochondrial metabolism independently of d-glucose metabolism, e.g. by overexpressing rate-limiting mitochondrial proteins, efficiently reduces both anchorage-dependent and -independent growth, as well as tumor growth in nude mice (15). However and unlike in cultured cells, selective activation of mitochondrial metabolism in vivo is difficult to achieve.
Recently established in silico methods, and in particular the so-called elementary mode analysis (EMA) of metabolic networks, can be used to identify novel pathways and alternate biochemical routes, including those that may selectively promote mitochondrial metabolism. EMA is capable of predicting so-called elementary modes (EMs), which are the smallest possible subsets of biochemical reactions connecting to points of a steady-state metabolic network (16–18).
In the present study, we have used this method to identify biochemical pathways that may increase oxidative metabolism of cancer cells when inhibited by appropriate compounds. Specifically and by applying EMA, we have identified the conversion of l-pyruvate into l-alanine by the enzyme ALAT (l-alanine aminotransferase) as a putatively crucial step, and we provide experimental support for this hypothesis primarily generated in silico. By applying ALAT inhibitors to malignant cells we observe fundamental metabolic changes that culminate in increased oxidative metabolism and cell cycle arrest, leading to impaired anchorage-dependent and -independent cancer cell growth.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals used were obtained from Sigma-Aldrich. l-Cycloserine (Cyclo) and β-chloro-l-alanine (Cl-Ala) were dissolved in dimethyl sulfoxide to obtain 10, 50, and 250 mm stock solutions that were then aliquoted and stored at −80 °C prior to use. For cell treatment, stock solutions were diluted 1:1000 in medium to a final dimethyl sulfoxide concentration of 0.1%. If not stated otherwise, Cyclo and Cl-Ala were used at a final concentration of 250 μm.
Elementary Mode Analysis
Determination of energy-producing EMs was conducted as previously described (16–18). Noncommercial software YANAsquare 0.98 was used.
Cancer Cell Lines and Cell Culture Conditions
The murine Lewis lung carcinoma cell line LLC1 was obtained from the American Type Culture Collection. Human diploid foreskin fibroblast cell lines transduced with the indicated genes for immortalization and/or malignant transformation (BJ1, hTERT; BJ4, hTERT; simian virus ST and simian virus LT, H-ras) were obtained from William C. Hahn (Dana-Farber Cancer Institute, Boston) (19), and aliquots derived from the primary vials were generated and stored in liquid nitrogen. Experiments were performed with cells that were maintained in culture for fewer than 10 passages. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/liter d-glucose (Sigma-Aldrich), 10% fetal bovine serum (FBS) (Biochrom KG, Berlin, Germany) and 1% penicillin/streptomycin (Invitrogen) (10 kilounits of penicillin/ml, 10 mg of streptomycin/ml). However, all experiments were conducted using glucose-free DMEM (Cambrex, Vervies, Belgium) supplemented with 1.8 g/liter d-glucose (Applichem, Darmstadt, Germany), 10% FBS, and 1% penicillin/streptomycin, except when explicitly stated otherwise.
Amino Acid Metabolism
Amino acid and organic acid concentrations were determined using an amino acid analyzer LC 3000 (Biotronik, Maintal, Germany) following the instructions of the manufacturer. Briefly, 50 μl of sulfosalicylic acid was added to a 200-μl sample and incubated for 30 min at 5 °C. Subsequently, samples were centrifuged for 15 min at 16,600 × g. One hundred microliters of the supernatant was diluted 1:1 with sample buffer and analyzed by ion exchange chromatography. Postcolumn derivatization was performed with ninhydrin. l-Alanine and l-aspartate turnover was calculated as net change in concentration of the respective amino acid in culture medium normalized to integrated cellular protein content per well.
14C-labeled 2-DOG Uptake
Cells were washed with PBS and maintained in standard glucose-free Krebs-Ringer-Hepes buffer for 60 min. Thereafter, the buffer was replaced by Krebs-Ringer-Hepes buffer containing uniformly 14C-labeled 2-DOG with an activity of 0.25 μCi/ml and supplemented with 100 μm unlabeled 2-DOG. After 10 min cells were washed three times with ice-cold PBS and lysed with 0.5 m NaOH. Scintillation was measured according to the protocol of the manufacturer (Becton-Dickinson). Aliquots of NaOH lysates were used to determine cellular protein content per well for normalization.
d-Glucose/l-Lactate Ratio
d-Glucose uptake from supernatant media was determined as described previously (20). l-Lactate was measured according to the method of Sweetmann et al. (21) modified as described below. After deproteinization of samples by the addition of perchloric acid (1:1), 500 μl of H2O, 50 μl of internal standard (4 mmol/liter 2-hydroxybutyric acid), and 500 μl of saturated NH4Cl solution were added to 100 μl of the deproteinized supernatant. Subsequently, sample extraction was performed by addition of 2 × 5 ml of ethyl acetate. The organic phase was collected, dried over anhydrous Na2SO4, evaporated to dryness under a gentle stream of nitrogen in a new tube, and derivatized (100 μl of BSTFA, 30 min, 60 °C). One microliter of the derivatized solution was subjected to analysis by gas chromatography-mass spectrometry using a ZB-5 column (Phenomenex, Aschaffenburg, Germany) and a Shimadzu QP2010 GC-MS (Shimadzu, Duisburg, Germany).
Cellular ATP Content
Cellular ATP content was measured by using a luciferin/luciferase-based bioluminescence assay (CellTiter-Glo, Promega, Madison, WI) as described before (20). Briefly, cells were seeded in a 96-well plate (2 × 103/well), washed with PBS following treatment, and lysed (CellTiter-Glo buffer), and aliquots were taken for protein determination prior to the addition of bioluminescent substrate/enzyme solution (CellTiter-Glo substrate). Luminescence was measured using a 96-well plate luminometer (Fluostar, BMG, Offenburg, Germany). Cellular ATP content was calculated by an ATP standard curve and normalized to cellular protein content/well.
Cellular Respiration
Briefly, 2 × 103 cells/well were seeded in a 96-well OxoPlate (PreSens, Regensburg, Germany), sealed using adhesive sealing foil (Thermo Fischer Scientific), kept at 37 °C, and fluorescence/phosphorescence was measured every 60 min up to 48 h according to the protocol of the manufacturer (PreSens). In parallel, equally treated plates for different time points were used to determine integrated cellular protein for normalization.
l-Glutamine Utilization
Briefly, 1 × 104 cells/well were seeded in a 24-well plate, and supernatant medium was collected after 48 h treatment. l-Glutamine and l-glutamate concentrations were determined in the supernatant by using an enzyme-based kit according to the protocol provided by the manufacturer (Glutamine and Glutamate Determination Kit GLN-1; Sigma-Aldrich). l-Glutamine utilization was calculated as l-glutamine uptake minus l-glutamate production both normalized to integrated cellular protein content/well.
Mitochondrial Reactive Oxygen Species (mtROS) Production
The measurement was performed according to the protocol provided by the manufacturer (CMXRos; Invitrogen). Briefly, 1 × 104 cells/well were seeded in a 24-well plate. After treatment cells were incubated with fresh medium containing 1 μm Mitotracker Red (CMXRos) for 30 min, washed twice with PBS, supplied with fresh medium, and fluorescence was measured after a 10-min incubation at 37 °C at 578 nm excitation and 599-nm emission wavelengths. CMXRos fluorescence was normalized to cellular protein content/well.
Protein Determination
Following lysis of cells with 1 m NaOH, protein contents were determined by using a bicinchoninic acid assay kit according to the protocol provided by the manufacturer (BCA Protein Assay, Thermo Fischer Scientific).
Immunoblotting and Western Blot Analysis
Methods for immunoblotting were performed as described previously (15) using the following primary antibodies: anti-phospho-AMP-activated kinase (anti-phospho-AMPK) (Thr172), anti-AMPK, anti-tubulin, anti-phospho-p38 (Thr180/Tyr182), anti-p38, anti-phospho-ERK1/2 (Thr202/Tyr204), anti-ERK1/2, anti-Cdk4, and anti-phospho-Rb (Ser780) supplied by Cell Signaling (Boston, MA), anti-Rb and anti-ALAT (anti-GPT) obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Western blots were tested for equal protein loading by both ponceau red staining of the membrane and anti-tubulin blotting (data not shown). Densitometric analysis of Western blots was conducted using ImageJ software (National Institutes of Health, Bethesda, MD) according to the program's manual.
Soft Agar Assay
Soft agar assays were performed in a semiautomated manner in 96-well plates using an epMotion 5075 Liquid Handling Work station (Eppendorf AG, Hamburg, Germany) as described previously (22). Briefly, in each well 100 μl of top agar containing 1 × 103 LLC1 cells and test compounds or solvent was placed on top of 100 μl of previously solidified base layer. After 6 days of incubation, Alamar Blue (resazurin) was added, and colony growth was quantified fluorometrically.
Animal Experiments
Nude mice (Crl:CD1-Foxn1nu, 5 weeks of age) were obtained from Charles River Laboratories. One million LLC1 cells (viability >90%) were resuspended in 1 ml of DMEM (without antibiotics and FBS) and injected subcutaneously in the left hind area using an insulin syringe (Becton-Dickinson). Starting on the next day, 10 μl of 0.9% sodium chloride solution/g of body weight (BW) with or without test compounds (Cyclo, 100 mg/kg BW; Cl-Ala, 20 mg/kg BW) was injected subcutaneously in the right hind area once a day at the same time. On day 13 mice were killed, tumors were excised, and the tumor mass was determined. Mice were kept in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals, and all experiments were approved by the corresponding institutional review boards.
Statistical Analyses
All calculations were performed with SPSS, version 13. A Kolmogorov-Smirnov test was used to test for normal distribution, which was confirmed in all experiments. The unpaired Student's t test was used to determine the statistical significance of the inhibitor effects. A p value below 0.05 was considered statistically significant. If not indicated otherwise, significance of differences in treated groups compared with control groups is shown by asterisks located above the respective treatment group in the corresponding figures.
RESULTS
Identifying ALAT as a Putative Target to Force Cancer Cells into Mitochondrial Metabolism
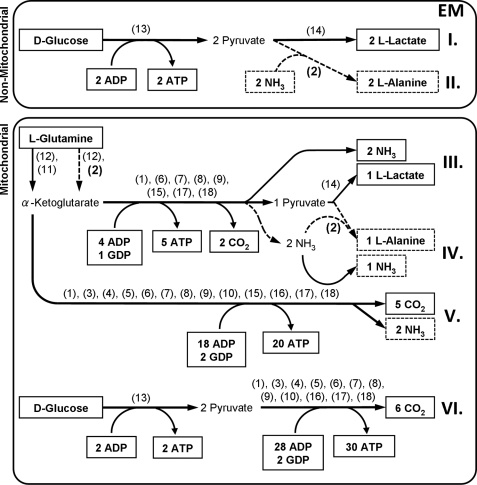
To identify a previously unidentified target for the metabolic inhibition of cancer cell growth we used software-based EMA. We constructed a stoichiometric metabolic model containing enzymes and metabolites likely to be related to the energy production in cancer cells (Fig. 1). We then used this metabolic model as input for an EMA software package named YANAsquare which proposed six major energy-producing EMs (Fig. 1, see also supplemental Fig. 1 for more details).
FIGURE 1.
Identification of energy-producing pathways in cancer cells using YANAsquare-based elementary mode analysis. I–VI, EM representing pathways with the least amount of enzymes involved to produce ATP from d-glucose or l-glutamine. All framed metabolites are intended to underlie a net production or consumption. Further involved metabolites and co-factors are considered to be compensated by anaplerotic and cataplerotic reactions to form a steady-state metabolite flux and therefore remain hidden, namely, acetyl-CoA, citrate, coenzyme Q, FAD, FADH2, fumarate, GDP, l-glutamate, GTP, d-isocitrate, l-malate, NAD+, NADH/H+, oxaloacetate, pyruvate, succinate, succinyl-CoA, and α-ketoglutarate. The following enzymes and simplified enzyme systems were used for EM determination: (1) ATP-GTP-mutase, (2) ALAT (dashed arrows), (3) citrate synthase, (4) aconitase, (5) isocitrate dehydrogenase, (6) α-ketoglutarate dehydrogenase, (7) succinyl-CoA synthase, (8) succinate dehydrogenase, (9) fumarase, (10) malate dehydrogenase, (11) glutamate dehydrogenase, (12) glutaminase, (13) glycolysis (simplified to an one-step reaction from d-glucose to pyruvate), (14) lactate dehydrogenase, (15) malic enzyme, (16) pyruvate dehydrogenase, (17) NADH/H+ = 2.5 ATP, (18) FADH2 = 1.5 ATP. All enzymes are intended to be unregulated and only dependent on energy-producing substrates.
Only two of these six pathways do not employ mitochondrial enzymes: anaerobic ATP generation from d-glucose-producing l-lactate (Fig. 1, EM I) or ATP generation from d-glucose-producing l-alanine (Fig. 1, EM II). Regarding anaerobic d-glucose metabolism, ample evidence exists that inhibiting this pathway both blocks cancer cell growth and induces mitochondrial energy conversion (see “Discussion”). The pathways utilizing mitochondrial enzymes correspond to the aerobic generation of ATP by converting l-glutamine into l-lactate (Fig. 1, EM III) or l-alanine (Fig. 1, EM IV) and the aerobic generation of ATP by completely oxidizing l-glutamine (Fig. 1, EM V) or d-glucose (Fig. 1, EM VI) along the tricarboxylic acid cycle. Conversely and as identified here by EMA, inhibiting the conversion of l-lactate into l-alanine (EM II and IV) may shift ATP production toward mitochondrial pathways (Fig. 1, EM V and VI) and may thereby inhibit cancer growth.
Inhibition of ALAT Impairs l-Alanine Production of Cancer Cells
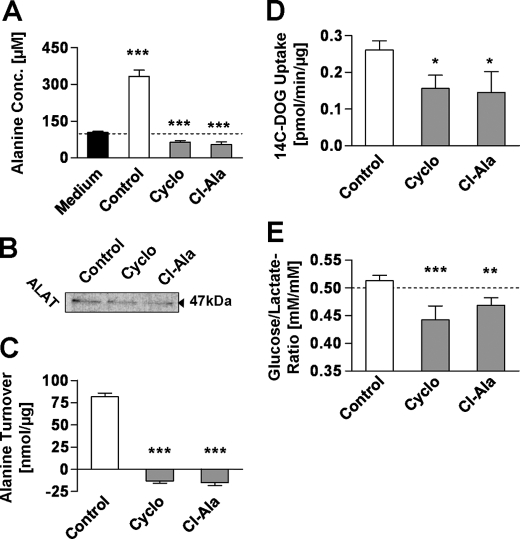
The cytosolic enzyme ALAT catalyzes the terminal step in l-alanine production from d-glucose or other l-pyruvate-contributing carbon sources. By using two different and previously established inhibitors of ALAT (23, 24), namely Cyclo and Cl-Ala, we were able to abolish the production of l-alanine in the highly transformed lung cancer cell line LLC1 (Fig. 2A). However, the expression levels of ALAT in LLC1, revealed by immunoblotting, were found to be unaltered by inhibitor treatment (Fig. 2B). It should be noted that untreated cells produce rather large amounts of l-alanine (Fig. 2C), as previously described for cultured cancer cells as well as human cancers in situ (25–27), Moreover and in contrast to untreated cells, inhibitor-treated LLC1 cells showed only minor consumption of the l-alanine present in the cell culture medium at the beginning of the experiment (Fig. 2C).
FIGURE 2.
Inhibitors of ALAT prevent l-alanine production and reduce d-glucose metabolism in cancer cells. A, l-alanine concentration in supernatant medium after a 48-h treatment with inhibitors. B, Western blot of ALAT after a 24-h treatment. C, l-alanine turnover defined as changes in medium l-alanine referred to integral of cellular protein over a 48-h treatment (n = 4). D, 14C-labeled 2OG uptake per μg of protein after a 48-h treatment with inhibitors (n = 4). E, ratio of d-glucose uptake and l-lactate production (n = 4). Inhibitors were used at a final concentration of 250 μm; error bars represent S.D.; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Cyclo and Cl-Ala have previously been reported to potentially exhibit inhibitory effects on other enzymes than ALAT, especially aspartate aminotransferase, in particular at significantly higher concentrations compared with those used in this study. However, in clear contrast to the reduction of l-alanine production after treatment (Fig. 2A), Cyclo and Cl-Ala did not inhibit l-aspartate turnover in LLC1 cells when used at a concentration of 250 μm, indicating the lack of any inhibitory effect on aspartate aminotransferase activity (supplemental Fig. 2).
Inhibition of ALAT Alters d-Glucose Metabolism of Cancer Cells
Excretion of l-alanine from cancer cells causes a net loss of energy for the individual cell. Hence, blocking l-alanine synthesis from l-lactate may reduce the need for exogenous energy equivalents, particularly d-glucose. To investigate whether the inhibition of l-alanine production from l-lactate and hence d-glucose may indirectly inhibit d-glucose import into the cell, we determined uptake of 14C-labeled 2-DOG. Both ALAT inhibitors entailed a significant decrease in d-glucose uptake (Fig. 2D), thus indicating that a decrease in overall d-glucose metabolism does in fact occur.
Although control cells showed a nearly stoichiometric conversion of d-glucose into l-lactate (i.e. 1 mol of d-glucose being converted into 2 mol of l-lactate, leading to a d-glucose/l-lactate ratio of approximately 0.5), we unexpectedly found that inhibitor-treated cells exhibit d-glucose/l-lactate ratios significantly below 0.5 (Fig. 2E). This strongly suggests that ALAT inhibition causes a relative increase in l-lactate production from carbon sources other than d-glucose.
ALAT Inhibitors Cause an Initial Energy Deficit in Cancer Cells
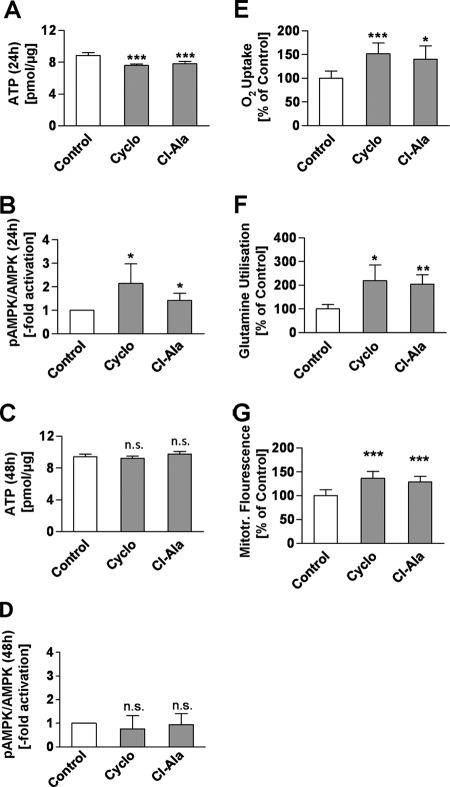
Glycolysis in cancer cells provides an adequate supply of ATP even in states of severely reduced respiration. Conversely, we questioned whether impairing both the disposal of l-lactate into l-alanine (Fig. 2A and supplemental Fig. 3) as well as d-glucose uptake (Fig. 2D) would subsequently cause a cellular energy deficit. Not surprisingly, after a 24-h treatment with either ALAT inhibitor we observed cellular ATP levels to be decreased (Fig. 3A).
FIGURE 3.
Inhibitors of ALAT modulate energy metabolism and promote respiration and l-glutamine utilization. A, cellular ATP concentration after a 24-h treatment with inhibitors. B, densitometric analysis of Western blots of phospho-AMPKα protein (pAMPKα) and basal AMPKα protein (n = 5) (A and B after a 24-h treatment with inhibitors). C, cellular ATP concentration/μg of protein (n = 8). D, densitometric analysis of Western blots of phospho-AMPKα protein (pAMPKα) and basal AMPKα protein (n = 3) (C and D after a 48-h treatment). E, cumulative oxygen uptake during a 36-h treatment with inhibitors referred to integral of cellular protein (n = 8). F, l-glutamine utilization, defined as l-glutamine uptake minus l-glutamate excretion during a 48-h treatment referred to integral of cellular protein (n = 4). G, mtROS-related fluorescence after a 24-h treatment normalized for cellular protein content (n = 8). For all panels, inhibitors were used at a final concentration of 250 μm; error bars represent S.D.; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
Inhibitor-initiated Energy Deficit Activates AMPK and Replenishes ATP
Decreased ATP availability is known to culminate in activation of a key energy sensor of the cell, AMPK (28), which subsequently induces mitochondrial metabolism. Because decreased availability of ATP following inhibitor treatment causes an energy deficit (Fig. 3A), we quantified phosphorylation of the α-subunit of AMP-activated kinase (AMPKα) at Thr172, which is known to be indicative of AMPK activity. Immunoblotting of both the basal as well as the phosphorylated forms of AMPKα indicated that this energy sensor is activated after a 24-h treatment with Cyclo or Cl-Ala (Fig. 3B). In accordance with these findings, ATP content in inhibitor-treated cells was identical to control cells 48 h after addition of the inhibitor (Fig. 3C), thus indicating that the initial ATP deficit (Fig. 3A) is transient and efficiently compensated by the subsequent activation of AMPK. Concurrent with the observed ATP restoration (Fig. 3C) we also found the initial AMPK activation to be abolished after 48 h (Fig. 3D).
Inhibitor-initiated Activation of Mitochondrial l-Glutamine Metabolism
The findings so far indicate that inhibitor-treated LLC1 cells generate amounts of ATP similar to control cells (Fig. 3C). This occurs despite the fact that d-glucose uptake is reduced (Fig. 2D) and l-alanine turnover is reduced (Fig. 2C). Also given the previously established role for AMPK in activating mitochondrial metabolism, we quantified cellular oxygen uptake, which was found to be dramatically increased in inhibitor-treated LLC1 cells (Fig. 3E).
However, the relative increase in l-lactate accumulation suggests that d-glucose-derived l-pyruvate may not be the predominant mitochondrial substrate compensating for d-glucose deficiency. To find out whether increased respiration is possibly due to mitochondrial oxidation of carbon sources other than l-pyruvate, l-lactate, and d-glucose, we quantified the turnover of l-glutamine. l-Glutamine is known to be an important fuel for fast growing cells in vitro and is therefore the second most abundant nutrient in cell culture media (supplemental Fig. 3). Utilization of this amino acid was found to be strongly increased following inhibitor treatment (Fig. 3F). Because ALAT inhibitors appear to impair d-glucose uptake and to promote l-glutamine turnover in parallel, we conclude that l-glutamine is the main substrate of increased mitochondrial metabolism in states of ALAT inhibition.
Inhibitor-initiated Activation of Respiration Promotes Mitochondrial Formation of ROS
Activation of mitochondrial oxygen metabolism, particularly in cancer cells, has been frequently connected to increased production of mtROS. We therefore used a rhodamine-based, redox-sensitive, fluorescent and cell-permeable dye to investigate mtROS levels in inhibitor-treated cells, and we observed increased mtROS-related fluorescence after a 24-h treatment with ALAT inhibitors (Fig. 3G). Taken together, the findings so far indicate that inhibition of ALAT leads to increased mitochondrial activity in an AMPKα-dependent manner to replenish ATP levels while also increasing mtROS formation.
ALAT Inhibition Promotes Several Growth-inhibiting Signaling Pathways
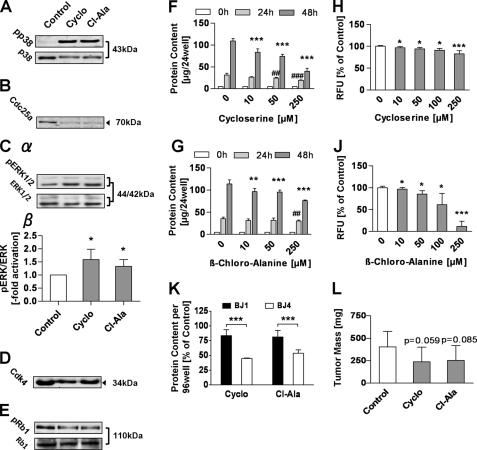
Activation of AMPKα, increased mitochondrial activity, and increased mtROS levels have independently been shown to impair cancer cell growth. These three metabolic states have also been shown to activate p38 MAP kinase signaling (15, 29, 30). Accordingly, we here observed increased phosphorylation and hence activation of p38 following treatment with ALAT inhibitors for 24 h (Fig. 4A). In line with this observation, expression of the phosphatase Cdc25a, known to be regulated by p38, was found to be decreased (Fig. 4B). Cdc25a-dependent dephosphorylation of mitogenic stress kinase ERK was consistently found to be decreased (Fig. 4C), suggesting increased degradation of the protein kinase Cdk4. Correspondingly, expression of protein kinase Cdk4, which in addition has been previously established to be regulated by Cdc25a, was also decreased after treatment with ALAT inhibitors (Fig. 4D). Consistent with the above-mentioned alterations, a decreased phosphorylation of retinoblastoma protein (Rb1) at the Cdk4-specific phosphorylation site Tyr780 was lastly observed (Fig. 4E). Taken together, these findings suggest activation of a signaling cascade that might cause growth inhibition of LLC1 cells treated with either Cyclo or Cl-Ala.
FIGURE 4.
Inhibitors of ALAT activate stress kinase-dependent pathways and inhibit growth of highly malignant cancer cells. A, Western blots of basal and phosphorylated p38 protein after a 24-h treatment with inhibitors. B, representative Western blot of Cdc25a protein after a 24-h treatment with inhibitors (n = 3). Cα, representative Western blots of basal and phosphorylated ERK protein. Cβ, densitometric analysis of basal and phosphorylated ERK (n = 4) (both after a 24-h treatment with inhibitors). D, representative Western blot of Cdk4 protein after a 24-h treatment with inhibitors (n = 3). E, representative Western blots of basal and phosphorylated Rb protein after a 24-h treatment with inhibitors (n = 3). F and G, protein content per well of anchorage-dependent growing cells after a 0, 24-, and 48-h treatment with Cyclo (F) and Cl-Ala (G). H and J, relative fluorescence units measured per soft agar well of anchorage-independent growing cells treated for 6 days with Cyclo (H) and Cl-Ala (J). K, protein content per well of anchorage-dependent growing BJ1 versus BJ4 cells relative to respective control after a 48-h inhibitor treatment. L, tumor masses in nude mice after 2 weeks of treatment with a daily dose of Cyclo (100 mg/kg of BW) or Cl-Ala (20 mg/kg of BW). A–E and K, inhibitors used at a final concentration of 250 μm; error bars represent S.D.; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
ALAT Inhibitors Impair Growth of Cancer Cells in Vitro and in Vivo
The aforementioned alterations of growth signaling may cause reduced growth rates following treatment with ALAT inhibitors. In a first step we investigated the effects of Cyclo and Cl-Ala on anchorage-dependent growth after a 0, 24-, and 48-h treatment. Cyclo showed stronger inhibitory effects than Cl-Ala, whereas both substances significantly reduced protein content after 48 h in a concentration-dependent manner in the range from 10 to 250 μm (Fig. 4, F and G).
We subsequently questioned whether the inhibitors would similarly affect anchorage-independent growth. Using a recently developed, semiautomated soft agar assay (22), we quantified colony formation by LLC1 cells in the absence and presence of ALAT inhibitors at different concentrations. Both inhibitors were capable of reducing colony formation in a concentration-dependent manner in the range from 10 to 250 μm (Fig. 4, H and J).
To test whether the observed growth inhibitory effects of Cyclo and Cl-Ala are specific for malignant cells, and not simply due to an unspecific toxicity, we simultaneously treated nonmalignant BJ fibroblasts (BJ1) and highly malignant BJ fibroblasts (BJ4) with the respective inhibitors. After a 48-h treatment, both inhibitors exerted significantly stronger reduction of cellular protein content of malignant BJ4 cells compared with a relatively mild effect on BJ1 cells, indicating a malignancy-dependent growth inhibitory effect of Cyclo and Cl-Ala (Fig. 4K).
Lastly, we injected LLC1 cells into immune-compromised nude mice and quantified absolute tumor masses after 2 weeks of exposure to ALAT inhibitors. In the groups treated with inhibitors we found tumor masses to be reduced by approximately 40% compared with tumor masses in saline-injected control mice. The statistical analysis revealed a trend toward a significantly reduced LLC1-derived cancer growth in nude mice (Cyclo, p = 0.059; Cl-Ala, p = 0.085) (Fig. 4L). Taken together, these findings indicate that inhibition of ALAT impairs malignant growth by inducing mitochondrial metabolism.
DISCUSSION
In the current study we have used in silico EMA to identify a previously unknown biochemical approach to reinstate mitochondrial metabolism in a highly malignant cancer cell line, culminating in the inhibition of both anchorage-dependent and -independent growth and thereby reducing the malignancy of such cells. Cancer cells are known to exhibit extremely high rates of glycolysis and concurrently reduced mitochondrial activity, as shown repeatedly in the past. These observations are supported by our findings in untreated LLC1 cells (Fig. 2E), which indicate that incorporated d-glucose is mainly converted into l-lactate. Based on EMA predictions, we have tested the possibility to impair glycolysis indirectly by preventing transamination of highly acidic l-lactate into less acidic l-alanine (Fig. 1), which, by applying the corresponding inhibitors, indeed causes reduced l-alanine production and turnover (Fig. 2, A and C). Notably and as predicted, this inability to convert l-lactate into l-alanine would cause excessive and therefore potentially detrimental acidification of the cellular environment, would subsequently reduce d-glucose uptake (Fig. 2D), presumably to prevent excess l-lactate production, and would lead to corresponding decreases in cellular pH. Consistently, both l-lactate production as well as l-alanine content of tumor tissues have previously been shown to correlate positively with tumor malignancy (27, 31). Moreover, high amounts of nutritive d-glucose may cause lactic acidosis in humans suffering from cancers (32), and, most interestingly, alanine is the only amino acid produced by human colon carcinomas in situ (25). Furthermore, the nonoxidative conversion of d-glucose and the oxidation of l-glutamine both contribute to the massive production of l-lactate and l-alanine in cancer cells (9, 33–35).
In cells that are highly dependent on glycolysis, decreased d-glucose uptake should initially cause an energy deficit, as reflected by the decreased intracellular ATP content observed 24 h after addition of the inhibitors (Fig. 3A). However, 48 h after addition of these inhibitors no energy deficit was detected anymore (Fig. 3C), thus suggesting that the cell compensates for the initial deficit by activation of AMPK, as experimentally shown in Fig. 3B. Notably, this kinase has repeatedly been reported to be involved in the control of cancer cell growth (36, 37). Given the initially shown reduction of glycolytic energy conversion, it appears that the cell initiates a compensatory switch to mitochondrial energy conversion, as reflected by increased oxygen consumption rates (Fig. 3E), increased l-glutamine consumption (Fig. 3F), and lastly increased production of mtROS (Fig. 3G). This activation of mitochondria is paralleled by the activation of previously established pathways (Fig. 4, A–E) and inhibits cancer cell growth in an anchorage-dependent (Fig. 4, F and G) and anchorage-independent (Fig. 4, H and J) manner. As shown previously, other anticancer agents, including dichloroacetate (12, 38) and 2-DOG (39, 40), appear to promote mitochondrial metabolism in a similar way. Moreover, genetic approaches to stimulate mitochondrial activity similarly impair cancer growth (15). Finally, there is limited evidence that ALAT inhibitors may affect prokaryotic (41) or malignant cell growth (42–45). These latter effects, however, have been linked to impaired ceramide synthesis, which may occur independently from our current findings. Taken together, we here provide evidence that in silico predictions of inhibitor-based alterations of nutrient metabolism are capable of anticipating their effects on cancer cell metabolism and growth, namely ALAT inhibition to cause an induction of mitochondrial metabolism and subsequently reduced malignancy.
Supplementary Material
Acknowledgments
We thank William C. Hahn at the Dana-Farber Cancer Institute Boston, MA, for providing the BJ-hTERT and BJ-hTERT-st-LT-Hras cell lines; and Beate Laube for excellent technical assistance and Daniel Scharlau and Michael Glei for helpful advice.
This work is part of the research program of the Jena Centre for Systems Biology of Ageing, JenAge, funded by the German Ministry for Education and Research (Bundesministerium für Bildung und Forschung) Grant 0315581.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- 2-DOG
- 2-deoxy-d-glucose
- ALAT
- l-alanine aminotransferase
- AMPK
- AMP-activated protein kinase
- BSTFA
- N,O-bis(trimethylsilyl)trifluoroacetamide
- BW
- body weight
- Cdc25a
- cell division cycle 25 homolog A
- Cdk4
- cyclin-dependent kinase 4
- Cl-Ala
- β-chloro-l-alanine
- Cyclo
- l-cycloserine
- EM
- elementary mode
- EMA
- elementary mode analysis
- LLC1
- Lewis lung carcinoma cells
- mtROS
- mitochondrial reactive oxygen species
- p38 MAPK
- mitogen-activated protein kinase 14
- Rb1
- retinoblastoma 1.
REFERENCES
- 1. Warburg O., Posener K., Negelein E. (1924) Biochem. Zeitschrift. 152, 319–344 [Google Scholar]
- 2. Warburg O. (1956) Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 3. Semenza G. L., Artemov D., Bedi A., Bhujwalla Z., Chiles K., Feldser D., Laughner E., Ravi R., Simons J., Taghavi P., Zhong H. (2001) Novartis Found. Symp. 240, 251–264 [PubMed] [Google Scholar]
- 4. Wallace D. C. (2005) Cold Spring Harbor Symp. Quant. Biol. 70, 363–374 [DOI] [PubMed] [Google Scholar]
- 5. Kim J. W., Dang C. V. (2006) Cancer Res. 66, 8927–8930 [DOI] [PubMed] [Google Scholar]
- 6. Ristow M. (2006) Curr. Opin. Clin. Nutr. Metab. Care 9, 339–345 [DOI] [PubMed] [Google Scholar]
- 7. Pedersen P. L. (2007) J. Bioenerg. Biomembr. 39, 1–12 [DOI] [PubMed] [Google Scholar]
- 8. Ortega A. D., Sánchez-Aragó M., Giner-Sánchez D., Sánchez-Cenizo L., Willers I., Cuezva J. M. (2009) Cancer Lett. 276, 125–135 [DOI] [PubMed] [Google Scholar]
- 9. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pfeiffer T., Schuster S., Bonhoeffer S. (2001) Science 292, 504–507 [DOI] [PubMed] [Google Scholar]
- 11. Sun R. C., Fadia M., Dahlstrom J. E., Parish C. R., Board P. G., Blackburn A. C. (2010) Breast. Cancer Res. Treat. 120, 253–260 [DOI] [PubMed] [Google Scholar]
- 12. Michelakis E. D., Webster L., Mackey J. R. (2008) Br. J. Cancer 99, 989–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu Z., Jiang W., McGinley J. N., Thompson H. J. (2005) Cancer Res. 65, 7023–7030 [DOI] [PubMed] [Google Scholar]
- 14. Chen Z., Lu W., Garcia-Prieto C., Huang P. (2007) J. Bioenerg. Biomembr. 39, 267–274 [DOI] [PubMed] [Google Scholar]
- 15. Schulz T. J., Thierbach R., Voigt A., Drewes G., Mietzner B., Steinberg P., Pfeiffer A. F., Ristow M. (2006) J. Biol. Chem. 281, 977–981 [DOI] [PubMed] [Google Scholar]
- 16. Schuster S., Fell D. A., Dandekar T. (2000) Nat. Biotechnol. 18, 326–332 [DOI] [PubMed] [Google Scholar]
- 17. Schwarz R., Musch P., von Kamp A., Engels B., Schirmer H., Schuster S., Dandekar T. (2005) BMC Bioinformatics 6, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pfeiffer T., Sánchez-Valdenebro I., Nuño J. C., Montero F., Schuster S. (1999) Bioinformatics 15, 251–257 [DOI] [PubMed] [Google Scholar]
- 19. Hahn W. C., Counter C. M., Lundberg A. S., Beijersbergen R. L., Brooks M. W., Weinberg R. A. (1999) Nature 400, 464–468 [DOI] [PubMed] [Google Scholar]
- 20. Zimmermann S., Zarse K., Schulz T. J., Siems K., Müller-Kuhrt L., Birringer M., Ristow M. (2008) Horm. Metab. Res. 40, 29–37 [DOI] [PubMed] [Google Scholar]
- 21. Sweetmann L. (1991) in Techniques in Diagnostic Human Biochemical Genetics: A Laboratory Manual (Hommes F. ed) pp. 143–176, Wiley-Liss, New York [Google Scholar]
- 22. Thierbach R., Steinberg P. (2009) Anal. Biochem. 387, 318–320 [DOI] [PubMed] [Google Scholar]
- 23. Cornell N. W., Zuurendonk P. F., Kerich M. J., Straight C. B. (1984) Biochem. J. 220, 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Golichowski A., Jenkins W. T. (1978) Arch. Biochem. Biophys. 189, 109–114 [DOI] [PubMed] [Google Scholar]
- 25. Holm E., Hagmüller E., Staedt U., Schlickeiser G., Günther H. J., Leweling H., Tokus M., Kollmar H. B. (1995) Cancer Res. 55, 1373–1378 [PubMed] [Google Scholar]
- 26. Neermann J., Wagner R. (1996) J. Cell. Physiol. 166, 152–169 [DOI] [PubMed] [Google Scholar]
- 27. Ziegler A., von Kienlin M., Décorps M., Rémy C. (2001) Cancer Res. 61, 5595–5600 [PubMed] [Google Scholar]
- 28. Hardie D. G. (2011) Am. J. Clin. Nutr. 93, 891S–896S [DOI] [PubMed] [Google Scholar]
- 29. Bulavin D. V., Fornace A. J., Jr. (2004) Adv. Cancer Res. 92, 95–118 [DOI] [PubMed] [Google Scholar]
- 30. Fruman D. A., Edinger A. L. (2008) Biochem. J. 412, e3–5 [DOI] [PubMed] [Google Scholar]
- 31. Cornel E. B., Smits G. A., Oosterhof G. O., Karthaus H. F., Deburyne F. M., Schalken J. A., Heerschap A. (1993) J. Urol. 150, 2019–2024 [DOI] [PubMed] [Google Scholar]
- 32. Goodgame J. T., Jr., Pizzo P., Brennan M. F. (1978) Cancer 42, 800–803 [DOI] [PubMed] [Google Scholar]
- 33. Márquez J., Sánchez-Jiménez F., Medina M. A., Quesada A. R., Núñez de Castro I. (1989) Arch. Biochem. Biophys. 268, 667–675 [DOI] [PubMed] [Google Scholar]
- 34. DeBerardinis R. J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., Thompson C. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dröge W., Eck H. P., Kriegbaum H., Mihm S. (1986) J. Immunol. 137, 1383–1386 [PubMed] [Google Scholar]
- 36. Kato K., Ogura T., Kishimoto A., Minegishi Y., Nakajima N., Miyazaki M., Esumi H. (2002) Oncogene 21, 6082–6090 [DOI] [PubMed] [Google Scholar]
- 37. Ashrafian H. (2006) Lancet 367, 618–621 [DOI] [PubMed] [Google Scholar]
- 38. Stacpoole P. W., Greene Y. J. (1992) Diabetes Care 15, 785–791 [DOI] [PubMed] [Google Scholar]
- 39. Cay O., Radnell M., Jeppsson B., Ahrén B., Bengmark S. (1992) Cancer Res. 52, 5794–5796 [PubMed] [Google Scholar]
- 40. Ralser M., Wamelink M. M., Struys E. A., Joppich C., Krobitsch S., Jakobs C., Lehrach H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17807–17811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Manning J. M., Merrifield N. E., Jones W. M., Gotschlich E. C. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laske R., Schönenberger H., Holler E. (1989) Arch. Pharm. 322, 857–862 [DOI] [PubMed] [Google Scholar]
- 43. Riboni L., Prinetti A., Bassi R., Caminiti A., Tettamanti G. (1995) J. Biol. Chem. 270, 26868–26875 [DOI] [PubMed] [Google Scholar]
- 44. Cinatl J., Jr., Cinatl J., Kotchetkov R., Pouckova P., Vogel J. U., Rabenau H., Michaelis M., Kornhuber B. (1999) Anticancer Res. 19, 5349–5354 [PubMed] [Google Scholar]
- 45. Charles A. G., Han T. Y., Liu Y. Y., Hansen N., Giuliano A. E., Cabot M. C. (2001) Cancer Chemother. Pharmacol. 47, 444–450 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.