Abstract
Sphingolipids such as ceramide are recognized as vital regulators of many biological processes. Neutral sphingomyelinase 2 (nSMase2) is one of the key enzymes regulating ceramide production. It was previously shown that the enzymatic activity of nSMase2 was dependent on anionic phospholipids (APLs). In this study, the structural requirements for APL-selective binding of nSMase2 were determined and characterized. Using lipid-protein overlay assays, nSMase2 interacted specifically and directly with several APLs, including phosphatidylserine and phosphatidic acid. Lipid-protein binding studies of deletion mutants identified two discrete APL binding domains in the N terminus of nSMase2. Further, mutagenesis experiments pinpointed the core sequences and major cationic amino acids in the domains that are necessary for the cooperative activation of nSMase2 by APLs. The first domain included the first amino-terminal hydrophobic segment and Arg-33, which were essential for nSMase2 to interact with APLs. The second binding domain was comprised of the second hydrophobic segment and Arg-92 and Arg-93. Moreover, mutation of one or both domains decreased APL binding and APL-dependent catalytic activity of nSMase2. Further, mutation of both domains in nSMase2 reduced its plasma membrane localization. Finally, these binding domains are also important for the capability of nSMase2 to rescue the defects of yeast lacking the nSMase homologue, ISC1. In conclusion, these data have identified the APL binding domains of nSMase2 for the first time. The analysis of interactions between nSMase2 and APLs will contribute to our understanding of signaling pathways mediated by sphingolipid metabolites.
Keywords: Enzymes, Lipid-binding Protein, Phosphatidic Acid, Phosphatidylserine, Sphingolipid, Ceramide, nSMase2, Sphingomyelinase
Introduction
Sphingolipids such as ceramide are currently recognized as vital regulators of many biological processes (1–4), and the levels and turnover of these bioactive sphingolipids are regulated by several enzymes. The hydrolysis of sphingomyelin (SM)2 is a major pathway for ceramide production, and this reaction is catalyzed by the sphingomyelinases (SMases). Currently, six different SMases have been identified and grouped based upon their catalytic pH optima, including an acid SMase, an alkaline SMase and four neutral SMases (N-SMases) (5–8). Activation of N-SMases has been implicated in various biological responses, such as growth arrest, inflammation and aging (9, 10). Recently, considerable progress has been made regarding the specific roles of the identified N-SMase isoforms, particularly neutral sphingomyelinase 2 (nSMase2), in signal transduction and a variety of cell processes (11).
Mammalian nSMase2 was first identified in 2000 based on similarity to bacterial SMase (6). The human nSMase2 protein is a membrane-bound 655 amino acid protein consisting of a C-terminal catalytic domain, two N-terminal hydrophobic segments (HSs), and a 200-residue collagen-like domain between the N and C termini (6). More recently, nSMase2 was also found to be palmitoylated via thioester bonds at two major sites, one in the N terminus and the other in the catalytic region of the enzyme. Notably, nSMase2 palmitoylation is important for correct subcellular localization and stability of this enzyme (12). In addition, nSMase2 was found to localize to the inner leaflet of the plasma membrane (PM) (13).
A number of studies have implicated nSMase2 in growth arrest, the cellular response to cytokines and oxidative stress. Prior to the cloning of nSMase2, it was first identified as a confluent cell-associated gene in rat 3Y1 cells (14) and was subsequently implicated in confluence-induced G0/G1 cell cycle arrest in MCF-7 breast cancer cells (15). Many studies have reported activation of nSMase2 by tumor necrosis factor in a variety of cell types including smooth muscle cells, HUVECs and A549 cells, although regulation of nSMase2 appears to be cell type dependent with protein kinase C-delta and p38 MAPK involved in A549 cells (16, 17), and matrix metalloproteinases, furin and integrins involved in smooth muscle cells (18). In addition, nSMase2 was implicated in aging-associated inflammation, particularly in the response to interleukin-1β (19). Finally, in vivo studies have demonstrated that nSMase2 is essential for late embryonic and postnatal bone development. In particular, nSMase2 knock-out mice develop a form of dwarfism and have delayed puberty as part of a hypothalamus-induced pituitary hormone deficiency (20). Interestingly, a distinct mutant mouse strain (fro/fro) with inactive nSMase2 developed osteogenesis and dentinogenesis imperfecta (21). Finally, a recent study of human cancers found mutations of nSMase2 in human leukemias (22). Taken together, these studies reveal a multitude of roles for nSMase2 in both physiological and pathological states. Consequently, further understanding of nSMase2 regulation is essential.
Protein-phospholipid interactions are important for regulation of both protein localization and activity and, by extension, can have effects on cell signaling. In particular, anionic phospholipids (APLs) such as phosphatidylserine (PS) and phosphatidic acid (PA) play important roles in these interactions (23). PS has been implicated in a number of cell signaling pathways. Indeed, several signature domains for PS binding have been identified such as the C2 domain in PKC (24) and the Gla domain in some coagulation factors (25) while other less conserved PS binding motifs have also been identified (26). Importantly, the majority of PS binding motifs contain both positively charged and hydrophobic amino acid residues. Another APL, PA, has also been implicated in cell signaling. PA can be generated rapidly via hydrolysis of PC by phospholipase D or by phosphorylation of diacylglycerol by diacylglycerol kinases. Notably, PA has been shown to regulate both localization and function of several effectors (26), which include important signaling regulators such as Raf-1 (27), protein kinase C epsilon (28) and sphingosine kinase 1 (29). However, although several PA binding sequences have been identified, a signature binding motif for PA has yet to be identified (26, 30). Thus, the identification of PA/PS-interacting proteins as well as their binding domains is of interest, and the information will provide further insight into the lipid interaction and regulation of these effectors. Previous studies have shown that nSMase2 activity can be stimulated by APLs (6, 31). However, as nSMase2 does not contain a previously characterized phospholipid binding domain, it is unclear if and where APLs interact with this enzyme.
In this study, we find that nSMase2 directly interacts with APLs, in particular with PS and PA. In addition, we have identified two discrete APL selective binding sites in the N terminus of nSMase2, nSM2-APLB1 (nSMase2 APL binding type 1) and nSM2-APLB2 (nSMase2 APL binding type 2) domains. Further mutagenesis experiments allowed us to pinpoint the major cationic amino acids in the domains that are necessary for the cooperative activation of nSMase2 by APLs. Moreover, both domains are important for both activity and subcellular localization of nSMase2 in mammalian cells. Finally, these binding domains are also important for the capability of nSMase2 to rescue the defects of yeast lacking the nSMase homologue, ISC1. Taken together, these results offer insight into the interactions between nSMase2 and APLs. Moreover, these data strongly establish the N terminus of nSMase2 as a crucial component in regulating its activity.
EXPERIMENTAL PROCEDURES
Materials
Anti-GFP antibody and anti-V5 antibody were purchased from Invitrogen. Goat anti-rabbit and anti-mouse antibodies were acquired from Santa Cruz Biotechnology, Inc. [choline-methyl-14C]SM was provided by the Lipidomics Core Facility at the Medical University of South Carolina. All other lipids were purchased from Avanti Polar Lipids. All other reagents were purchased from Sigma unless otherwise stated.
Cell Culture and cDNA Transfection
HEK293 cells were cultured in MEM (Invitrogen) supplemented with 10% fetal bovine serum at 37 °C in a humidified 5% CO2 incubator. Cells were transfected using Lipofectamine 2000 (Invitrogen) or Effectene reagent (Qiagen) according to the manufacturer's instruction.
Mutagenesis of nSMase2 Constructs
Using a QuikChange site-directed mutagenesis kit (Stratagene), point and deletion mutations were introduced into the nSMase2 vectors generated previously (12, 31), including GFP-fused mouse nSMase2 in a pEGFP-N2 vector (Clontech), nSMase2(aa 1–123)-GFP, nSMase2(aa 1–52)-GFP, and nSMase2 (aa 1–123/Δ12–59)-GFP, nSMase2 tagged with V5 tag at the C terminus in a pEF6 vector (Invitrogen) (12) and FLAG-tagged nSMase2 in pYES2 yeast expression vector containing a galactose-inducible promoter (31). All oligonucleotides used in this study are listed in supplemental Table S1. The PCR products and constructs were subsequently sequenced to verify the desired mutations.
Western Blot Analysis
The HEK293 or MCF-7 cells were collected in lysis buffer (1× PBS, protease inhibitor mixture (Roche)) and homogenized by brief sonication. The homogenate was centrifuged for 10 min at 800 × g at 4 °C, and the supernatant was taken for protein determination using BCA Assay (Bio-Rad). Equal amount total proteins from each lysate were loaded onto 4–20% gradient SDS-polyacrylamide gels, subjected to electrophoresis, and then transferred to polyvinylidene difluoride membranes. The blots were probed using 1:2000 dilution of primary (anti-GFP, FLAG, or β-actin) antibody followed by horseradish peroxidase labeled secondary antibody (1:5000 dilution) (Santa Cruz Biotechnology, Inc.). The signals were detected using Enhanced Chemiluminescence reagents (Pierce).
Expression and Immunoprecitation of nSMase2 in Yeast
The Saccharomyces cerevisiae strain JK9–3dα/isc1Δ (MATa trp1 leu2–3 his4 ura3 ade2 rme1 ISC1::KanMX) (32) was used in this study. Synthetic minimal medium, containing galactose, and uracil dropout supplement were purchased from Clontech. Yeast extract and peptone were from Difco. Plasmids were transformed into yeast cells as previously described (33), and the expression of nSMase2 was induced by incubating the cells in synthetic complete-uracil dropout medium containing 2% galactose overnight and then harvested. Cells were lysed by vortexing with glass beads in lysis buffer (20 mm Tris pH 7.4, 0.5% Triton X-100, 1 mm EDTA, protease inhibitor mixture), and then spun for 100,000 × g for 60 min. The supernatant was subjected to immunoprecipitation using a FLAGIPT1-FLAG immunoprecipitation kit according to the manufacturer's instruction and the nSMase-FLAG was finally eluted from beads by adding 3×FLAG peptide solution in the presence of 0.5% Triton X-100.
Lipid-Protein Overlay Assay
Lipid-protein overlay assays were performed as described previously (34). Briefly, equimolar amounts of the indicated dioleoyl-lipids were spotted onto Hybond C extra nitrocellulose membrane (GE Healthcare). The membranes were allowed to dry for 1 h and were then wetted by floating on purified water. The membranes were equilibrated in Tris-buffered saline containing 0.1% (v/v) Tween 20 (TBS-T) for 5 min, followed by blocking with 1.5% fatty acid free bovine serum albumin (BSA)/TBS-T (blocking reagent) for 1 h at room temperature. Cell lysates containing nSMase2 were diluted into blocking reagent to a final concentration of 10 μg/ml. The membranes were then incubated in the presence of the enzyme overnight at 4 °C. The protein was identified by incubating with a 1:1500 dilution of anti-GFP or FLAG antibody, followed by incubation with secondary antibody (1:5000 dilution). Finally, the protein was visualized using Enhanced Chemiluminescence with exposure to Biomax MR film (Eastman Kodak Co.). Quantitative densitometry was performed using ImageQuant TL software (GE Healthcare).
Assays of Neutral Sphingomyelinase Activities
In vitro neutral SMase enzymatic assays were performed as described previously with modifications (31). Briefly, cells were collected and lysed as described above. Each reaction mixture contained 100 mm Tris (pH 7.4), 5 mm MgCl2, 0.1% Triton X-100, 2.5 mm dithiothreitol, and 100 μm [choline-methyl-14C]SM. For kinetics determination, 12.5 μm (0.8 mol%) to 400 μm (25.8 mol%) SMs were used as substrate for the activity assays. To determine the PA or PS effects on the nSMase activity, 3.1 μm (0.2 mol%) to 25 μm (1.6 mol%) PA or 12.5 μm (0.8 mol%) to 400 μm (25.8 mol%) PS were included in the buffer. After incubation for 0.5 h at 37 °C, the reaction was stopped by adding 1.5 ml of chloroform/methanol (2:1), followed by adding 400 μl of water. Phases were separated by centrifugation at 2000 × g for 5 min. By subjecting 500 μl of the upper phase to scintillation counting, SMase activity was determined by quantification of the amount of released radioactive phosphocholine.
Immunofluorescence and Confocal Microscopy
HEK293 or MCF-7 cells were plated onto 35-mm confocal dishes (MatTek) and transfected with V5 tagged nSMase2 vectors (1 μg/dish). Twenty-four hours post-transfection, cells were treated and fixed with 3.7% formaldehyde for 10 min. Following fixation, cells were permeabilized for 5 min with 100% methanol at −20 °C and then blocked in 2% human serum (Jackson) in PBS for 30 min at room temperature. Cells were blocked in 2% human serum for 1 h at room temperature. Anti-giantin (Covance) or anti-calnexin antibodies were co-incubated with anti-V5 antibody. Incubations were performed in 2% human serum at 1:300 dilutions for 1 h at room temperature. Following incubation with the primary antibody, cells were washed three times with PBS, probed with fluorescent secondary antibody for 1 h at room temperature. Cells were viewed on a Zeiss LSM 510 Meta Confocal Microscope.
Spot Test
Yeast cells from colonies transformed with pYES2 vector containing ISC1, ISC1-K168A, nSMase2, or nSMase2 mutants, including R33A/Δ45–48, R92A/R93A, and R33A/R92A/R93A/Δ45–48 were inoculated into liquid minimum medium without uracil containing 2% dextrose. When the cells reached log phase, the cultures were diluted to an absorbance of A600 of 0.3. Three dilutions of 1:10 were performed consecutively for each transformant, cells from each dilution were spotted on uracil dropout minimal medium plates containing galactose and raffinose as carbon source. The plates were made with or without hydroxyurea (10 mg/ml). Spots were checked for growth after 3 to 4 days of incubation at 30 °C.
Statistical Methods
Results are expressed as the mean ± S.D. The mean value is calculated from a single experiment. The data are representative of at least two independent experiments.
RESULTS
Binding of nSMase2 to APLs
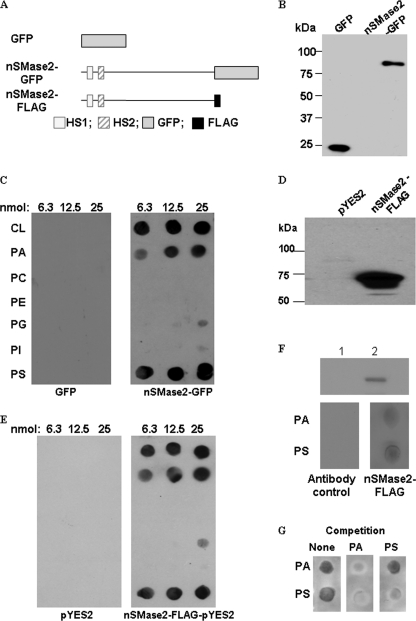
Previously, we demonstrated that nSMase2 activity is stimulated by several APLs, especially PS, cardiolipin (CL), and PA (31). To define the physical interactions between nSMase2 and APLs, binding studies were carried out using the lipid-protein overlay method (fat-blot assays) as described previously (35). Using this approach, several phospholipids, including CL, PA, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol, and PS, were immobilized on a nitrocellulose membrane, and binding was examined by incubation with cell lysates from HEK293 cells overexpressing nSMase2-GFP protein followed by immunostaining with anti-GFP antibody. As a control for nonspecific binding of protein to lipids, GFP overexpressor HEK293 cells were used (Fig. 1, A and B). It was observed that nSMase2-GFP bound strongly to PS, CL, and PA, with modest binding to phosphatidylglycerol and phosphatidylinositol. On the other hand, no binding was observed to the other immobilized phospholipids. The binding of nSMase2-GFP to phospholipids was very prominent whereas that of GFP was hardly detectable (Fig. 1C). Similar results were further observed when using lyates with overexpressed FLAG-tagged nSMase2 in the isc1Δ strain of Saccharomyces cerevisiae (Fig. 1, D and E). Because nSMase2 localizes predominantly to the PM and partly to the Golgi and CL is predominantly a mitochondrial lipid, we chose to focus the subsequent studies on PA and PS. To demonstrate direct binding of the protein to phospholipids, we evaluated the ability of immunoprecipitated nSMase2 to interact with APLs. As shown in Fig. 1F, the interaction was also detected between APLs and immunoprecipitated nSMase2-FLAG expressed from yeast, confirming the direct binding. In addition, we have performed a competition experiment by adding PA or PS to nSMase2-FLAG prior to overlay assay (Fig. 1G). Interestingly, while preincubation with PS in solution specifically inhibited the PS binding, a preincubation with PA was able to inhibit the binding signals of both PA and PS, suggesting that more than one APL binding domain may exist. Altogether, these results demonstrate specific interaction of nSMase2 with its specific activating APLs.
FIGURE 1.
Interaction of nSMase2 with APLs. A, schematic diagram of nSMase2-GFP in pEGFP-N2 and nSMase2-FLAG in pYES2 expression vectors. Mouse nSMase2 was fused to GFP or FLAG tag at the C terminus. B, HEK293 cells were transfected with pEGFP vector or an expression construct of nSMase2-GFP. The immunoblot analysis of nSMase2-GFP and GFP was performed with anti-GFP antibody as described under “Experimental Procedures.” C, lipid-protein overlay assay showed nSMase2 binding to immobilized lipids. Equimolar amounts of lipids were immobilized on nitrocellulose membranes and probed with HEK293 cell lysates. The lipid-protein binding was identified by immunostaining with an anti-GFP monoclonal antibody. The following dioleoyl-lipids were immobilized on the membrane as indicated: CL, PA, PC, PE, PG, PI, PS, D, the immunoblot analysis of nSMase2-FLAG expressed in yeast was performed with anti-FLAG antibody. nSMase2-FLAG was expressed in ISC1Δ S. cerevisiae and detected as described under “Experimental Procedures”. E, lipid-protein overlay assay showed nSMase2 expressed in yeast binding to immobilized lipids. F, interactions between APLs and immunoprecipitated nSMase2-FLAG. The nSMase-FLAG expressed in the yeast was immunoprecipitated as described under “Experimental Procedures.” The same percentage of the supernatant of immunoprecipitation (lane 1) or eluted nSMase2 (lane 2) was analyzed by Western blot analysis (upper panel). Lipid-protein overlay assays were performed using eluted nSMase2-FLAG (lower panel). G, competition overlay assays were performed. 50 μm PA (middle panel) or (right panel) PS was incubated with lysates overexpressing nSMase2-FLAG for 1 h prior to performing the overlay assays. The lysate together with PA or PS was then added to the blot for lipid-protein overlay assay. The lipid binding signals were detected using an antibody against FLAG. The results are representative experiments of at least two independent experiments.
The N-terminal Is Essential for nSMase2 Binding to APLs
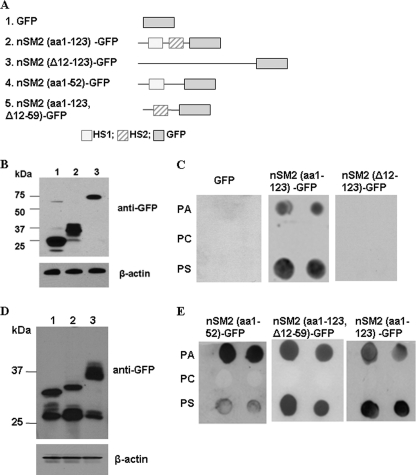
As the N terminus of nSMase2 has two HSs, this region may also contain the APL binding domain. To test this hypothesis, we determined the APL binding capability of the N terminus of nSMase2 (aa 1–123)-GFP and a deletion nSMase2 mutant (Δ12–123) without the HSs (Fig. 2, A and B). As shown in Fig. 2C, only the nSMase2 (aa 1–123)-GFP displayed a robust interaction with PA and PS. In contrast, the N-terminal deletion (Δ12–123) mutant showed no detectable binding. Although the Δ12–123 mutant displayed a lower level of expression (Fig. 2B), no interaction signal detected even at a 5-fold higher concentration for this mutant (supplemental Fig. 2A). These results demonstrate that the N-terminal (aa 1–123) of nSMase2 is both necessary and sufficient for binding to APLs.
FIGURE 2.
nSMase2 N-terminal domain is essential for nSMase interaction with APLs. A, schematic diagram of the four mutants of nSMase2. The nSMase2 mutants were fused to GFP at the C terminus. B, immunoblot analysis of deletion mutants of nSMase2 with anti-GFP antibody. Lane 1, GFP; lane 2, nSMase2 (aa1–123)-GFP; lane 3, nSMase2(Δ12–123)-GFP. C, lipid-protein overlay assay showing the binding capabilities of various nSMase2 constructs to APLs. D, immunoblot analysis of deletion mutants of nSMase2 with anti-GFP antibody. Lane 1, nSMase2 (aa 1–52)-GFP; lane 2, nSMase2 (aa 1–123/Δ12–59)-GFP; lane 3, nSMase2 (aa 1–123)-GFP. E, lipid-protein overlay assay showing the binding capabilities of various nSMase2 constructs to APLs. The results are representative experiments of at least two independent experiments.
Two APL Binding Domains Exist in the N Terminus of nSMase2
Next, we examined the binding location by separating the N-terminal of nSMase2 into two segments tagged by GFP at C terminus, nSMase2 (aa 1–52)-GFP containing the 1st HS, and nSMase2 (aa 1–123/Δ12–59)-GFP containing only the 2nd HS (Fig. 2, A and D). Importantly, each of the segments showed binding capability to APLs (Fig. 2E). Interestingly, while the 1st segment showed a higher PA-binding signal, the 2nd segment bound to PS and PA with similar intensity. Thus, two distinct APL-binding domains exist in the N terminus of nSMase2, with preferential interactions with PA.
Characterization of the nSM2-APLB1 Domain
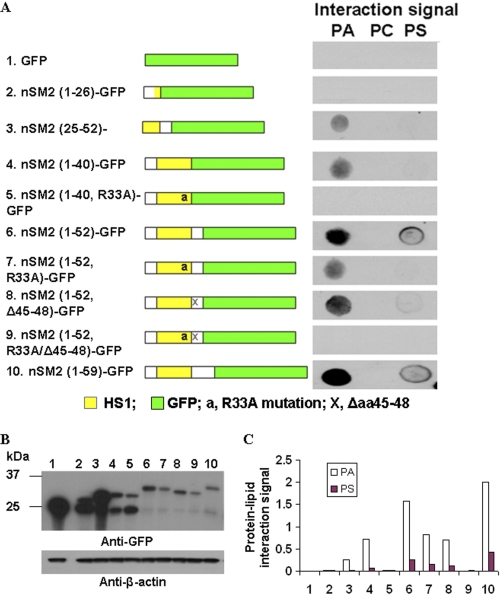
As there are no significant matches between the sequence of nSMase2 and other defined lipid-binding domains, it became important to identify the novel APL binding domains in nSMase2. A mutagenesis approach was applied to identify the key determinants of the 1st APL binding domain in nSMase2. Therefore, a series of GFP-tagged deletion or point mutants were made from mutants within aa 1–59 in nSMase2 tagged by GFP at the C terminus (Fig. 3A). The mutated vectors were then transformed and overexpressed in HEK293 cells successfully according to the Western blot results (Fig. 3B). The PA binding signal on the fat blot was examined by incubation with HEK293 lysates expressing the truncated or point mutants of nSMase2 (aa 1–59)-GFP. As shown in Fig. 3, A and C, while significant binding to PA was observed using the mutant nSMase2(aa 1–52)-GFP, decreased binding was found when deleting the hydrophobic segment as only weak signals were detected from nSMase2 (aa 25–52)-GFP and no binding signals from nSMase2(aa 1–26)-GFP. Because positively charged amino acid have been shown to be important for APL and protein interaction (36), we next determined the effects of mutation of Arg-33 and/or deletion of aa 45–48 (KRQR) on binding to APL. Interestingly, a point mutation of Arg-33 to Ala was enough to eliminate the PA binding signal of nSMase2 (aa 1–40)-GFP. For nSMase2 (aa 1–52)-GFP, mutation of R33A or deletion of aa 45–48 resulted in decreased PA binding. However, only the combination of R33A and deleting aa 45–48 could abolish the interaction, suggesting these positive charged amino acid residues are important for interaction between nSMase2 and APLs via electrostatic interactions. Interestingly, all mutants of the first domain showed substantially less PS binding, and mutation of Arg-33 or deletion of aa 45–48 nearly abolished all detected PS binding. Taken together, the results identify the first domain necessary for APL binding in nSMase2, with a conserved “FPCYWXXDRLXASXXXTXXEKRXR” motif as the binding core sequence (supplemental Fig. S4A).
FIGURE 3.
Identification of the first APL binding determinants of N terminus mutants of nSMase. A, a series of GFP-tagged deletion or point mutants were made from mutants within aa 1–59 in nSMase2 tagged by GFP at C terminus. Schematic diagram of deletion mutants is shown in the left panel. The APL binding signal on the fat blot was examined by incubation with HEK293 lysates expressing the truncated or point mutants of nSMase2 (aa1–59)-GFP. B, immunoblot analysis of expression of the deletion mutants of nSMase2 with anti-GFP antibody. C, densitometry analyses were performed using ImageQuant TL software (GE Healthcare). The relative value of each lipid-protein interaction signal was calculated by subtracting the background and normalized to its western signals from the upper band (the correct sized band). Similar results were obtained in two separate experiments.
Characterization of the nSM2-APLB2 Domain
To localize the second APL-specific binding domain in nSMase2, a series of GFP-tagged deletion or point mutants were generated from mutant nSMase2 (aa 1–123/Δ12–59)-GFP (which harbors the second HS) (supplemental Fig. S1A) and overexpressed in HEK293 cells (supplemental Fig. S1B). The lysates were further subjected to fat-blot binding assays (supplemental Fig. S1, A and C). Mutation of either Arg-92 or Arg-93 to Ala decreased APL binding signals whereas mutation of Arg-99 to Ala showed no effects on the interaction signal. Moreover, mutating both Arg-92 and Arg-93 resulted in complete loss of the interaction for nSMase2 (aa 1–123/Δ12–59)-GFP, suggesting that Arg-92 and Arg-93 are the essential amino acid residues to interact with APLs. Further, when part of the 2nd HS was truncated (nSMase2 (aa 68–123)-GFP), the binding signal decreased, and further deletion of this HS (nSMase2 (aa 82–123)-GFP) eliminated interaction, proving that the 2nd HS in nSMase2 is also essential to this APL interaction domain. Therefore, the results define the 2nd binding domain with motif “LXLLXXXLPFAXXGFXXWXPXQXXR(R/K)” as the binding core, which is capable of binding PA and PS (supplemental Fig. S4A).
Effects of PS and PA on nSMase2 Enzymatic Activity
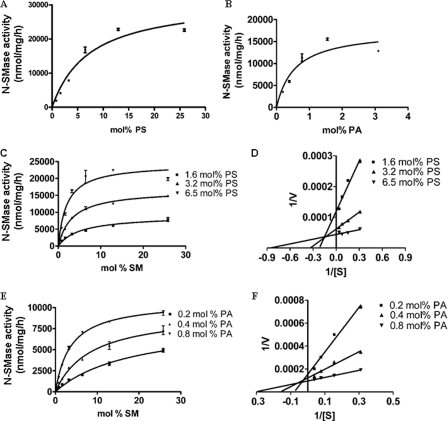
The identification of two distinct domains that interact with APLs suggested more complex interactions of PA and PS with the enzyme. Therefore, we investigated how PA and PS affect nSMase2 enzymatic activity. The FLAG-tagged full-length nSMase2 was overexpressed in isc1Δ cells, which are deleted in the yeast sphingomyelinase Isc1p, thus, providing negligible background of SMase activity. As shown in Fig. 4, A and B, nSMase2 was significantly stimulated by PS and PA in a dose-dependent manner, confirming that PA and PS can activate nSMase2. Interestingly, PA was significantly more effective in activating nSMase2 than PS, although the extent of activation was more pronounced with PS.
FIGURE 4.
Effects of PS and PA on nSMase2 enzymatic activity. FLAG-tagged nSMase2 was expressed in isc1Δ S. cerevisiae and analyzed as described under “Experimental Procedures.” A, N-SMase activity of nSMase2 was measured at various concentrations of PS. B, N-SMase activity of nSMase2 was measured at various concentrations of PA. C-D, PS effects on kinetics of nSMase2 activity. Under three various PS concentrations (1.6, 3.2, and 6.5 mol%), enzymatic kinetics for N-SMase activity were analyzed using 0.8–25.8 mol % SM as substrate. E-F, PA effects on kinetics of nSMase2 activity. Under different PA concentrations (0.2, 0.4, and 0.8 mol%), enzymatic kinetics for N-SMase activity were analyzed. The data are the averages of duplicates. The values are expressed as the mean ± S.D. The data are representative of at least two independent experiments.
Next, kinetic studies were undertaken to define the interdependence of PS and PA, and therefore nSMase2 activity was evaluated in the presence of various concentrations of PA and PS. Double-reciprocal plots showed that the Km was decreased whereas the Vmax was increased at higher concentrations of PS (Fig. 4, C and D). Similar effects was also observed using PA with the primary effects of PA being on decreasing the Km of the enzyme (Fig. 4, E and F). These results demonstrate that PA and PS can activate nSMase2 by affecting both the Vmax and Km of nSMase2.
Effects of Mutations of the APL Binding Domains on Lipid Binding Capability of Full-length nSMase2
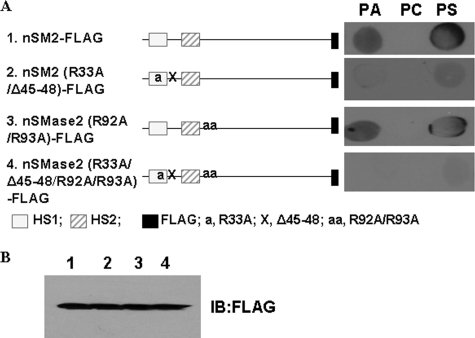
Upon identification of the APL binding sequence using the various mutants, we next examined if these domains are important to the whole nSMase2 protein. Several mutants were generated, including nSMase2(R33A/Δ45–48)-FLAG (nSM2-APLB1 mutant), nSMase2(R92A/R93A)-FLAG (nSM2-APLB2 mutant) and nSMase2(R33A/R92A/R93A/Δ45–48)-FLAG (combined mutant). The wild type and mutated nSMase2 were transformed and expressed in isc1Δ cells. As shown in the fat-blot assays (Fig. 5A), mutating the key amino acids in the nSM2-APLB1 domain or the nSM2-APLB2 domain caused a decrease of interaction signal for PA and PS. However, when mutating both domains, almost all the binding signals were abolished, confirming that the identified domains are important for the full-length nSMase2 binding to APLs. By Western blot analysis, nSMase2 and its mutant were expressed at similar levels (Fig. 5B). These results suggest a complementary mechanism between the two APL binding domains. Thus, these results further prove that both domains are essential to the interaction between nSMase2 and APLs.
FIGURE 5.
Effects of mutations of the APL binding domains on lipid binding capability of full-length nSMase2. A, several mutants of full-length nSMase2 were generated, including nSMase2(R33A/Δ45–48)-FLAG (nSM2-APL1 mutant), nSMase2(R92A/R93A)-FLAG (nSM2-APL2 mutant) and nSMase2(R33A/R92A/R93A/Δ45–48)-FLAG (combined mutant). The wild type and mutated nSMase2 were transformed and expressed in isc1Δ S. cerevisiae cells. The APL binding signal on the fat blot was examined by incubation with cell lysates expressing nSMase2-FLAG and its mutants. B. immunoblot analysis of nSMase2 and its mutants were performed using anti-FLAG antibody. The data are representative of at least two independent experiments.
Effects of Mutations of the APL Binding Domains on APL-dependent Activity of nSMase2
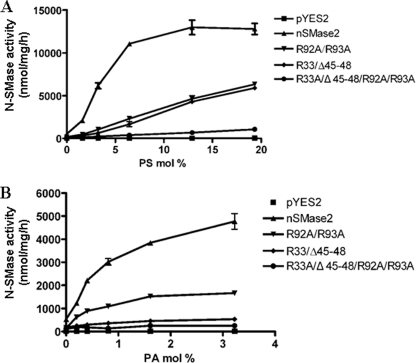
The above studies provided specific tools to determine the roles of the two APL binding domains in mediating activation of nSMase2 by PA and PS. Consistent with the binding results, weaker activation was observed among these mutants. As shown in Fig. 6A, at various concentrations of PS, a significant decrease of activity was observed for both nSM2-APLB1 and nSM2-APLB2 mutants. Importantly, only minimal stimulation by PS was detected when both domains were mutated. Similarly, the activation of nSMase2 by PA was also abolished when mutating both domains (Fig. 6B). However, a much more pronounced decrease of activation was observed in the nSM2-APLB1 domain mutant than the nSM2-APLB2 domain mutant under PA stimulation. These results demonstrate that the two APL binding domains are essential for the APL-dependent activation of nSMase2.
FIGURE 6.
Effects of the mutations in APL binding domains of full-length nSMase2 on enzymatic activity. The wild type and mutated nSMase2 were transformed and expressed in isc1Δ cells, and the cell lysates were subjected to N-SMase activity assay as described in “Experimental Procedures.” A, effects of mutations in APL binding domains on PS activation of nSMase2. B, effects of mutations in APL binding domains on PS activation of nSMase2. The data are the averages of duplicates. The values are expressed as the mean ± S.D. The data are representative of at least two independent experiments.
APL Binding Domains Are Important for nSMase2 to Localize at the PM
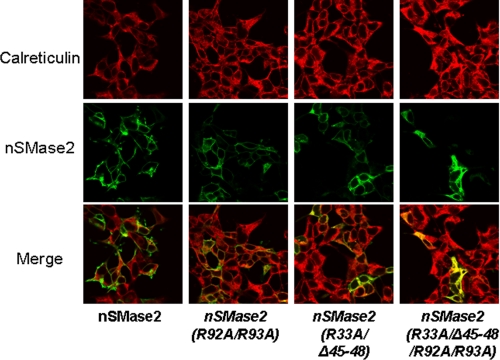
Thus far, results have indicated that binding of APL to nSMase2 is important for its activity, consistent with PS as an activator of nSMase2. A previous study showed that nSMase2 locates to the PM in confluent cells (15). Thus, we wanted to determine if the APL binding domains are also important for the subcellular localization of nSMase2. For this, the effects of the specific mutations on the location of nSMase2 in HEK293 cells were examined. As can be seen (Fig. 7), wild-type V5-tagged nSMase2 localized predominantly to the PM, consistent with the previous study. The nSM2-APLB2 mutant nSMase2-(R92A/R93A)-V5 also showed a predominant PM pattern, while mutation of nSM2-APLB1 resulted in a decreased PM location. However, the double APL binding domain mutant nSMase2-(R33A/R92A/R93A/Δ45–48)-V5 had a strikingly different localization, and co-localized primarily with the ER marker, calreticulin. Similar results were also observed in MCF-7 cells (data not shown). These results indicate that binding of APL to nSMase2 is necessary for correct trafficking and localization of nSMase2 to the PM. Moreover, it is suggested that the first APL binding domain plays a more prominent role in this process.
FIGURE 7.
APL binding domains are important for nSMase2 to localize at the PM. HEK293 cells were transfected with nSMase-V5 and mutants (R33A/Δ45–48, R92A/R93A and R33A/Δ45–48/R92A/R93A) expression vectors. After 24 h, the cells were fixed and co-stained with an antibody against calreticulin (red, upper panel) and antibody against V5 (green, middle panel) for nSMase2 signal and then subjected to confocal microscopy evaluation. The colocalization signals were observed as yellow or orange (lower panel). The data are representative of at least two independent experiments.
Mutation of the nSMase2 in APL Binding Domains Abolishes Its Capability to Correct the Sensitivity to Hydroxyurea in isc1Δ
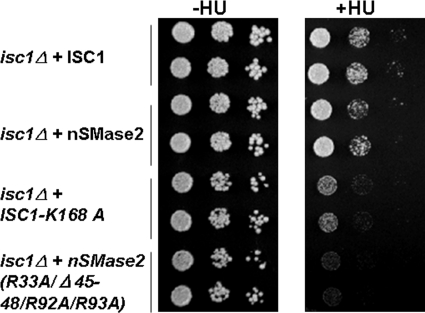
To further confirm if the APL binding domains are important for nSMase2 function, a yeast system was further applied. It has been recently shown that the deletion of ISC1, the budding yeast homologue of nSMase, renders cells sensitive to the genotoxic agent hydroxyurea (37). To examine if the APL binding domains are important for nSMase2 function, wild-type nSMase2 and the APL mutants cloned under a GAL promoter in yeast expression plasmid pYES2 were used in a spot test. Plasmid containing ISC1, ISC1-K168A (a non-functional ISC1 mutant (38)), nSMase2 and nSMase2 mutant (R33A/R92A/R93A/Δ45–48) were used. As shown in Fig. 8 (right panel), overexpressing ISC1 or wild-type nSMase2 enhanced the growth of isc1Δ cells in the presence of hydroxyurea compared with the overexpression of a nonfunctional ISC1-K168A mutant. The overexpression of the nSMase2 (R33A/R92A/R93A/Δ45–48) mutant failed to correct the defect in isc1Δ cell. In addition, decreased rescue effects were also observed for nSM-APLB1 or nSM-APLB2 mutant (supplemental Fig. S3). These results show that the APL binding domains are important for the nSMase2 functions.
FIGURE 8.
APL binding domain mutants fail to correct the defect of isc1Δ S. cerevisiae cells. nSMase2 APL binding domain mutant of nSMase2 (R33A/Δ45–48/R92A/R93A), nSMase2, ISC1, or its nonfunctional mutant ISC1-K168A was expressed in isc1Δ S. cerevisiae cells, respectively. Exponentially growing cells were first diluted to an A600 of 0.3, and then serially diluted and spotted onto agar Raf/Galactose uracil minus plates with or without 10 mg/ml hydroxyurea (HU). The plates were incubated for 3–4 days at 30 °C. This experiment was performed two times with similar results.
DISCUSSION
The interactions of proteins with APLs can have implications both for cellular localization and enzymatic activity. In this study, we have identified the interactions between nSMase2 and APLs. We find that nSMase2 interacts strongly with PS and PA, defining two discrete domains in the N terminus of nSMase2 as APL selective binding sites. These domains proved to be both necessary and sufficient for binding of PA or PS and were able to impart APL-dependent stimulation of activity. In addition, the results pinpoint the major cationic amino acids that are necessary for the cooperative activation of nSMase2 by PA and PS. Functionally, these domains are important for the correct subcellular localization of nSMase2 and the ability of nSMase2 to rescue the defects of yeast deficient in the nSMase homologue ISC1. Taken together, these results provide insight into the regulation of nSMase2 and strong evidence that the N-terminal region of nSMase2 functions as a crucial regulatory domain.
Previous studies from our laboratory reported in vitro activation of nSMase2 by APLs (31). Extending these findings, the current study focused on which APLs interact with nSMase2 and further determining how nSMase2 is activated by these lipids. Utilizing lipid-protein overlay assays, nSMase2 was found to interact with several APLs, including PA, PS, and CL (Fig. 1). This is consistent with our earlier data where these three lipids, particularly PS and CL showed the greatest activation of nSMase2 activity. It should be noted that CL is predominantly localized to the mitochondria (39). However, nSMase2 does not appear to possess a mitochondrial targeting peptide and so does not localize to mitochondria. Thus, as nSMase2 primarily localizes to the PM, Golgi, and endosomal system (40), it is more likely that PA and PS are the primary lipids that interact with nSMase2 in the cell. Indeed, both nSMase2 and PS are localized on the inner leaflet of the PM (13, 41).
Analysis of the nSMase2 sequence revealed no previously characterized APL-binding motifs, suggesting the possibility that novel APL binding motifs may exist in nSMase2. To probe this, studies utilizing a wide range of deletions within the nSMase2 sequence were undertaken. Initial deletion studies revealed that the N-terminal region between residues 12 and 123 was both necessary and sufficient for binding to PS and PA (Fig. 2). Upon further investigation, we identified two distinct APL-binding domains in the N-terminal region. Mutagenesis analyses and sequence alignments revealed that the conserved motif “FPCYWXXDRLXASXXXTXXEKRXR” is the core sequence of the nSM2-APLB1 domain, while the motif “LXLLXXXLPFAXXGFXXWXPXQXXR(R/K)” reflects the core sequence of the nSM2-APLB2 domain. Of these sequences, the amino acids Arg-33, Arg-45, and Arg-48 were crucial for lipid binding to the first domain whereas Arg-92 and Arg-93 were critical residues in the second domain. In previous studies, the motif “FXFXLKXXXKXR” was found in the APL-binding C2 domain in PKC and other enzymes (42). Notably, a similar peptide sequence “FLFGRSEIR” was found in the C terminus of Isc1p and was also demonstrated to be important for PS binding (36). Although no such motif was identified in nSMase2, the pattern of a hydrophobic segment containing phenylalanine and leucine residues which in turn are followed by multiple basic amino acids is largely conserved between the sequences. This provides further support that the core sequences identified here are important for nSMase2-APL interactions. This also suggests that for many lipid-protein interactions, there may not always be a defined motif among all proteins, but that the overall biochemical properties of the core amino acid sequence are the more important factor. Thus, for PS and PA binding, there appear to be the three determinants of a hydrophobic domain, Phe/Leu residues, and cationic amino acids.
It should be also noted that the binding signals may be derived from indirect interactions through membrane-associate partner proteins of nSMase2. To examine this possibility of indirect effects, lipid-protein binding assays were also performed using nSMase2 overexpressed in the S. cerevisiae, which is less likely to harbor an endogenous partner protein of nSMase2. Similar binding signals were detected in both mammalian and yeast expression systems. The direct interactions were further confirmed between APLs and immunoprecipitated nSMase2 expressed in the yeast. Moreover, according to the topology of nSMase2 (13), the identified domains are partly embedded in and partly on the surface of membrane. As a result, the key positively charged amino acid residues of these binding domains can form electrostatic bonds with the negative changed heads of APLs while the HSs inserts into the membrane, resulting direct and specific interaction between nSMase2 and membrane. Finally, the identified domains required for APL binding resemble those of other PS-acting enzymes such as PKC and phosphatidylserine decarboxylase (42). Altogether, the above evidences support that the direct interactions exist between nSMase2 and APLs.
The current results also suggest that the two domains are cooperative in their binding to APLs. Within this context, the most important information can be deduced by comparing the full length nSMase2 with the nSMase mutants containing one or two non-functional domains. In lipid-protein overlay assays, while mutating the key amino acids in either the nSM2-APLB1 or the nSM2-APLB2 domain caused a decrease of interaction signal for PA and PS whereas mutating both domains, almost all the binding signals were inhibited (Fig. 5). Similarly in cells, mutation of individual APL-binding domains resulted in moderate subcellular localization changes whereas mutation of both domains resulted in a stronger mislocalization of the enzyme. Finally, as the two domains have different binding specificities to PA and PS, the effects of both PA and PS on nSMase2 activity were determined and, interestingly, some additive effects were observed (data not shown). Taken together, these results suggest that there is cooperativity between the two APL binding domains. However, the limitations imposed by immunodetection of protein binding have prevented an accurate quantitation of kinetic binding parameters in this study. Notably, as nSMase2, is a hydrophobic protein, it does not lend itself to classical approaches to tease out specific from nonspecific membrane interactions. In future, it will be important to perform a more in-depth analysis of the kinetic binding parameters between nSMase2 and APLs.
It is also notable that the core binding sequence of nSMase2 are highly conserved across different species (supplemental Fig. S4A), suggesting that interactions with APLs are conserved among mammalian nSMase2. Interestingly, a novel member of the N-SMase family was identified in zebrafish and found to localize to mitochondria (43). Although distinct from zebrafish nSMase2, the mitochondrial SMase was strongly activated by APLs. Moreover, the APL-binding motifs in nSMase2 identified here share a high degree of homology with the zebrafish SMase. In particular, there is conservation of some of the essential amino acid residues in thee domains such as Arg-33, Arg-48, Arg-92, and Arg-93 (supplemental Fig. S4B). Thus, we speculate that these regions very likely are important for APL activation of the mitochondrial SMase. Although, given its distinct localization it is likely to be a direct in vivo target of mitochondrial CL rather than PS. Thus, besides nSMase2, the identified motifs may clearly play roles in other N-SMase family members and other proteins.
The enzymologic assessment of APL effects on nSMase2 suggested mechanisms by which APLs can activate nSMase2. Given the localization of the binding domains in the N terminus and the catalytic core in the C terminus, one potential mechanism would be that interaction of nSMase2 with APLs causes a conformational change in nSMase2 resulting in an active form. This hypothesis was supported by the effects of APL on nSMase2 kinetics showing that APLs decrease the Km of nSMase2 (Fig. 4), thus increasing the affinity of nSMase2 for the SM substrate. However, APLs also increased the Vmax of nSMase2 implying that they can also enhance catalytic activity of the enzyme. Nevertheless, this again suggests that the primary effects of APLs on nSMase2 conformation are to induce optimal interaction with the substrate. This was further substantiated by the fact that mutations in the APL binding domains decreased the capacity for APL to activate nSMase2 (Fig. 6). Previous studies from our laboratory put forward a model of APL activation of Isc1p. In this tether and pull model, the C terminus of Isc1p. has greater affinity for membranes in the presence of PS, CL, or PG (the tether), and intramolecular interactions with the N-terminal catalytic domain serve to “pull” the active site to interact with lipid substrates. However, as the domain structures of nSMase2 and Isc1p are opposite (nSMase2 has N-terminal phospholipid binding and C-terminal catalytic core whereas Isc1p is the other way round), it is uncertain if the same activation model would hold true for nSMase2. Moreover, as the structures of both nSMase2 and Isc1p are unknown, the orientation of the active site and the N-terminal phospholipid binding sites are unclear. Thus, further investigation and, indeed, identification of the crystal structure of nSMase2 would shed light on the exact mechanism for APL activation of nSMase2.
In cells, nSMase2 has been observed to localize to several organelles and a number of studies have suggested that the trafficking of nSMase2 is important for nSMase2 functions. For example, the PM localization of nSMase2 could be enhanced upon stimulation (e.g. TNF-α, H2O2) or cell confluence (11, 15, 16, 44). A recent study also demonstrated that nSMase2 localization and activity were regulated by endocytosis (45). In this study, we have shown that, in addition to effects on enzymatic activity, mutation of APL binding domains also seem to be important for subcellular localization of nSMase2 (Fig. 7). Mechanistically, the reasons for this are unclear. One possibility is that mutations in APL-nSMase2 interaction inhibit the trafficking of nSMase2 to Golgi and PM. Alternatively, as mutation of nSM2-APL binding domains decreases the proteins capability of anchoring on the PM. It is also possible these mutants are subjected to quick endocytosis at the PM, resulting in decrease PM localization. Another possibility is that interaction with APLs is required for correct insertion of nSMase2 into the membrane as has been reported for other enzymes (46). Consequently, the topology of the enzyme is altered and it is unable to move beyond the ER and/or Golgi owing to masking of targeting signals. Overall, the exact mechanism for the roles of APL binding in regulating nSMase2 trafficking warrants further investigation.
Both PA and PS have been implicated in a large range of cellular processes including cell growth, development, and stress responses (26). Indeed, PA has emerged as a pivotal lipid messenger and can be produced by both phospholipase D and diacylglycerol kinase pathways. The identification of PA effectors can provide valuable insights into the cellular roles of PA. Interestingly, some of the known nSMase2 activators have also been reported to generate PA. For example, H2O2 has been reported to induce nSMase2 activity in bronchial epithelial cells (44) and has also been shown to stimulate phospholipase D activity and generation of PA (47). Additionally, hypoxia can increase the intracellular level of PA through the action of DGK (48) and can also stimulate nSMase2 activity (49). A recent study showed that PLD1 can play important roles in mediating signaling responses to TNF-α (50). Finally, PA generation has been reported to occur in the caveolae/lipid raft domains in response to some stimuli (16) and N-SMase activity was also localized to these domains (51, 52). Indeed, this was reported to be due to nSMase2 (53). Thus, nSMase2 may be ideally localized for activation in response to rapid generation of PA. Similarly, like nSMase2, PS is also present on the cytoplasmic side of the PM where it provides anchorage for signaling proteins.
In conclusion, the results from this study have identified two distinct phospholipid binding domains in nSMase2. These domains are important for both activity and subcellular localization of nSMase2 and were essential for the capacity of nSMase2 to rescue the defects of yeast deleted in the N-SMase homologue ISC1. The identification of these domains confirms the importance of the N terminus of nSMase2 for full enzymatic activity, and provides further insight into the regulation of nSMase2 at the molecular level.
Supplementary Material
Acknowledgment
We thank Dr. Motohiro Tani for initial generation of nSMase2 vectors.
This study was supported, in whole or in part, by National Institutes of Health Grant GM 43825 (to Y. A. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- SM
- sphingomyelin
- nSMase2
- neutral sphingomyelinase 2
- APL
- anionic phospholipids
- SMase
- sphingomyelinase
- nSMase
- neutral sphingomyelinase
- PM
- plasma membrane
- HS
- hydrophobic segment
- PS
- phosphatidylserine
- PA
- phosphatidic acid
- CL
- cardiolipin. nSM2-APLB1 domain, nSMase2 APL binding type 1 domain
- nSM2-APLB2 domain
- nSMase2 APL binding type 2 domain
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- aa
- amino acids.
REFERENCES
- 1. Kim R. H., Takabe K., Milstien S., Spiegel S. (2009) Biochim. Biophys. Acta. 1791, 692–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stancevic B., Kolesnick R. (2010) FEBS Lett. 584, 1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 4. Bartke N., Hannun Y. A. (2009) J. Lipid Res. 50, suppl., S91–S96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tomiuk S., Hofmann K., Nix M., Zumbansen M., Stoffel W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3638–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hofmann K., Tomiuk S., Wolff G., Stoffel W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5895–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krut O., Wiegmann K., Kashkar H., Yazdanpanah B., Krönke M. (2006) J. Biol. Chem. 281, 13784–13793 [DOI] [PubMed] [Google Scholar]
- 8. Wu B. X., Rajagopalan V., Roddy P. L., Clarke C. J., Hannun Y. A. (2010) J. Biol. Chem. 285, 17993–18002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nikolova-Karakashian M., Karakashian A., Rutkute K. (2008) Subcell. Biochem. 49, 469–486 [DOI] [PubMed] [Google Scholar]
- 10. Clarke C. J., Snook C. F., Tani M., Matmati N., Marchesini N., Hannun Y. A. (2006) Biochemistry 45, 11247–11256 [DOI] [PubMed] [Google Scholar]
- 11. Clarke C. J., Hannun Y. A. (2006) Biochim. Biophys. Acta. 1758, 1893–1901 [DOI] [PubMed] [Google Scholar]
- 12. Tani M., Hannun Y. A. (2007) J. Biol. Chem. 282, 10047–10056 [DOI] [PubMed] [Google Scholar]
- 13. Tani M., Hannun Y. A. (2007) FEBS Lett. 581, 1323–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hayashi Y., Kiyono T., Fujita M., Ishibashi M. (1997) J. Biol. Chem. 272, 18082–18086 [DOI] [PubMed] [Google Scholar]
- 15. Marchesini N., Osta W., Bielawski J., Luberto C., Obeid L. M., Hannun Y. A. (2004) J. Biol. Chem. 279, 25101–25111 [DOI] [PubMed] [Google Scholar]
- 16. Clarke C. J., Truong T. G., Hannun Y. A. (2007) J. Biol. Chem. 282, 1384–1396 [DOI] [PubMed] [Google Scholar]
- 17. Clarke C. J., Guthrie J. M., Hannun Y. A. (2008) Mol. Pharmacol. 74, 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tellier E., Nègre-Salvayre A., Bocquet B., Itohara S., Hannun Y. A., Salvayre R., Augé N. (2007) Mol. Cell. Biol. 27, 2997–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karakashian A. A., Giltiay N. V., Smith G. M., Nikolova-Karakashian M. N. (2004) Faseb. J. 18, 968–970 [DOI] [PubMed] [Google Scholar]
- 20. Stoffel W., Jenke B., Blöck B., Zumbansen M., Koebke J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4554–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aubin I., Adams C. P., Opsahl S., Septier D., Bishop C. E., Auge N., Salvayre R., Negre-Salvayre A., Goldberg M., Guénet J. L., Poirier C. (2005) Nat. Genet. 37, 803–805 [DOI] [PubMed] [Google Scholar]
- 22. Kim W. J., Okimoto R. A., Purton L. E., Goodwin M., Haserlat S. M., Dayyani F., Sweetser D. A., McClatchey A. I., Bernard O. A., Look A. T., Bell D. W., Scadden D. T., Haber D. A. (2008) Blood 111, 4716–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Klompenburg W., Nilsson I., von Heijne G., de Kruijff B. (1997) EMBO. J. 16, 4261–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stahelin R. V., Digman M. A., Medkova M., Ananthanarayanan B., Rafter J. D., Melowic H. R., Cho W. (2004) J. Biol. Chem. 279, 29501–29512 [DOI] [PubMed] [Google Scholar]
- 25. McDonald J. F., Shah A. M., Schwalbe R. A., Kisiel W., Dahlbäck B., Nelsestuen G. L. (1997) Biochemistry 36, 5120–5127 [DOI] [PubMed] [Google Scholar]
- 26. Stace C. L., Ktistakis N. T. (2006) Biochim. Biophys. Acta. 1761, 913–926 [DOI] [PubMed] [Google Scholar]
- 27. Rizzo M. A., Shome K., Watkins S. C., Romero G. (2000) J. Biol. Chem. 275, 23911–23918 [DOI] [PubMed] [Google Scholar]
- 28. Corbalán-Garcia S., Sánchez-Carrillo S., García-García J., Gómez-Fernández J. C. (2003) Biochemistry 42, 11661–11668 [DOI] [PubMed] [Google Scholar]
- 29. Delon C., Manifava M., Wood E., Thompson D., Krugmann S., Pyne S., Ktistakis N. T. (2004) J. Biol. Chem. 279, 44763–44774 [DOI] [PubMed] [Google Scholar]
- 30. Jones J. A., Rawles R., Hannun Y. A. (2005) Biochemistry 44, 13235–13245 [DOI] [PubMed] [Google Scholar]
- 31. Marchesini N., Luberto C., Hannun Y. A. (2003) J. Biol. Chem. 278, 13775–13783 [DOI] [PubMed] [Google Scholar]
- 32. Sawai H., Okamoto Y., Luberto C., Mao C., Bielawska A., Domae N., Hannun Y. A. (2000) J. Biol. Chem. 275, 39793–39798 [DOI] [PubMed] [Google Scholar]
- 33. Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 34. Jones J. A., Hannun Y. A. (2002) J. Biol. Chem. 277, 15530–15538 [DOI] [PubMed] [Google Scholar]
- 35. Stevenson J. M., Perera I. Y., Boss W. F. (1998) J. Biol. Chem. 273, 22761–22767 [DOI] [PubMed] [Google Scholar]
- 36. Okamoto Y., Vaena De Avalos S., Hannun Y. A. (2002) J. Biol. Chem. 277, 46470–46477 [DOI] [PubMed] [Google Scholar]
- 37. Matmati N., Kitagaki H., Montefusco D., Mohanty B. K., Hannun Y. A. (2009) J. Biol. Chem. 284, 8241–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Okamoto Y., Vaena de Avalos S., Hannun Y. A. (2003) Biochemistry 42, 7855–7862 [DOI] [PubMed] [Google Scholar]
- 39. van Meer G., Voelker D. R., Feigenson G. W. (2008) Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Milhas D., Clarke C. J., Hannun Y. A. (2010) FEBS Lett. 584, 1887–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vance J. E., Steenbergen R. (2005) Prog. Lipid Res. 44, 207–234 [DOI] [PubMed] [Google Scholar]
- 42. Igarashi K., Kaneda M., Yamaji A., Saido T. C., Kikkawa U., Ono Y., Inoue K., Umeda M. (1995) J. Biol. Chem. 270, 29075–29078 [DOI] [PubMed] [Google Scholar]
- 43. Yabu T., Shimuzu A., Yamashita M. (2009) J. Biol. Chem. 284, 20349–20363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Levy M., Castillo S. S., Goldkorn T. (2006) Biochem. Biophys. Res. Commun. 344, 900–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Milhas D., Clarke C. J., Idkowiak-Baldys J., Canals D., Hannun Y. A. (2010) Biochim. Biophys. Acta. 1801, 1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lomize A. L., Pogozheva I. D., Lomize M. A., Mosberg H. I. (2007) BMC. Struct. Biol. 7, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Natarajan V., Taher M. M., Roehm B., Parinandi N. L., Schmid H. H., Kiss Z., Garcia J. G. (1993) J. Biol. Chem. 268, 930–937 [PubMed] [Google Scholar]
- 48. Aragonés J., Jones D. R., Martin S., San Juan M. A., Alfranca A., Vidal F., Vara A., Mérida I., Landázuri M. O. (2001) J. Biol. Chem. 276, 10548–10555 [DOI] [PubMed] [Google Scholar]
- 49. Cogolludo A., Moreno L., Frazziano G., Moral-Sanz J., Menendez C., Castañeda J., Gonzalez C., Villamor E., Perez-Vizcaino F. (2009) Cardiovasc. Res. 82, 296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sethu S., Mendez-Corao G., Melendez A. J. (2008) J. Immunol. 180, 6027–6034 [DOI] [PubMed] [Google Scholar]
- 51. Czarny M., Liu J., Oh P., Schnitzer J. E. (2003) J. Biol. Chem. 278, 4424–4430 [DOI] [PubMed] [Google Scholar]
- 52. Veldman R. J., Maestre N., Aduib O. M., Medin J. A., Salvayre R., Levade T. (2001) Biochem. J. 355, 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goswami R., Ahmed M., Kilkus J., Han T., Dawson S. A., Dawson G. (2005) J. Neurosci. Res. 81, 208–217 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.