Abstract
Warburg effect is a hallmark of cancer manifested by continuous prevalence of glycolysis and dysregulation of oxidative metabolism. Glycolysis provides survival advantage to cancer cells. To investigate molecular mechanisms underlying the Warburg effect, we first compared oxygen consumption among hFOB osteoblasts, benign osteosarcoma cells, Saos2, and aggressive osteosarcoma cells, 143B. We demonstrate that, as both proliferation and invasiveness increase in osteosarcoma, cells utilize significantly less oxygen. We proceeded to evaluate mitochondrial morphology and function. Electron microscopy showed that in 143B cells, mitochondria are enlarged and increase in number. Quantitative PCR revealed an increase in mtDNA in 143B cells when compared with hFOB and Saos2 cells. Gene expression studies showed that mitochondrial single-strand DNA-binding protein (mtSSB), a key catalyst of mitochondrial replication, was significantly up-regulated in 143B cells. In addition, increased levels of the mitochondrial respiratory complexes were accompanied by significant reduction of their activities. These changes indicate hyperactive mitochondrial replication in 143B cells. Forced overexpression of mtSSB in Saos2 cells caused an increase in mtDNA and a decrease in oxygen consumption. In contrast, knockdown of mtSSB in 143B cells was accompanied by a decrease in mtDNA, increase in oxygen consumption, and retardation of cell growth in vitro and in vivo. In summary, we have found that mitochondrial dysfunction in cancer cells correlates with abnormally increased mitochondrial replication, which according to our gain- and loss-of-function experiments, may be due to overexpression of mtSSB. Our study provides insight into mechanisms of mitochondrial dysfunction in cancer and may offer potential therapeutic targets.
Keywords: Cell Metabolism, Gene Regulation, Mitochondria, Mitochondrial DNA, Respiratory Chain, Cancer, Mitochondrial Replication, mtSSB, Warburg Effect
Introduction
One of the landmark phenotypical characteristics attributed to cancer cells is their predominant reliance on glycolysis. In the 1920s, Otto Warburg demonstrated that tumor cells metabolize glucose at a much higher rate when compared with their non-malignant counterparts. This phenomenon has been referred to as the “Warburg effect” (1, 2). It is manifested by dysregulation of the biochemical pathway that mediates oxidative metabolism of glucose in the cell. Furthermore, recent studies have shown that cells predominantly relying on glycolysis to satisfy their energy needs exhibit increased resistance to pro-apoptotic signals and do not respond well to anti-cancer treatment (3–5). The molecular mechanisms underlying the Warburg effect in tumorigenesis remain poorly understood. Initially, it has been suggested that a unique defect in oxidative metabolism in tumor cells results in a corresponding shift to the glycolytic pathway (2). However, subsequent observations revealed similar patterns in normal cells under conditions of rapid proliferation, where the majority of glucose was converted to lactate, effectively excluding the possibility that aerobic glycolysis is a feature unique to malignant cells or that it is indicative of any unique defect in the oxidative metabolism (6, 7). In contrast, several highly proliferative tumor cell lines have been shown to exhibit normal characteristics of oxidative metabolism (8).
The present report attempts to elucidate the role of mitochondria in the Warburg phenomenon. Here, we hypothesize that in the most aggressive human osteosarcoma cell line, 143B, defects in mitochondrial morphogenesis directly contribute to decreased oxygen consumption and abnormal proliferation of the cells. Based on this hypothesis, we have analyzed parameters of mitochondrial function in different osteosarcoma cells that demonstrate varying degrees of proliferative capacity and the invasion potential.
EXPERIMENTAL PROCEDURES
Materials
Chemicals were from Sigma unless otherwise noted. Cell culture media, ingredients, and antibiotics were from Invitrogen. Oligonucleotides were custom made by IDT (Coralville, IA). Primary antibodies against human mtSSB2 and human β-actin were from Novus Biologicals (Littleton, CO) and Sigma, respectively. Secondary antibodies, Precision Plus molecular weight markers, dry milk, and buffer ingredients for immunoblotting were from Bio-Rad. shRNA against mtSSB was from Santa Cruz (Santa Cruz, CA). FuGENE® HD transfection reagent was from Roche Applied Science.
Cell Culture
Human fetal osteoblasts transformed with SV40 T antigen, hFOB 1.19, as well as osteosarcoma cells, Saos2 and 143B, were obtained from ATCC (Manassas, VA). hFOB cells were cultured in DMEM/F-12 medium, and Saos2 and 143B cells were cultured in DMEM medium at 37 °C. All media were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin mixture.
Cell Growth Assay
Cells were initially plated on 6-well plates at a density of 200,000 per well. After 48 h of incubation, cells were trypsinized, resuspended in 1 ml of PBS, and counted using a Cellometer® automated cell counter from Nexcelom Bioscience (Lawrence, MA).
Matrigel Invasion Assay
Cell invasion was assayed using Matrigel Invasion Chambers manufactured by BD Bioscience. Cells were seeded in control and Matrigel chambers at a density of 5 × 104 cells per chamber in 0.5 ml of serum-free DMEM. Chambers were placed in wells with DMEM containing 10% FBS. After 48 h of incubation at 37 °C, chambers were removed, and membrane tops were scrubbed with a wet cotton swab to remove non-invading cells. Chambers were then placed in methanol for 2 min, incubated for 2 min in 1% toluidine blue solution containing 1% borax, rinsed with water, and air-dried. Membranes were removed from chambers with a scalpel and mounted on glass slides. The number of cells migrated through Matrigel membranes was counted and divided by the number of cells migrated through control membranes to determine the cell invasion index.
Oxygen Consumption
Rates of oxygen consumption in cell suspensions were measured with a Clark-type oxygen electrode (Microelectrodes, Inc; Bedford, NH) that was placed in a custom-made air-tight temperature-controlled chamber. After 300 μl of medium at 37 °C containing re-suspended cells at 2 × 106 per ml was added, the chamber was sealed. After signal stabilization, oxygen consumption was recorded for 15 min, and then a reducing agent, dithionite, was added to calibrate for 0% oxygen content. The signal trace was digitized on a computer connected to the electrode via a data acquisition device (DATAQ® Instruments; Akron, OH) using version 2.54 of WinDaqTM Waveform Browser software (DATAQ® Instruments; Akron, OH).
Electron Microscopy
Samples were prepared as described previously (9). Briefly, pelleted cultured cells were fixed in 2.5% glutaraldehyde in 0.1 m phosphate buffer, post-fixed in 1.0% OsO4, trapped in 4% agarose, dehydrated with ethanol, and embedded in Spurr epoxy resin. Tissue pieces of xenografted tumors were fixed in 2.5% glutaraldehyde in 0.1 m sodium cacodylate buffer, post-fixed in 1.0% OsO4, dehydrated with ethanol, and transitioned into propylene oxide for infiltration and embedding into EPON/Araldite epoxy resin. Thin sections were cut onto grids, stained with uranyl acetate and lead citrate, examined with a H-7650 Hitachi transmission electron microscope, and digitally photographed. Archival human osteosarcoma epoxy-embedded tissue blocks were thin-sectioned and photographed. Cell area occupied by either mitochondria or the cytosol was outlined and measured with ImageJ software. A ratio of the mitochondrial area to the cytosolic area was calculated. A total of 30 cells per sample were analyzed.
Real-time RT-PCR
Total cellular RNA was isolated using the RNeasy Mini kit by Qiagen (Valencia, CA) and reverse-transcribed into cDNA using iScript cDNA synthesis kit by Bio-Rad according to the manufacturer's instructions. One μg of cDNA was subjected to real-time quantitative PCR using primer pairs for the following human genes: mtSSB (5′-TGA ATC GTG TGC ACT TAC TTG GGC-3′ and 5′-GAT TGT TGT TGC TTG TCG CCT CAC-3′), polymerase G (5′-GTG GCC ATG AAG TGG CTG TTT GAA-3′ and 5′-TCA TTC AGA CCC AGC TTG TAG GCA-3′), and GAPDH (5′-GAG TCA ACG GAT TTG GTC GT-3′ and 5′-GAC AAG CTT CCC GTT CTC AG-3′). Real-time PCR was performed using the RotorGene® real-time DNA amplification system (Qiagen®). SYBR Green reagent produced by Abgene (Rockford, IL) was used to monitor DNA synthesis. The expression of proteins of interest was normalized to the expression of GAPDH.
Mitochondrial DNA Quantitation
Total cellular DNA was isolated using the Wizard® Genomic DNA purification kit by Promega (Madison, WI), according to the manufacturer's instructions. 250 ng of DNA per sample was subjected to real-time quantitative PCR analysis using SYBR Green reagent and RotorGene® real-time DNA amplification system as described above. To detect mtDNA levels, we used a primer pair for the mtDNA-encoded human gene, ND1 (5′-TGG GTA CAA TGA GGA GTA GG-3′ and 5′-GGA GTA ATC CAG GTC GGT-3′). To detect nuclear DNA levels, we used a primer pair for the nuclear-encoded human β-actin (5′-TCA CCC ACA CTG TGC CCA TCT ACG A-3′ and 5′-CAG CGG AAC CGC TCA TTG CCA ATG G-3′). mtDNA was normalized to nuclear DNA.
Western Blotting
Cells were lysed, and the protein concentration in lysates was measured using the Bradford assay. Twenty-five μg of total protein per sample was mixed 1:1 with 2× Laemmli buffer, boiled, and subsequently subjected to electrophoresis using NuPage® pre-cast 4–12% gradient polyacrylamide gels (Invitrogen). The samples were electroblotted onto polyvinylidene difluoride membranes. Blots were blocked in 5% dry milk dissolved in PBS containing 0.01% of Tween 20 (PBST), probed with primary antibody resuspended in 2.5% dry milk dissolved in PBST at 2 μg/ml, incubated with horseradish peroxidase-conjugated secondary antibody resuspended in 2.5% dry milk dissolved in PBST at 0.2 μg/ml, developed using SuperSignal WestPico chemiluminescent substrate produced by Thermo Scientific (Rockford, IL), and photographed. To verify equal loading, blots were stripped in Re-Blot Plus stripping buffer produced by Millipore (Billerica, MA), re-probed with β-actin antibody resuspended in 2.5% dry milk in PBST at 0.5 μg/ml, and developed as described above. Band intensities were measured using densitometry and Adobe® Photoshop® software.
Isolation of Mitochondria
Cells were resuspended in MS buffer (195 mm mannitol, 65 mm sucrose, 2 mm HEPES, pH 7.4, 0.05 mm EGTA, 10 μm MgCl2, 0.5 mg/ml BSA, 0.05 mm KPi), homogenized in a glass Teflon homogenizer (15 strokes), and centrifuged in Hermle centrifuge at 3500 rpm at 4 °C for 3 min. The resulting supernatant was further centrifuged at the same temperature at 10,500 rpm for 9 min. After the spin-down, the pellet was resuspended in the MS buffer, centrifuged at 9000 rpm for 9 min, and finally resuspended in 100 μl of the MS buffer with/without BSA. Mitochondrial protein concentration was determined using the Bradford method.
Non-denaturing Clear Native Gel Separation of Mitochondrial Proteins
Isolated mitochondria were incubated for 20 min in clear native extraction buffer consisting of dodecyl maltoside (1%), NaCl (50 mm), imidazole (50 mm), aminocaproic acid (2 mm), and EDTA (1 mm), pH 7.0, at 4 °C. Extractions were centrifuged at 10,000 × g for 5 min to pellet non-soluble proteins. The supernatant was loaded directly onto the gel using a 50% glycerol, 0.1% Ponceau loading buffer. The gel (5–12.5% gradient, 25 mm imidazole, 500 mm aminocaproic acid) was run at 100 V for the first hour and with a 15-mA limit for the remainder of the run. The anode buffer consisted of imidazole (25 mm), and the cathode buffer consisted of imidazole (7.5 mm), Tricine (50 mm), deoxycholate (0.05%), and dodecyl maltoside (0.02%), all at pH 7.0 at 4 °C. Gels were visualized with silver staining, photographed, and analyzed using Adobe software.
Activity of Mitochondrial Enzyme Complexes
For complex I and II enzyme activity assays, the electron transfer was monitored by observing colorimetric absorbance of 2,6-dichlorophenolindophenol, a dye that absorbs at 600 nm and becomes colorless by accepting electrons from ubiquinol. Ubiquinol production was dependent on the ability of complex I or complex II enzymes to reduce synthetic analogs of ubiquinone, ubiquinone-1 and -2, respectively. The solution for either complex activity assay contained 50 mm phosphate buffer, 0.1 mm EDTA, 45 μm 2,6-dichlorophenolindophenol, 1 mm KCN, and 2.5 mg/ml BSA. Mitochondrial protein was added at 100 μg/ml concentration. For complex I activity, an initial incubation to minimize substrate-dependent nonlinear rate was followed by monitoring absorbance at 600 nm for 5 min after the addition of ubiquinone-1 at 0.1 mm in the presence of complex-specific substrates, 10 mm glutamate, and 5 mm malate. 10 μm rotenone was then added, and the inhibited absorbance was monitored for additional 5 min.
For complex II activity, 0.05 mm ubiquinone-2, 1 μm rotenone, and complex-specific substrate, 20 mm succinate, were added to the assay solution, and the absorbance rate was monitored at 600 nm for 5 min. 1 mm thenoyltrifluoroacetone was then added, and the inhibited absorbance was monitored for an additional 5 min. The corrected absorbance rate was calculated by subtracting the inhibited rate from the initial rate for the corresponding enzyme complex and subsequently multiplying the obtained value by the extinction coefficient for 2,6-dichlorophenolindophenol.
For the complex III assay, production of reduced cytochrome c as a result of enzymatic activity of ubiquinol cytochrome c reductase was followed for 5 min at 550 nm in the presence of 100 μg/ml mitochondrial protein sample. Cyanide was added to prevent re-oxidation of cytochrome c by complex IV. The assay solution contained 50 mm phosphate buffer, 1 mm EDTA, 5 mm MgCl2, 20 mm KCN, 1 μm rotenone, 15 μm cytochrome c, and 15 μm ubiquinol. Non-enzymatic reduction of cytochrome c was determined initially and subtracted from the experimental absorbance values after addition of ubiquinol.
For complex IV assay, oxidation of 50 μm reduced cytochrome c was followed for 5 min at 550 nm in the presence of 100 μg/ml mitochondrial protein in 10 mm phosphate buffer. The slope of the absorbance curve was used to determine the rate constant for cytochrome oxidase activity.
Stable Overexpression of mtSSB
Human mtSSB cDNA was PCR-amplified from the human cDNA library, subcloned into the Topo II TA vector (Invitrogen), excised from the Topo II TA vector using EcoRI, and inserted into the EcoRI site of the pcDNA3.1+ vector. Correct orientation and nucleotide composition of mtSSB was verified by sequencing. Saos2 cells were transfected with the pcDNA-mtSSB construct using FuGENE® HD reagent (Roche Applied Science). After 48 h, selection was started with 750 μg/ml Geneticin (Invitrogen) and continued for 2 weeks. Stable mtSSB overexpression was confirmed with immunoblotting and persisted for at least five passages.
Stable Knock-down of mtSSB
MtSSB knock-down in 143B cells was achieved by transfection with the plasmid carrying shRNA against human mtSSB made by Santa Cruz (Santa Cruz, CA). Infected cells were selected using 2 μg/ml puromycin for 2 weeks, and stable mtSSB knock-down in 143B-KD-mtSSB was verified with immunoblotting and persisted for at least 10 passages. As a control, a subset of 143B cells was transfected with the control plasmid carrying scrambled sequence.
In Vivo Tumor Growth
Wild-type 143B or 143B-KD-mtSSB cells were detached from plates and mixed 1:1 with Matrigel, and 500,000 cells in a 100-μl suspension were injected subcutaneously into the right flanks of nude mice. Five mice per group were injected. Tumors were measured in two dimensions using digital calipers at days 7, 14, and 21. Tumor size was calculated using the formula Tumor Volume = Length × Width2 × π/6 (10).
Statistical Analysis
Experiments were repeated 3–5 times. Data were analyzed using Prism, Version 5.01 for Windows, by GraphPad Software (San Diego, CA). Mean values and S.E. of the mean were calculated, and the statistical significance was established using either Student's t test or one-way analysis of variance as appropriate. When analysis of variance revealed statistical significance, Dunnett's multiple comparison post hoc analysis was performed to confirm differences between experimental groups. Data with p < 0.05 were considered statistically significant.
RESULTS
Growth Rate and the Invasion Profile of Osteosarcoma Cells
For the experiments in this study, we used two cell lines derived from human osteosarcoma patients, Saos2 and 143B. Saos2 cells are neither tumorigenic nor metastatic, whereas 143B cells are tumorigenic and metastatic when injected into mice (11–13). For the non-malignant control, we included an immortalized human osteoblastic cell line, hFOB (14). hFOB cells are transformed with a temperature-sensitive SV40 T large antigen that is active at 33 °C (14). To avoid potential artifacts caused by the active SV40 T large antigen, we used hFOB cells at a non-permissive temperature of 37 °C. These conditions allow adequate growth of hFOB cells and have been used in previous studies (15).
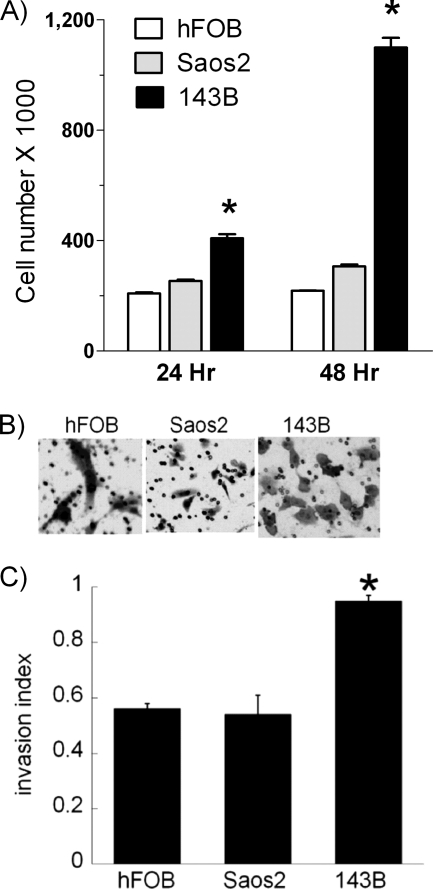
To assess the phenotype of Saos2 and 143B cells, we first compared their growth rate to hFOB cells after 24 or 48 h of incubation. As shown in Fig. 1A, growth rate of Saos2 cells was comparable with the growth rate of non-malignant hFOB cells, whereas143B cells grew almost twice as fast. To assess invasiveness of cells, we performed an invasiveness assay using Matrigel-coated membranes. The microphotograph in Fig. 1B demonstrates that the number of toluidine-positive Saos2 cells penetrating the Matrigel coating of the membrane is similar to that of hFOB cells, whereas a significantly larger number of 143B cells invade the membrane (right panel). Quantitative analysis of the cell invasion index confirms the above observation (Fig. 1C). These assays demonstrate that growth and invasiveness of Saos2 cells are similar to those of non-malignant hFOB cells, whereas 143B cells possess much higher proliferative and invasive capacity, confirming the benign and aggressive phenotypes of Saos2 and 143B osteosarcoma cells, respectively.
FIGURE 1.
Proliferation rate and invasive potential of osteosarcoma cells. A, hFOB, Saos2, and 143B cells were seeded in 6-well plates at the density of 200,000 cells per well. After 24 or 48 h of incubation, cells were trypsinized, harvested, and counted using an automatic cell counter. B, hFOB, Saos2, and 143B cells were seeded onto either control or Matrigel-coated membranes of the invasion chambers at the density of 50,000 cells per chamber. After 48 h of incubation, cells that have penetrated through the membranes were stained with toluidine blue to assess the invasive potential. C, quantitative analysis of the invasive potential was performed by calculating the invasion index, as described under “Experimental Procedures.” Values represent the means ± S.E.; n = 3–4 independent wells. *, p < 0.05, as compared with hFOB cells.
Respiratory Capacity of the Studied Cancer Cells
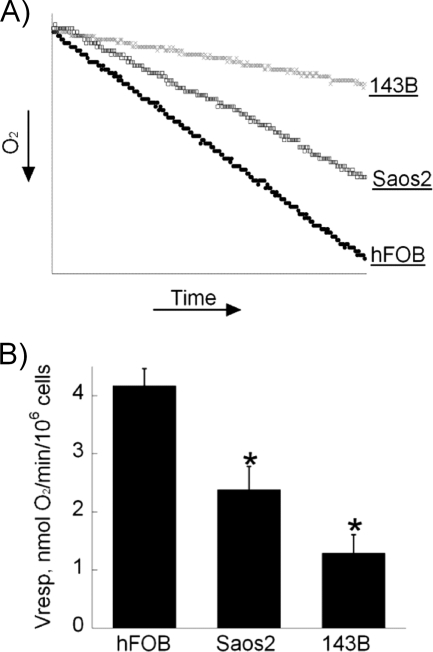
We next examined the rate of oxygen consumption by the Saos2 and 143B cancer cells (Fig. 2A). Using a Clark oxygen electrode, we detected that in comparison to hFOB cells, Saos2 and 143B osteosarcoma cells exhibited various degrees of decrease in the respiratory capacity (Fig. 2B). Whereas hFOB cells consumed more than 4 nmol of O2/min/106 cells, oxygen consumption by benign Saos2 cells was almost half that level, whereas aggressive 143B cells consumed only 1.3 nmol of O2/min/106 cells. These results suggest an apparent inhibition of the functioning of the respiratory chain in malignant cells in solid tumor cell lines. The degree of such inhibition appears to correlate with the malignant potential of the studied cells.
FIGURE 2.
Oxygen consumption by osteosarcoma cells is significantly decreased. A, a graphic representation of the oxygen consumption rate is shown. 600,000 cells were suspended in 300 μl of medium at 37 °C in a sealed chamber, and oxygen consumption (Vresp.) was measured for 15 min with a Clark-type electrode. B, respiratory capacity of the cells as expressed in nmol of oxygen/min/1 million cells is shown. Values represent the means ± S.E.; n = 3 independent measurements. *, p < 0.05, as compared with hFOB cells.
Mitochondria Increase in Size and Number in Malignant Cells
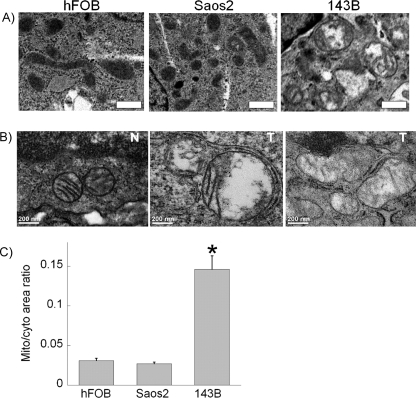
We next assessed the morphology of mitochondria in the studied cells using electron microscopy. As shown in Fig. 3A, mitochondria in hFOB and Saos2 cells show normal “orthodox” morphology, whereas mitochondria in 143B cells increase in size and number. To determine whether the observed phenomena persist in vivo and are not specific to cell culture, we xenografted 143B cells subcutaneously into nude mice and, after 3 weeks, excised developed tumors and processed them for electron microscopy. Fig. 3B shows that osteosarcoma tumors derived from 143B cells reveal mitochondrial morphology similar to that observed in vitro (middle panel), whereas mitochondrial morphology in the adjacent non-tumor tissue is normal and orthodox (left panel). We also examined archival specimens of osteosarcoma and observed changes in mitochondria similar to those in 143B cells and tumor xenografts (Fig. 3B, right panel). Quantitative analysis of the area occupied by mitochondria relative to the cytosolic area revealed that the ratio of mitochondria to the cytosol dramatically increased in aggressive 143B osteosarcoma cells compared with either non-malignant hFOB or benign Saos2 osteosarcoma cells (Fig. 3C). These experiments indicate an increase in the mitochondrial mass in aggressive cancer cells that correlates with the degree of inhibition of mitochondrial function.
FIGURE 3.
Changes in mitochondrial morphology of osteosarcoma cells in vitro and in vivo. Thin sections were prepared for electron microscopic analysis from pellets of hFOB, Saos2, and 143B cells (A) or tissue obtained from xenografted tumors (B, middle panel) and archival tissue blocks (B, right panel) as described under “Experimental Procedures.” In B, N and T stand for normal or tumor tissue, respectively. C, quantitative analysis of changes in mitochondrial morphology expressed as the ratio of the area occupied by mitochondria relative to the area of the cytosol is shown. Values represent the means ± S.E.; n = 30 cells per sample analyzed. *, p < 0.05, as compared with hFOB cells.
Increased Mitochondrial Replication in 143B Cells
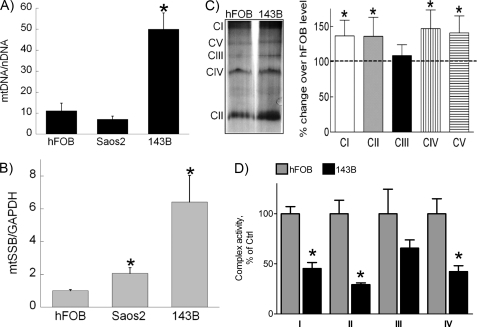
To determine whether the observed increase in mitochondrial mass is due to increased mitochondrial replication, we analyzed mtDNA levels in hFOB, Saos2, and 143B cells (Fig. 4A). Real-time quantitative PCR analysis revealed that the level of mtDNA was significantly increased in 143B cells, indicating a state of abnormally increased mitochondrial replication. To elucidate possible mechanisms of induced mitochondrial replication in cancer cells, we measured expression levels of known regulators of mitochondrial replication machinery, polymerase G and mtSSB (16–19). As shown in Fig. 4B, compared with hFOB cells, mtSSB expression in Saos2 cells was slightly higher and in 143B cells was significantly (6-fold) higher. Polymerase G expression was similar in all studied cells (data not shown). These results demonstrate that mtSSB, which is known to be a key catalyst of mitochondrial replication (16–19), is overexpressed in highly malignant 143B cells and accompanies increased mtDNA levels. Together the data presented in Figs. 3 and 4, such as an increase in mitochondrial mass, mtDNA, and mtSSB levels, indicate the state of abnormally increased mitochondrial replication in the most aggressive 143B cells that correlates with inhibition of respiratory capacity.
FIGURE 4.
Levels of mitochondrial DNA, mtSSB, and respiratory complexes in osteosarcoma cells. A, total cellular DNA was isolated and subjected to real-time PCR analysis to determine the amount of mtDNA in the studied cells. The data are expressed as a ratio between the amounts of ND1, an mtDNA-encoded gene, and β-actin, a nuclear-encoded gene. B, total cellular RNA was isolated and reverse-transcribed. Real-time PCR analysis was performed to determine expression levels of several genes implicated in mitochondrial biogenesis. Experimental data were normalized to the expression of GAPDH. C, mitochondria were isolated from hFOB and 143B cells, and mitochondrial proteins were separated using non-denaturing gel electrophoresis. The plot depicts densitometric changes in protein level of 143B cells compared with hFOB cells. D, isolated mitochondria was used for assay of enzyme activities of respiratory complexes I-IV in hFOB and 143B cells, as described under “Experimental Procedures.” The data are expressed as percentage of activity compared with hFOB cells. Values represent the means ± S.E.; n = 3 independent measurements. *, p < 0.05, as compared with hFOB cells.
Increased Protein Levels of Mitochondrial Respiratory Complexes Are Accompanied by Reduced Respiratory Chain Enzyme Activity
To assess how the described changes in the expression of mtSSB and mtDNA may affect mitochondrial function, we examined protein levels and activity of mitochondrial respiratory chain complexes. Gel separation of mitochondrial protein revealed that 143B cells express significantly larger amounts of the enzyme components that comprise complexes I, II, and IV (Fig. 4C). On the other hand, when enzymatic activity of complexes I through IV was assayed, we were able to detect significantly lower activity of complexes I, II, and IV with a 70, 54, and 57% decrease in catalytic activity, respectively (Fig. 4D). Even though the activity of mitochondrial complex III was reduced by 34%, it did not reach statistical significance.
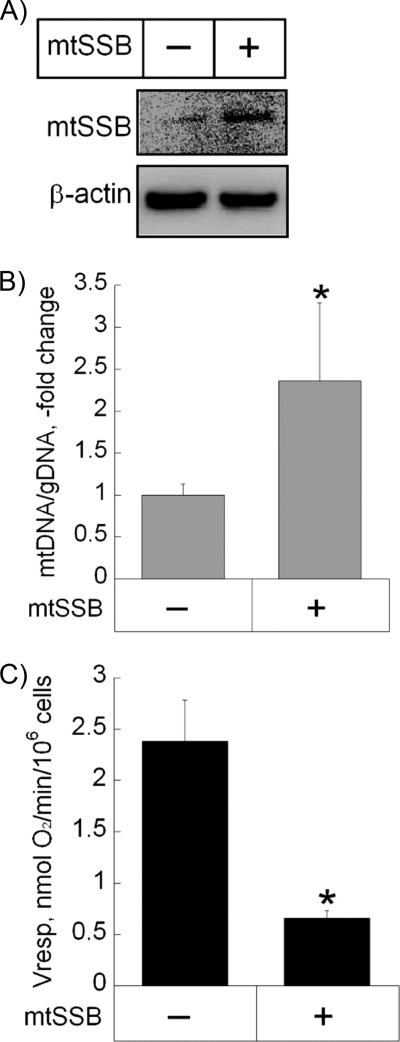
Effect of Gain of mtSSB Function on Cancer Cell Respiration
To determine the contribution of mtSSB and of mitochondrial replication to the observed dysregulation of mitochondrial function, we performed a stable transfection of benign osteosarcoma Saos2 cells with an expression vector containing cDNA for human mtSSB. Overexpression of mtSSB in Saos2-mtSSB cells was confirmed by immunoblotting (Fig. 5A). Increased levels of mtSSB were accompanied by a 2.4-fold increase in the mtDNA level (Fig. 5B), indicating increased mitochondrial replication. Additionally, overexpression of mtSSB was accompanied by a significant decrease in oxygen consumption (Fig. 5C). These data further support our hypothesis that mtSSB overexpression leads to abnormally increased mitochondrial replication accompanied by decreased respiratory function in cancer cells.
FIGURE 5.
Effect of gain of mtSSB function on mitochondrial mass and function. A, shown is a representative microphotograph of an immunoblot demonstrating stable mtSSB overexpression in Saos2 cells. Cells were transfected with the pcDNA-mtSSB construct, and the levels of mtSSB protein were confirmed for at least five passages. B and C, shown is the effect of mtSSB overexpression on the amount of mtDNA (B) and oxygen consumption (C) in Saos2 cells. gDNA, genomic DNA. Mitochondrial DNA content and the rate of oxygen consumption were determined as described under “Experimental Procedures.” Values represent the means ± S.E.; n = 3 independent measurements. *, p < 0.05, as compared with non-transfected Saos2 cells.
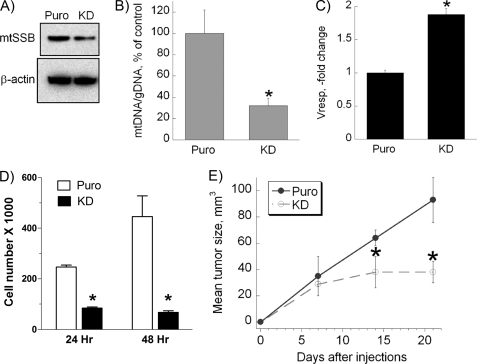
Effect of Loss of mtSSB Function on Respiration and Growth of Cancer Cells
To further assess significance of mtSSB in mitochondrial dysregulation in cancer, we conducted a loss of function study by performing a knockdown of mtSSB expression in 143B cells. As shown in Fig. 6A, using anti-mtSSB shRNA, we were able to achieve a 50% reduction in the level of mtSSB protein in 143B cells compared with the control. This down-regulation resulted in a 70% decrease in mtDNA level (Fig. 6B). Moreover, the decrease in mtSSB protein levels and mtDNA was accompanied by a corresponding 2-fold increase in oxygen consumption by 143B-KD-mtSSB cells, as demonstrated in Fig. 6C. The changes in mtDNA levels and respiratory capacity in the presence of decreased levels of mtSSB were also accompanied by a markedly slowed rate of growth of these cells both in vitro (Fig. 6D) and in subcutaneous tumor xenografts in nude mice in vivo (Fig. 6D). These data indicate that targeting mtSSB in highly malignant cells leads to slowdown of mitochondrial replication, normalization of mitochondrial function and suppression of tumor growth.
FIGURE 6.
Effect of loss of mtSSB function on mitochondrial mass, oxygen consumption, and cell growth. A, shown are representative microphotographs of an immunoblot demonstrating stable mtSSB knockdown (KD) in 143B cells that was achieved by using a viral vector containing shRNA against human mtSSB. The knockdown was confirmed for at least 10 passages. B and C, shown is the effect of mtSSB knockdown on the amount of mtDNA (B) and oxygen consumption (Vresp.) (C) in 143B cells. Mitochondrial DNA content and the rate of oxygen consumption were determined as described under “Experimental Procedures.” D and E, shown is the effect of mtSSB loss-of-function on the growth of cancer cells in vitro and in vivo. D, wild-type 143B or 143B-KD-mtSSB cells were seeded in 6-well plates at the density of 100,000 cells per well. After 24 or 48 h of incubation, the cells were trypsinized, harvested, and counted using an automatic cell counter. E, wild-type 143B or 143B-KD-mtSSB cells were injected subcutaneously into mice. Tumor size was measured at 7, 14, and 21 days. Values represent the means ± S.E.; n = 4 independent measurements. *, p < 0.05, as compared with non-transfected 143B cells.
DISCUSSION
In summary, our key findings indicate the following. 1) The 143B cell line, which represents an aggressive, poorly differentiated variety of human osteosarcoma, demonstrates a markedly increased proliferation rate and invasion capacity compared with non-malignant hFOB cells. At the same time, benign osteosarcoma cells, Saos2, reveal the degree of proliferation and invasiveness that is similar to hFOB cells. 2) Highly aggressive 143B osteosarcoma cells show the most pronounced inhibition of cell respiration. 3) Mitochondria in 143B cells increase in size and number and have elevated mtDNA levels as well as increased expression levels of mtSSB, a protein implicated in mitochondrial biogenesis. 4) Protein levels of mitochondrial respiratory chain complexes are elevated in 143B cells, a finding that is accompanied by a significant decrease in their enzymatic activity. 5) Gain-of-function experiments demonstrate that in the presence of overexpressed mtSSB, benign osteosarcoma Saos2 cells have increased mtDNA levels and become less dependent on oxidative phosphorylation. On the other hand, shRNA-mediated knockdown of mtSSB in 143B cells leads to a decrease in mtDNA, a noticeable improvement in the respiratory capacity of the cells, and a significant reduction in the growth rate both in vitro and in vivo. Based on these experimental data, we conclude that in highly aggressive cancer cells, mitochondrial dysfunction may be due to the abnormally increased mitochondrial replication due to overexpression of mtSSB, resulting in accumulation of large number of non-functional “immature” organelles. It is, however, important to note that the above changes cannot be explained exclusively by the aberrant mitochondrial replication because complex II, which is not coded by mtDNA, was also overexpressed in 143B cells, and, therefore, nuclear gene regulation was also somehow affected. This effect was beyond the scope of our study and needs further attention in future work.
It has been shown previously that oxidative phosphorylation may directly correlate with aggressiveness in cancer (20, 21). We provide additional evidence that in osteosarcoma, the most aggressive and poorly differentiated cell line, 143B, demonstrates the lowest rate of oxygen consumption and the most invasive phenotype compared with both non-malignant as well as more differentiated osteosarcoma cells.
Numerous reports describe important differences between mitochondria of normal and cancer cells. They include alterations of mitochondrial DNA, metabolic activity, and proteomic composition observed during malignant transformation (20, 22–24). Increased levels of mitochondrial DNA have been demonstrated in vitro. Additionally, high mtDNA content is detected in patients with aggressive ovarian, breast, and testicular cancer (25–27). Consistent with these findings, our study demonstrates that in highly proliferative and invasive osteosarcoma, there is marked elevation of mtDNA accompanied by a significant disruption of respiratory function as well as abnormalities in the number and ultrastructural composition of mitochondria.
Gel electrophoresis has been used previously to quantify levels of mitochondrial oxidative phosphorylation enzymes in muscle tissue (28). We used native polyacrylamide gel separation of mitochondrial respiratory chain complexes to show that compared with human osteoblasts, malignant 143B osteosarcoma cells express significantly higher levels of these multisubunit complexes. In contrast, enzymatic activity of mitochondrial complexes I, II, and IV in 143B cells was markedly inhibited, whereas enzymatic activity of complex III, despite being lower in 143B cells than in hFOB cells, did not reach statistical significance.
mtSSB has been described as an evolutionarily conserved crucial component in the process of replication and maintenance of mtDNA (11,17–19, 29–33). Arakaki et al. (17) discovered that mtSSB also maintains mitochondrial morphology and promotes cell survival in normal murine myoblasts. Our experimental data suggest that increased levels of mtSSB in 143B osteosarcoma cells may indeed facilitate changes in mitochondrial number and morphology, further validating the role mtSSB in promoting mitochondrial biogenesis. This finding is also confirmed by other studies that report elevated mtSSB content in other solid tumors (34). It remains to be seen whether up-regulation of mtSSB in osteosarcoma is accompanied by increased resistance of cancer cells to apoptosis.
Based on the reported findings, we suggest a mechanism of mitochondrial dysregulation in osteosarcoma where increased levels of mtSSB promote accelerated mitochondrial replication and, as a consequence, accumulation of a large number of immature non-functional organelles. Among possible causes of increased mtSSB expression may be transcriptional up-regulation of the protein by NFκB. Increased activity of NFκB has been reported in osteosarcoma (5, 35). Additionally, we believe that mtSSB promoter contains an NFκB binding site,3 thus potentially facilitating accelerated transcription of mtSSB in osteosarcoma cells.
In conclusion, our data demonstrate a novel dysregulatory mechanism of mitochondrial morphogenesis and metabolism in osteosarcoma that involves accelerated proliferation of the organelles in response to elevated levels of mtSSB. Therefore, choosing mtSSB as one of the therapeutic approaches for cancer treatment may represent a highly selective target for slowing tumor growth and, potentially, making cells more susceptible to pro-apoptotic signals.
Acknowledgments
We gratefully acknowledge Dr. T. Gunter, Dr. R. Rosier, and Dr. R. O'Keefe of the University of Rochester for expertise and fruitful discussions.
This work was supported, in whole or in part, by National Institutes of Health KL2 RR 024136 (Center for Research Resources; to R. A. E.). This work was also supported by the James P. Wilmot Foundation and the Karen D'Amico Foundation.
R. A. Eliseev, unpublished data.
- mtSSB
- mitochondrial single-strand DNA-binding protein.
REFERENCES
- 1. Warburg O. (1930) The Metabolism of Tumours, Constable Press, London [Google Scholar]
- 2. Warburg O. (1956) Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 3. Gatenby R. A., Gillies R. J. (2007) Int. J. Biochem. Cell Biol. 39, 1358–1366 [DOI] [PubMed] [Google Scholar]
- 4. Gatenby R. A., Gillies R. J. (2004) Nat. Rev. Cancer 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 5. Eliseev R. A., Zuscik M. J., Schwarz E. M., O'Keefe R. J., Drissi H., Rosier R. N. (2005) J. Cell. Biochem. 96, 1262–1273 [DOI] [PubMed] [Google Scholar]
- 6. Loos J. A., Roos D. (1973) Exp. Cell Res. 79, 136–142 [PubMed] [Google Scholar]
- 7. Roos D., Loos J. A. (1973) Exp. Cell Res. 77, 127–135 [DOI] [PubMed] [Google Scholar]
- 8. Moreno-Sánchez R., Rodríguez-Enríquez S., Marín-Hernández A., Saavedra E. (2007) FEBS J. 274, 1393–1418 [DOI] [PubMed] [Google Scholar]
- 9. Eliseev R. A., Gunter K. K., Gunter T. E. (2002) Mitochondrion 1, 361–370 [DOI] [PubMed] [Google Scholar]
- 10. Butler L. M., Agus D. B., Scher H. I., Higgins B., Rose A., Cordon-Cardo C., Thaler H. T., Rifkind R. A., Marks P. A., Richon V. M. (2000) Cancer Res. 60, 5165–5170 [PubMed] [Google Scholar]
- 11. Luu H. H., Kang Q., Park J. K., Si W., Luo Q., Jiang W., Yin H., Montag A. G., Simon M. A., Peabody T. D., Haydon R. C., Rinker-Schaeffer C. W., He T. C. (2005) Clin. Exp. Metastasis 22, 319–329 [DOI] [PubMed] [Google Scholar]
- 12. Thomas D. M., Johnson S. A., Sims N. A., Trivett M. K., Slavin J. L., Rubin B. P., Waring P., McArthur G. A., Walkley C. R., Holloway A. J., Diyagama D., Grim J. E., Clurman B. E., Bowtell D. D., Lee J. S., Gutierrez G. M., Piscopo D. M., Carty S. A., Hinds P. W. (2004) J. Cell Biol. 167, 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang P., Yang Y., Zweidler-McKay P. A., Hughes D. P. (2008) Clin. Cancer Res. 14, 2962–2969 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Harris S. A., Enger R. J., Riggs B. L., Spelsberg T. C. (1995) J. Bone Miner. Res. 10, 178–186 [DOI] [PubMed] [Google Scholar]
- 15. Nathan S. S., Pereira B. P., Zhou Y. F., Gupta A., Dombrowski C., Soong R., Pho R. W., Stein G. S., Salto-Tellez M., Cool S. M., van Wijnen A. J. (2009) Mol. Biol. Rep. 36, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tiranti V., Rocchi M., DiDonato S., Zeviani M. (1993) Gene 126, 219–225 [DOI] [PubMed] [Google Scholar]
- 17. Arakaki N., Nishihama T., Kohda A., Owaki H., Kuramoto Y., Abe R., Kita T., Suenaga M., Himeda T., Kuwajima M., Shibata H., Higuti T. (2006) Biochim. Biophys. Acta 1760, 1364–1372 [DOI] [PubMed] [Google Scholar]
- 18. Garesse R., Vallejo C. G. (2001) Gene 263, 1–16 [DOI] [PubMed] [Google Scholar]
- 19. Goffart S., Wiesner R. J. (2003) Exp. Physiol. 88, 33–40 [DOI] [PubMed] [Google Scholar]
- 20. Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S. J., Capaldi R. A. (2004) Cancer Res. 64, 985–993 [DOI] [PubMed] [Google Scholar]
- 21. Fantin V. R., St-Pierre J., Leder P. (2006) Cancer Cell. 9, 425–434 [DOI] [PubMed] [Google Scholar]
- 22. Brandon M., Baldi P., Wallace D. C. (2006) Oncogene 25, 4647–4662 [DOI] [PubMed] [Google Scholar]
- 23. Czarnecka A. M., Czarnecki J. S., Kukwa W., Cappello F., Scińska A., Kukwa A. (2010) J. Biomed. Sci. 17, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Modica-Napolitano J. S., Kulawiec M., Singh K. K. (2007) Curr. Mol. Med. 7, 121–131 [DOI] [PubMed] [Google Scholar]
- 25. Ellinger J., Albers P., Müller S. C., von Ruecker A., Bastian P. J. (2009) BJU Int. 104, 48–52 [DOI] [PubMed] [Google Scholar]
- 26. Kohler C., Radpour R., Barekati Z., Asadollahi R., Bitzer J., Wight E., Bürki N., Diesch C., Holzgreve W., Zhong X. Y. (2009) Mol. Cancer 8, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zachariah R. R., Schmid S., Buerki N., Radpour R., Holzgreve W., Zhong X. (2008) Obstet. Gynecol. 112, 843–850 [DOI] [PubMed] [Google Scholar]
- 28. Zerbetto E., Vergani L., Dabbeni-Sala F. (1997) Electrophoresis 18, 2059–2064 [DOI] [PubMed] [Google Scholar]
- 29. Longley M. J., Smith L. A., Copeland W. C. (2009) Methods Mol. Biol. 554, 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maier D., Farr C. L., Poeck B., Alahari A., Vogel M., Fischer S., Kaguni L. S., Schneuwly S. (2001) Mol. Biol. Cell. 12, 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Müller-Höcker J., Schäfer S., Li K. (2001) Appl. Immunohistochem. Mol. Morphol. 9, 276–280 [DOI] [PubMed] [Google Scholar]
- 32. Shadel G. S., Clayton D. A. (1997) Annu. Rev. Biochem. 66, 409–435 [DOI] [PubMed] [Google Scholar]
- 33. Tomáska L., Nosek J., Kucejová B. (2001) Biol. Chem. 382, 179–186 [DOI] [PubMed] [Google Scholar]
- 34. Verma M., Kagan J., Sidransky D., Srivastava S. (2003) Nat. Rev. Cancer 3, 789–795 [DOI] [PubMed] [Google Scholar]
- 35. Naugler W. E., Karin M. (2008) Curr. Opin. Genet. Dev. 18, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]